Introduction
Papillary thyroid carcinoma (PTC) is the most common
type of thyroid cancer, accounting for ~80% of all thyroid cancers.
With appropriate treatment, including surgery and radioiodine
ablation, the majority of PTCs have excellent prognoses (1). Examining genetic factors could aid
early detection and also facilitate the treatment and prevention of
PTC. However, the ideal genetic marker for PTC detection has not
yet been identified.
Although originally considered to be spurious
transcriptional noise, long non-coding RNAs (lncRNAs) are now
recognized as regulators in tumorigenesis and tumor progression
(2,3). Functional lncRNAs have the potential
to be used for diagnosing cancer and determining prognosis, as well
as being a potential therapeutic target that could become a
valuable novel diagnostic and therapeutic tool (4). BRAF-activated lncRNA (BANCR) is a
693-bp transcript on chromosome 9, which is frequently
overexpressed and has a possible functional role in the migration
of melanoma cells (5,6). BANCR is strongly linked with
V600EBRAF, which is the most prevalent mutation of the
BRAF gene. V600EBRAF mutations are exhibited in 70% of
malignant melanomas, 36–53% of papillary thyroid cancers and 5–22%
of CRCs (7). The
V600EBRAF mutation is considered to be a putative
prognostic marker for the aggressiveness of PTC (8), but the expression pattern and
biological functions of BANCR in PTC remain to be elucidated.
Autophagy is a lysosome-mediated intracellular
catabolic process by which cells remove damaged organelles and
long-lived proteins to maintain cellular homeostasis (9). Autophagy is activated in cancer cells
and contributes to tumor cell survival (10). High oncogenic BRAF levels have been
shown to initiate autophagy, which is possibly involved in tumor
progression (11). Since a close
association exists between the presence of the BRAF gene and
autophagy, it has been speculated that BANCR could be involved in
the regulation of autophagy.
The aims of the present study were to detect the
expression levels of BANCR and to investigate the function and
molecular mechanisms of BANCR in PTC.
Materials and methods
Tissue samples and cell culture
In total, six specimens of human PTC and adjacent
normal tissues were obtained, with informed consent, from surgeries
performed between March and June 2013 at the First Affiliated
Hospital of Nanjing Medical University (Nanjing, Jiangsu, China).
The protocol used in this study was approved by the hospital’s
Protection of Human Ethics Committee. The diagnosis of PTC was
histopathologically confirmed. The resected tissue samples were
immediately frozen in liquid nitrogen and stored at −80°C until RNA
extraction. The human PTC-derived cell line, IHH-4, was provided by
Professor Congyou Lu (The Chinese University of Hong Kong, Hong
Kong, China). The IHH-4 cells were routinely cultured at 37°C in
RPMI 1640 medium (Wisent, Inc., QC, Canada) with 10% fetal bovine
serum (Wisent, Inc.) and 5% carbon dioxide.
Quantitative polymerase chain reaction
(PCR)
Total RNA from tissues and cells was extracted using
RNAiso Plus (Takara Biotechnology (Dalian) Co., Ltd., Dalian,
China), and reverse transcription (RT) reactions were performed
using a PrimeScript RT reagent kit (Takara Biotechnology (Dalian)
Co., Ltd.) according to the manufacturer’s instructions.
Quantitative PCR reactions were prepared at a final volume of 20 μl
using a standard protocol and the SYBR Green PCR kit (Roche
Diagnostics Co., Indianapolis, IN, USA), and the reactions were
performed on the StepOnePlus Real-Time PCR System (Applied
Biosystems, Inc., CA, USA). Each reaction was performed in
triplicate. The 2−ΔΔCT method was used to determine the
relative gene expression levels, using β-actin as the endogenous
control to normalize the data. The primers used in this study were
as follows: BANCR forward, 5′-ACAGGACTCCATGGCAAACG-3′ and reverse,
5′-ATGAAGAAAGCCTGGTGCAGT-3′; β-actin forward,
5′-AGAAAATCTGGCACCAACC-3′ and reverse, 5′-TAGCACAGCCTGGATAGCAA-3′;
LC3 forward, 5′-CCACACCCAAAGTCCTCACT-3′ and reverse, 5′-CAC
TGCTGCTTTCCGTAACA-3′. PCR was performed at 95°C for 30 sec, 40
cycles of 95°C for 5 sec, 60°C for 31 sec, and then, for
dissociation, at 95°C for 15 sec, 60°C for 1 min and 95°C for 15
sec.
