Introduction
Human gliomas originate from neural mesenchymal
cells and account for 4–50% of nervous system tumors (1). The 2007 World Health Organization
(WHO) classification (2) classifies
astrocytomas as well-differentiated low-grade diffuse astrocytoma
(WHO grade I-II), anaplastic astrocytoma (WHO grade III) and
glioblastoma multiforme (GBM; WHO grade IV). Despite the use of
aggressive surgery combined with radiation, chemotherapy and
biological therapy (3), glioma
remains a notable therapeutic challenge. There is an acknowledged
requirement for novel therapeutic approaches based on an increased
understanding of the biological and molecular nature of glioma
(4).
microRNAs (miRNAs) are a class of short, non-coding
single-stranded RNA molecules that are 22–25 nucleotides in length.
miRNAs negatively regulate gene expression through the
post-transcriptional silencing of target messenger RNAs (mRNAs),
which occurs due to complementary binding (5,6). An
increasing quantity of evidence has indicated an important role for
miRNAs in the development of various cancers, including gliomas,
and miRNAs have been associated with tumor suppressor and oncogenic
activities (7,8). Among these miRNAs, miRNA 218 (miR-218)
has been demonstrated to be downregulated in human GBM specimens
compared with the adjacent tumor-free brain tissue (9–12).
Accumulated evidence has revealed that upregulation of miR-218 can
inhibit tumor cell invasion and proliferation in glioma cells by
altering the expression of multiple target genes (12–15).
Previous studies have found that miR-218 inhibited
the invasion and metastasis of gastric cancer by targeting the
roundabout, axon guidance receptor, homolog 1 (Robo1) receptor and
suppressing nasopharyngeal cancer progression through the
downregulation of survivin and the Slit homolog 2 (Slit2)-Robo1
pathway (16,17). In the present study we examined how
miR-218 affects the migration and invasion of glioma cells and the
mechanism for miRNA-mediated direct suppression of the Slit2-Robo1
pathway in gliomas.
Materials and methods
Clinical samples
Tumor specimens were obtained from patients who
underwent positive debulking surgery in the Neurosurgery Department
of the The First Affiliated Hospital of Soochow University (Suzhou,
China) between 2011 and 2013. The diagnosed gliomas were reviewed
by an experienced neuropathologist, using histological slides,
according to the 2007 WHO classification. The present study
comprised 20 grade I–II, 20 grade III and 20 grade IV glioma
samples. In addition, 10 normal brain tissue samples were obtained
from internal decompression of patients with cerebral injury. This
study complied with the requirements of the ethics committee of The
First Affiliated Hospital of Soochow University and informed
consent was obtained from all participants.
Cell lines and transfection
Primary normal human astrocytes (NHA) were purchased
from Sciencell Research Laboratories (Carlsbad, CA, USA). The U251,
U87, SNB19 and LN229 glioma cell lines were obtained from the
Institute of Biochemistry and Cell Biology (Shanghai Institutes for
Biological Sciences, Chinese Academy of Science, Shanghai, China).
The cells were maintained in RPMI-l640 medium containing 10% FBS,
50 units/ml penicillin G, and 250 μg/ml streptomycin (all purchased
from Invitrogen Life Technologies, Carlsbad, CA, USA) in a
humidified atmosphere containing 5% CO2, at 37°C.
Transfections with miR-218 were performed in serum-free medium 24 h
subsequent to plating, using Lipofectamine 2000 (Invitrogen Life
Technologies). After 6 h, the cells were placed in complete medium
and maintained at 37°C in a 5% CO2 atmosphere.
Small interfering (si)RNA and
transfection assays
Robo1-specific siRNA (forward, 5′-GGAUGUAUUUGCAAC
AAGATT-3′ and reverse, 5′-UCUUGUUGCAAAUACAUC CTT-3′) was chemically
synthesized by Qiagen (Hilden, Germany). The U87 cells were
transfected with siRNA (HiPerFect Transfection Reagent; Qiagen)
according to the manufacturer’s instructions. Briefly, the original
stock of the siRNA was suspended in the siRNA suspension buffer
provided by the manufacturer and stored at −20°C until use. On the
day of transfection, 1×105 cells were seeded in six-well
plates (Corning Inc., Corning, NY, USA) with a total volume of
1,000 μl per well. siRNA was then gently introduced into the cells
by adding 4 μl Oligofectamine™ Transfection Reagent (Invitrogen
Life Technologies) and 5 μl siRNA (20 nM) per well. Non-silencing
siRNA (GE Dharmacon, Lafayette, CO, USA) (forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′) was used to control any effects of the
transfection reagent and siRNA. The in vitro assays described
herein were performed 24 h after transfection.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA, including miRNA, was extracted from the
cultured cells or fresh glioma cancer tissues using TRIzol reagent
(Invitrogen Life Technologies), according to the manufacturer’s
instructions. The expression level of miR-218 was quantified using
an miRNA-specific TaqMan miRNA Assay kit (Applied Biosystems,
Foster City, CA, USA). U6 small nuclear RNA was used as an internal
control. The mRNA expression of Slit2 and Robo1 was analyzed by
qPCR, using the SYBR-Green method. All protocols were performed
according to the manufacturer’s instructions and the results were
normalized to the expression of GAPDH. The primer sequences were as
follows: Slit2 forward, 5′-ACCTCT TGGCCAATCCTTTT-3′ and reverse,
5′-GAAGTCCTGAAT GGCCACAT-3′; Robo1 forward, 5′-GCCACCAGCAAGGAT
GTATT-3′ and reverse, 5′-CCTGTAACATGGGCTGGAGT-3′; and GAPDH
forward, 5′-TCGGAGTCAACGGATTTGG-3′ and reverse,
5′-CATGGGTGGAATCATATTGGA-3′.
Western blotting
The cells were lysed in 1% Nonidet P-40 lysis buffer
for 48 h following exposure to LY294002 or vehicle treatment. The
homogenates were clarified by centrifugation at 20,000 × g for 15
min, at 4°C, and the protein concentrations were determined with a
bicinchoninic acid protein assay kit (Pierce Biotechnology, Inc.,
Rockford, IL, USA). SDS-PAGE was performed on 40 μg of protein from
each sample, the gels were transferred to polyvinylidene fluoride
membranes (Merck Millipore, Darmstadt, Germany) and incubated with
primary polyclonal goat anti-human Slit2 and rabbit anti-human
Robo1 antibodies (1:200 dilution; sc-26599 and sc-25672,
respectively, Santa Cruz Biotechnology, Inc., Dallas, TX, USA)
followed by incubation with horseradish peroxidase-conjugated
monoclonal goat anti-rabbit and donkey anti-goat secondary
antibodies (1:1,000 dilution; cat nos. sc-2004 and sc-2020,
respectively; Zymed Life Technologies, Carlsbad, CA, USA). The
membranes were stripped using 1X phosphate-buffered saline with
Tween 20 buffer (Wuhan Boster Biological Technology, Ltd., Wuhan,
China) and reprobed with a primary monoclonal mouse anti-rabbit
antibody against GAPDH (1:1,000 dilution; Bioworld, Nanjing,
China). The protein bands were quantitated by densitometry using
the gel analysis software ImageJ (National Institutes of Health,
Bethesda, MA, USA. The values were normalized to GAPDH
expression.
Transwell assay and scratch-wound
assay
Cell invasion was determined using Transwell and
scratch-wound assays. For the Transwell assay, the appropriate
oligonucleotides were transfected into the cells according to the
aforementioned protocol. Subsequent to incubation for 48 h,
3×104 cells were transferred to the top of the
Matrigel-coated invasion chambers (BD Biosciences, San Jose, CA,
USA) in a serum-free Dulbecco’s modified Eagle’s medium (DMEM).
DMEM containing 10% fetal bovine serum was added to the lower
chamber. After 24 h, the non-invading cells were removed, and the
invading cells were fixed using 95% ethanol, stained with 0.1%
crystal violet and images were captured under a ×100 magnification.
The tests were repeated in three independent experiments. For the
scratch-wound assay, the appropriate oligonucleotides were
transfected into the cells in six-well plates. The cell layers were
then scratched using a 200 μl sterile pipette tip to form wound
gaps. The wound location in the six-well plates was marked. Images
of the cells were captured to record the wound width at 0 and 24 h,
and the images were captured at the marked wound location to
measure the migratory ability of the cells.