Generation of stable infected cell
lines
Recombinant lentiviruses containing short hairpin
(sh)RNA-323 (LV-BANCR-323, GGA GTGGCGACTATAGCAAAC), shRNA-540
(LV-BANCR-540, GGACTCCATGGCAAACGTTGT), human full-length BANCR cDNA
(LV-BANCR) and a negative control (LV-NC) were purchased from
GenePharma Co., Ltd. (Shanghai, China). The IHH-4 cells were
infected with LV-BANCR-323, LV-BANCR-540, LV-BANCR and LV-NC
(multiplicity of infection, 20). The supernatant was removed after
24 h and fresh culture medium was added to the cells. The infection
efficiency was confirmed by RT-PCR at 72 h post-infection, and the
cells were treated with 2 μg/ml puromycin for 2 weeks.
Cell proliferation assays
Cell proliferation assays were performed using Cell
Counting Kit-8 (CCK8; Beyotime Institute of Biotechnology, Jiangsu,
China). The cells were plated in triplicate in 96-well plates at
~2×103 cells per well and cultured in the growth medium.
The number of cells per well was measured by the absorbance at 450
nm at the indicated time-points, according to the
manufacturer’s instructions.
Flow cytometric analysis
The cells (4×105) were seeded in 6-well
plates. After 24 h, the cells were collected and incubated with
Annexin V-fluorescein isothiocyanate and 7-amino-actinomycin D
(Biolegend, Inc., San Diego, CA, USA) for 15 min in the dark and
apoptosis was analyzed using a flow cytometer. The cell cycle was
also analyzed subsequent to propidium iodide staining for 30
min.
Transwell migration assay
In total, 4×104 cells were plated in
medium without serum on a non-coated membrane in the top chamber
(24-well insert; 8-mm pore size; Corning Costar; Corning, Inc.,
Corning, NY, USA). Medium supplemented with serum was used as a
chemotactic agent in the lower chamber. The cells were incubated
for 24 h, and those cells that did not migrate through the pores
were removed with a cotton swab. The cells on the lower surface of
the membrane were stained with crystal violet (Beijing Solarbio
Science & Technology Co., Ltd., Beijing, China). The cell
numbers were determined by counting the penetrating cells under a
microscope (Nikon, Kobe, Japan) in random fields, with five fields
per chamber. Each experiment was performed in triplicate.
Western blot analysis
Proteins were extracted with
radioimmunoprecipitation assay (Beyotime Institute of
Biotechnology) and equal amounts of protein were electrophoresed on
a 12 or 15% sodium dodecyl sulfate-polyacrylamide gel and
subsequently transferred to polyvinylidene fluoride membranes
(Millipore, Boston, MA, USA). The membranes were blocked with 5%
skimmed milk in Tris-buffered saline containing 0.1% Tween-20
(TBST), at room temperature for 2 h. The membranes were incubated
with the rabbit polyclonal LC3-I, LC3-II (1:1,000; Cell Signaling
Technology, Inc., Danvers, MA, USA) and rabbit polyclonal GAPDH
(1:10,000; Beijing Biosynthesis Biotechnology Co., Ltd., Beijing,
China) primary antibodies at 4°C overnight. The membranes were then
washed three times with TBST and incubated with horseradish
peroxidase-conjugated secondary anti-rabbit antibody (1:5,000;
Beijing Biosynthesis Biotechnology Co., Ltd.) at room temperature
for 2 h. Following three washes with TBST, the membranes were
developed using ECL Plus (EMD Millipore, Billerica, MA, USA), and
exposed to X-ray film. GAPDH was used as an internal loading
control.
Statistical analysis
Data were analyzed using SPSS 19.0 software (SPSS,
Chicago, IL, USA), and are expressed as the mean ± standard
deviation of data from at least three independent experiments. The
differences between the groups were analyzed using Student’s
t-test, Pearson’s χ2-test or one-way analysis of
variance, as appropriate. P<0.05 was considered to indicate a
statistically significant difference.
Results
BANCR levels are significantly
upregulated in PTC
To investigate the role of BANCR in PTC development,
BANCR RNA expression levels in PTC tissue samples were examined
first. The RT-PCR results showed that BANCR expression was
significantly higher in five out of six of the tumor tissues
compared with the adjacent normal tissues. Additionally, BANCR
expression in the PTC IHH-4 cell line was upregulated compared with
the mean expression level of the adjacent normal tissues
(P<0.05; Fig. 1).
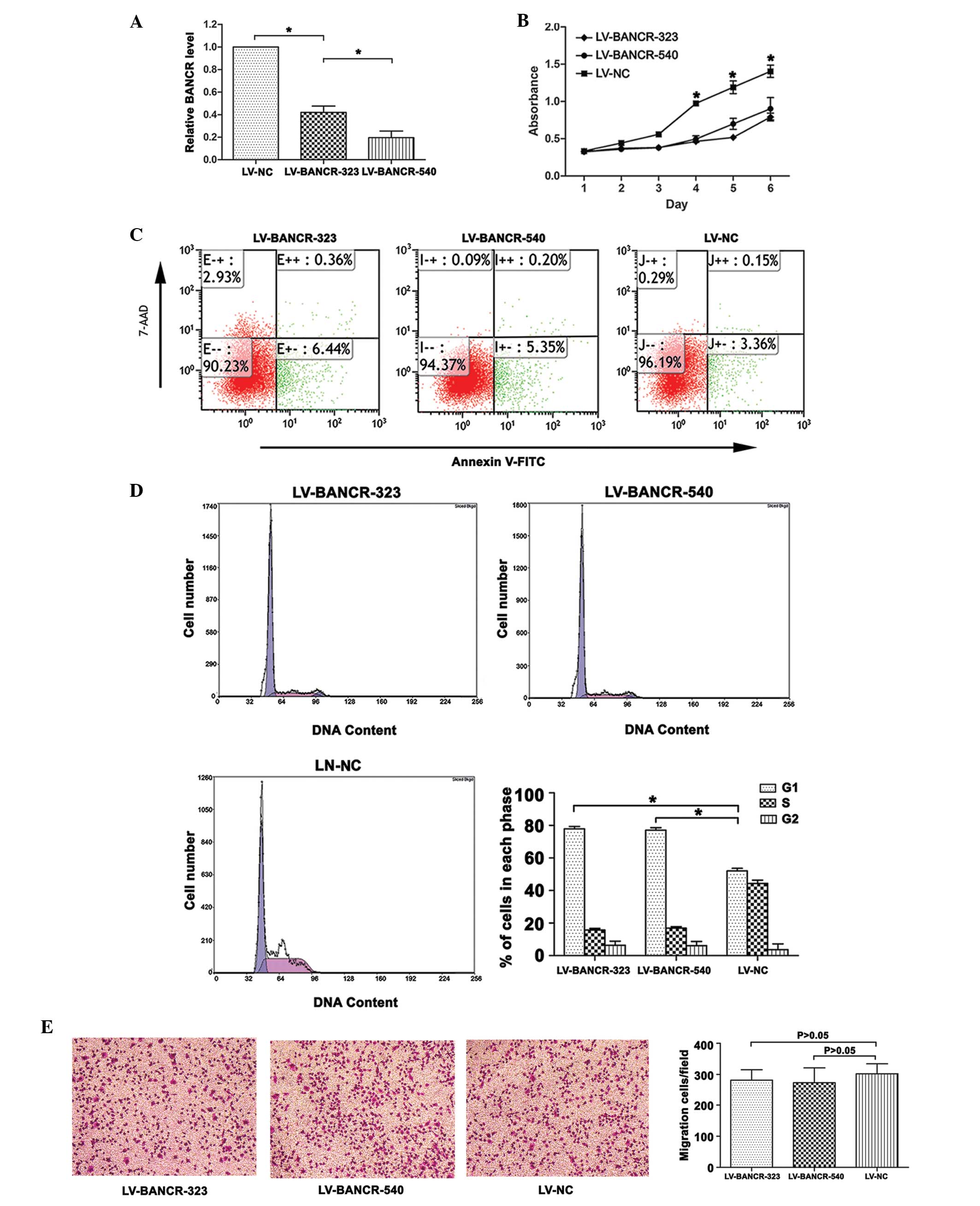
BANCR-knockdown inhibits proliferation
and increases apoptosis of PTC IHH-4 cells
Following infection with LV-BANCR-323 and
LV-BANCR-540, the BANCR expression level was significantly
downregulated compared with the LV-NC (P<0.05; Fig. 2A). As shown in Fig. 2C, the knockdown of BANCR induced
cell apoptosis. The results were presented as the percentage of
apoptotic cells in the total number of counted cells. Signals from
apoptotic cells were localized in the lower right quadrant
(F=603.832, P<0.05). The CCK8 assays revealed that the knockdown
of BANCR inhibited the proliferation of the IHH-4 cells (P<0.05;
Fig. 2B). Further investigation
into the role of BANCR in the regulation of cell proliferation