Luciferase assay
The 3′-UTR sequence of Robo1 predicted to interact
with miR-218 or a mutated sequence with the predicted target sites
was synthesized and inserted into the XbaI and FseI
sites of a pGL3 control vector (Promega Corporation, Madison, WI,
USA). These constructs were named pGL3-Robo1–3′-UTR and
pGL3-Robo1–3′-UTR-mut, respectively. For the reporter assay, the
U87 cells were plated onto 24-well plates and transfected with
pGL3-Robo1–3′-UTR or pGL3-Robo1–3′-UTR-mut and P-miR-218 or
P-miR-control vectors using the FuGENE HD transfection reagent
(Promega Corporation). A Renilla luciferase vector pRL-SV50
(Promega Corporation) was cotransfected to normalize the
differences in transfection efficiency. Subsequent to 48 h
transfection, the cells were harvested and assayed using the
Dual-Luciferase Reporter Assay System (Promega Corporation),
according to the manufacturer’s instructions. Transfection was
repeated in triplicate in three independent experiments.
Target prediction and network
analysis
Robo1 was identified as a notable novel target of
miR-218 using the conventional prediction tool, TargetScan
(www.targetscan.org; Whitehead Institute for Biomedical
Research, Cambridge, MA, USA).
Statistical analysis
Experimental data were presented as the mean ±
standard deviation. All analyses were performed using a two-tailed
Student’s t-test performed on SPSS software, version 12.0
(SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-218 is downregulated in glioma
tissues
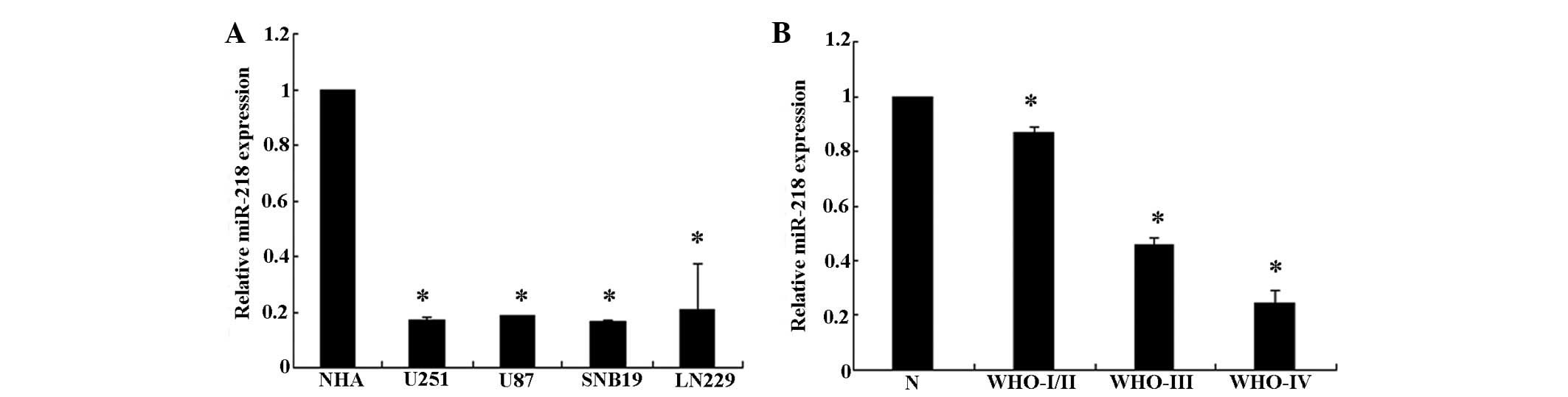
To analyze the expression levels of miR-218, a qPCR
analysis of miR-218 expression was conducted in the U251, U87,
SNB19 and LN229 glioma cell lines. The present results revealed
that miR-218 is downregulated in all glioma cell lines compared
with the NHA cells (P<0.05). In addition, when miR-218
expression was measured in 10 normal brain tissue and 60 glioma
tissue samples, it was observed that the expression level of
miR-218 was significantly decreased in the glioma tissues,
particularly in grade III/IV glioma tissues, compared with the
normal brain tissues (Fig. 1). This
result demonstrates that miR-218 expression decreases markedly
between normal brain tissue and low grade to GBM tissue. Overall,
the present results indicate that miR-218 is downregulated in
glioma cell lines and glioma tissues.