revealed that the knockdown of BANCR resulted in an increase in the
cell population in the G1 phase (P<0.05; Fig. 2D). The Transwell migration assays
showed that BANCR-knockdown had no significant effect on the
migration of the IHH-4 cells (P>0.05; Fig. 2E).
Overexpression of BANCR increases
autophagy activation in PTC IHH-4 cells
To explore the mechanism by which BANCR regulates
cell proliferation, the present study investigated whether BANCR
regulates cell autophagy. The IHH-4 cells were treated with
LV-BANCR, with or without 3-methyladenine (3-MA), an inhibitor of
autophagy. The results revealed that following infection with
LV-BANCR, the BANCR expression level was significantly upregulated
compared with the LV-NC (P<0.05; Fig. 3A). The overexpression of BANCR
inhibited the apoptosis of the IHH-4 cells, promoted cell growth
and decreased the cell population in the G1 phase,
whereas autophagy inhibition increased cell apoptosis (F=167.557,
P<0.05; Fig. 3C), inhibited cell
growth (P<0.05; Fig. 3B) and
increased G1 arrest (P<0.05; Fig. 3D) in the BANCR-overexpressed IHH-4
cells. The western blotting results demonstrated that BANCR
overexpression resulted in an increase in the ratio of
LC3-II/LC3-I, a marker for autophagy, while knockdown of BANCR and
treatment with 3-MA decreased the ratio of LC3-II/LC3-I (P<0.05;
Fig. 3E). The RT-PCR results
indicated that the level of LC3 mRNA had increased in the
BANCR-overexpressed cells, while it had decreased following
BANCR-knockdown and in the 3-MA-treated cells (P<0.05; Fig. 3F).
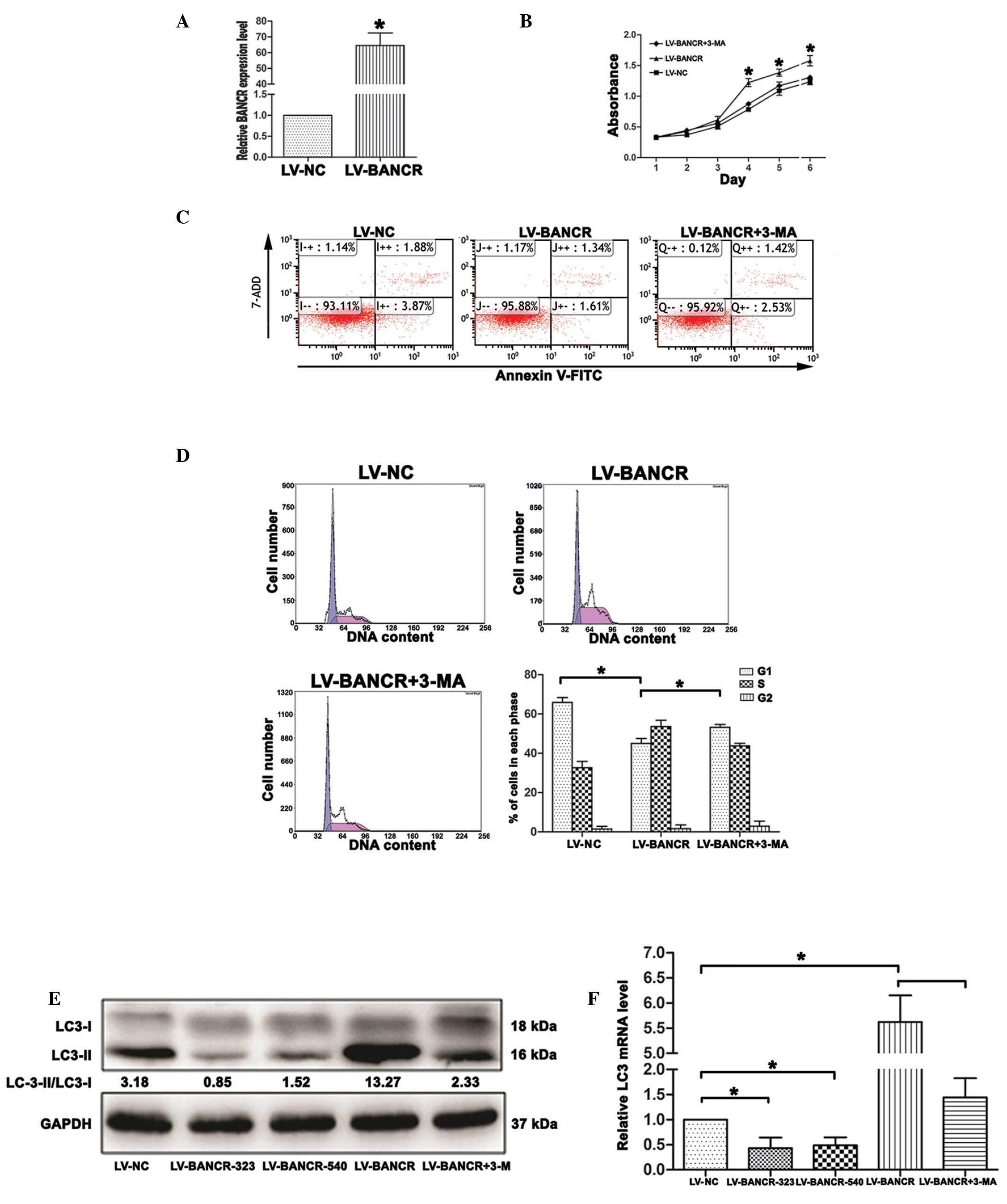 | Figure 3Overexpression of BANCR increases
autophagy activation in papillary thyroid carcinoma IHH-4 cells.(A)
Following treatment with LV-BANCR, BANCR expression in IHH-4 cells
was upregulated compared with the cells treated with LV-NC. (B)
Cell Counting Kit-8 assays showed that the overexpression of BANCR
promoted cell growth, whereas the inhibition of autophagy inhibited
the proliferation of the IHH-4 cells. (C) Overexpression of BANCR
inhibited cell apoptosis, whereas autophagy inhibition increased
apoptosis in the IHH-4 cells, which was detected by flow cytometry.
(D) Overexpression of BANCR decreased the cell population in the
G1 phase, whereas autophagy inhibition increased the
cell population in the G1 phase. The data represent one
of at least three independent experiments. (E) Western blotting
results revealed that BANCR overexpression resulted in an increase
in the ratio of LC3-II/LC3-I, while knockdown of BANCR and
treatment with 3-MA decreased the ratio of LC3-II/LC3-I. (F)
Reverse transcription polymerase chain reaction results showed that
BANCR overexpression resulted in an increase in the LC3 mRNA level,
while knockdown of BANCR and treatment with 3-MA decreased LC3 mRNA
level. The results are presented as the mean ± standard deviation
(*P<0.05). BANCR, BRAF-activated long non-coding RNA;
LV-BANCR, lentivirus containing human full-length BANCR cDNA;
LV-BANCR-323, LV containing short hairpin (sh)RNA-323;
LV-BANCR-540, LV containing shRNA-540; LV-NC, LV negative control;
3-MA, 3-methyladenine; 7-AAD, 7-amino-actinomycin D; FITC,
fluorescein isothiocyanate. |
Discussion
Advances in molecular techniques have led to the
identification of a novel type of gene regulators called lncRNA.
These lncRNAs are >200 nucleotides and do not code for proteins.
However, they can interact with proteins and can likely act as
regulators of other genes (12).
Although lncRNAs are not as well-characterized as small non-coding
microRNAs, they play a critical role in the regulation of diverse
cellular processes (13,14). In thyroid cancer, one such example
of oncogenic lncRNA is papillary thyroid carcinoma susceptibility
candidate 3 (PTCSC3). Using quantitative PCR, PTCSC3 expression was
revealed to be strongly downregulated in thyroid tumor tissues, and
it was demonstrated that the restoration of PTCSC3 expression in
PTC cells inhibited cell growth and affected the expression of
numerous genes (15). Another
classic oncogenic lncRNA is non-coding RNA associated with the
mitogen-activated protein (MAP) kinase pathway and growth arrest
(NAMA), which is weakly expressed in thyroid cancer tissues.
Knockdown of BRAF has been revealed to induce inhibition of the MAP
kinase pathway, growth arrest and DNA damage in thyroid cancer cell
lines (16).
BANCR is recurrently overexpressed in melanoma. In
previous studies, shRNA-mediated knockdown of BANCR in melanoma
cells was revealed to alter the expression levels of 88 genes,
several of which are involved in cell migration and chemotaxis.
BANCR depletion impaired the migration of the melanoma cells in
vitro (5,6). Mutation of BRAF is hypothesized to be
a putative prognostic marker for the aggressiveness of PTC
(17). Based on these findings, it
was hypothesized that BANCR could play a critical role in PTC. In
the present study, it was found that BANCR expression levels were
upregulated in five out of six PTC tumor tissues compared with
their adjacent normal tissues. Although samples from only six
patients were used in the present study and the results may not be
entirely accurate due to type I or II errors, the present data
suggest a possible oncogenic role of BANCR in several human
cancers. Furthermore, in vitro examination of the potential
role of BANCR in PTC IHH-4 cells demonstrated that the knockdown of
BANCR in the IHH-4 cells was associated with the inhibition of
proliferation and the promotion of apoptosis, but exhibited no
significant effect on cell migration. This observation was in
contrast with previous studies regarding the role of BANCR in
regulating cell migration (5),
thereby suggesting that the function of BANCR could be
tissue-specific.
Autophagy is a self-degradative process through
which the cytoplasmic materials within the lysosome are degraded.
The process acts as a dynamic system that provides the building
blocks of a cell and the energy for cellular homeostasis and
regeneration (18). Maddodi et
al observed that the presence of high levels of mBRAF triggers
the hyperactivation of extracellular-signal-regulated kinase (ERK),
a senescence-like phenotype and initiates autophagy through the
inhibition of mammalian target of rapamycin complex signaling
(11). BANCR was considered to play
a role in controlling cell proliferation by regulating autophagy
activation, although there was no direct evidence to support this
hypothesis. In the present study, it was demonstrated that the
overexpression of BANCR induced autophagy activation, whereas
BANCR-knockdown decreased autophagy activation in the PTC IHH-4
cells. Autophagy activation was evaluated by observing the ratio of
LC3-II/LC3-I. Overexpression of BANCR inhibited the apoptosis of
the IHH-4 cells, promoted cell growth and decreased the cell
population in the G1 phase; all these effects could be
suppressed by 3-MA, an inhibitor of autophagy. These findings
suggest that BANCR may increase PTC cell proliferation by
activating autophagy.
To the best of our knowledge, this is the first
study to report that BANCR is highly expressed in PTC and that
BANCR is likely to be a useful biomarker of this disease.
Additionally, the fact that BANCR increases PTC cell proliferation
by activating autophagy adds to our understanding of the molecular
mechanisms governing BANCR. Significantly, BANCR could be used as a
potential molecular target to treat human PTC.
Acknowledgements
This study was funded by the ‘333 project’ of
Jiangsu Province (grant no. BK20131448).
References
|
1
|
Sherman SI: Thyroid carcinoma. Lancet.
361:501–511. 2003.
|
|
2
|
Nakagawa T, Endo H, Yokoyama M, et al:
Large noncoding RNA HOTAIR enhances aggressive biological behavior
and is associated with short disease-free survival in human
non-small cell lung cancer. Biochem Biophys Res Commun.
436:319–324. 2013.
|
|
3
|
Zhu L and Xu PC: Downregulated LncRNA-ANCR
promotes osteoblast differentiation by targeting EZH2 and
regulating Runx2 expression. Biochem Biophys Res Commun.
432:612–617. 2013.
|
|
4
|
Qi P and Du X: The long non-coding RNAs, a
new cancer diagnostic and therapeutic gold mine. Mod Pathol.
26:155–165. 2013.
|
|
5
|
Flockhart RJ, Webster DE, Qu K, et al:
BRAFV600E remodels the melanocyte transcriptome and induces BANCR
to regulate melanoma cell migration. Genome Res. 22:1006–1014.
2012.
|
|
6
|
McCarthy N: Epigenetics. Going places with
BANCR. Nat Rev Cancer. 12:4512012.
|
|
7
|
Davies H, Bignell GR, Cox C, et al:
Mutations of the BRAF gene in human cancer. Nature. 417:949–954.
2002.
|
|
8
|
Howell GM, Nikiforova MN, Carty SE, et al:
BRAF V600E mutation independently predicts central compartment
lymph node metastasis in patients with papillary thyroid cancer.
Ann Surg Oncol. 20:47–52. 2013.
|
|
9
|
Amelio I, Melino G and Knight RA: Cell
death pathology: cross-talk with autophagy and its clinical
implications. Biochem Biophys Res Commun. 414:277–281. 2011.
|
|
10
|
Ding WX, Chen X and Yin XM: Tumor cells
can evade dependence on autophagy through adaptation. Biochem
Biophys Res Commun. 425:684–688. 2012.
|
|
11
|
Maddodi N, Huang W, Havighurst T, Kim K,
Longley BJ and Setaluri V: Induction of autophagy and inhibition of
melanoma growth in vitro and in vivo by
hyperactivation of oncogenic BRAF. J Invest Dermatol.
130:1657–1667. 2010.
|
|
12
|
Zhang YC and Chen YQ: Long noncoding RNAs:
new regulators in plant development. Biochem Biophys Res Commun.
436:111–114. 2013.
|
|
13
|
Luo M, Li Z, Wang W, Zeng Y, Liu Z and Qiu
J: Long non-coding RNA H19 increases bladder cancer metastasis by
associating with EZH2 and inhibiting E-cadherin expression. Cancer
Lett. 333:213–221. 2013.
|
|
14
|
Cheng W, Zhang Z and Wang J: Long
noncoding RNAs: new players in prostate cancer. Cancer Lett.
339:8–14. 2013.
|
|
15
|
Fan M, Li X, Jiang W, Huang Y, Li J and
Wang Z: A long non-coding RNA, PTCSC3, as a tumor suppressor and a
target of miRNAs in thyroid cancer cells. Exp Ther Med.
5:1143–1146. 2013.
|
|
16
|
Yoon H, He H, Nagy R, et al:
Identification of a novel noncoding RNA gene, NAMA, that is
downregulated in papillary thyroid carcinoma with BRAF mutation and
associated with growth arrest. Int J Cancer. 121:767–775. 2007.
|
|
17
|
Liu D, Liu Z, Condouris S and Xing M: BRAF
V600E maintains proliferation, transformation, and tumorigenicity
of BRAF-mutant papillary thyroid cancer cells. J Clin Endocrinol
Metab. 92:2264–2271. 2007.
|
|
18
|
Zhou S, Zhao L, Kuang M, et al: Autophagy
in tumorigenesis and cancer therapy: Dr. Jekyll or Mr. Hyde? Cancer
Lett. 323:115–127. 2012.
|