Upregulation of miR-218 inhibited cell
invasion in U87 cells
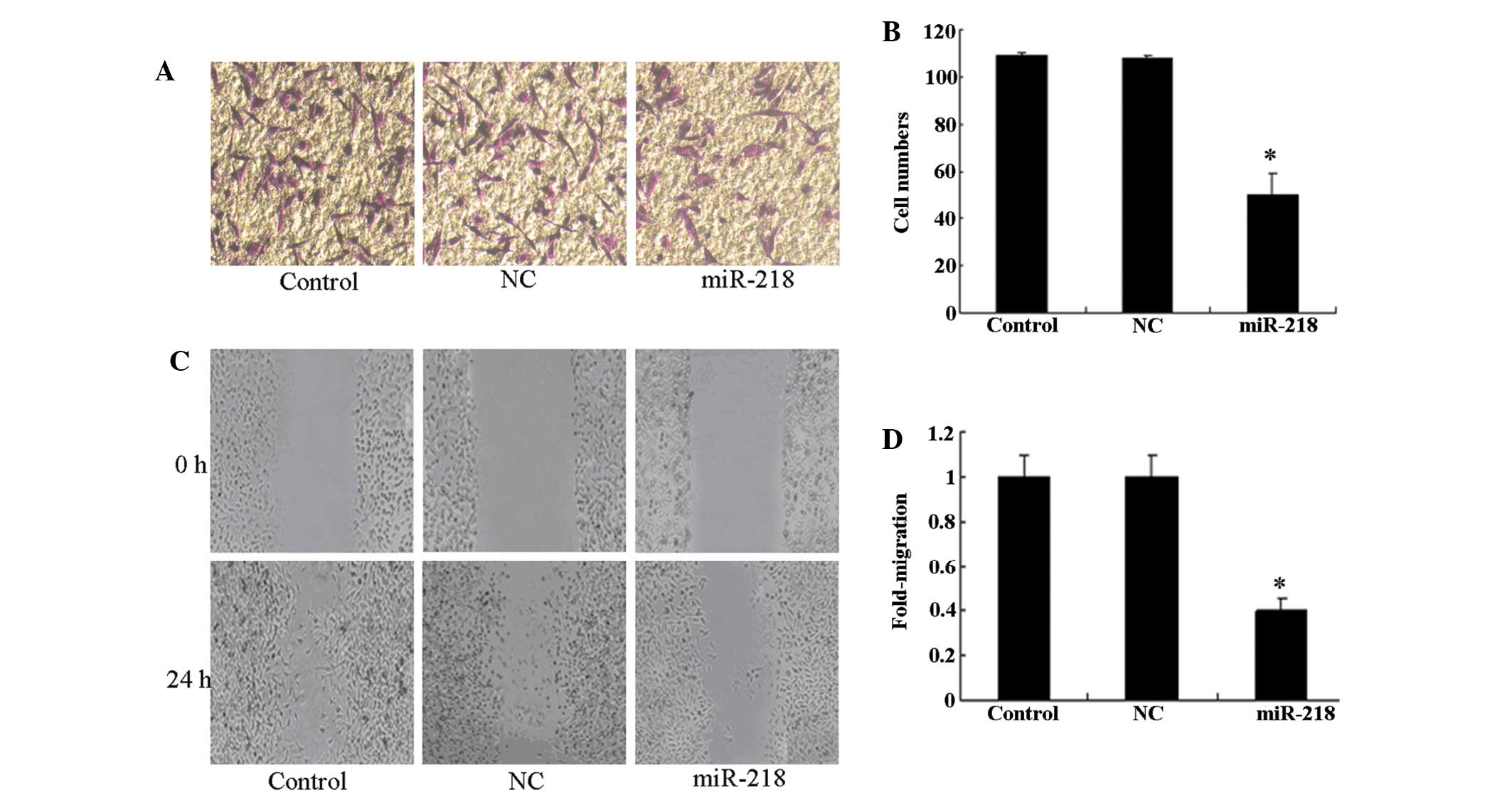
As invasiveness is one of the pathophysiological
features of human malignant gliomas, the effects of miR-218 on the
invasiveness and migration of glioma cells were checked by
Transwell and scratch-wound assays. An in vitro Matrigel
invasion assay revealed that the invasiveness of U87 cells
transfected with the miR-218 mimic was suppressed compared with
control and NC groups (Fig. 2A and
B). The results of the in vitro wound healing assay
revealed that miR-218 significantly attenuated the migration of U87
cells compared with control and NC groups (Fig. 2C and D). This finding indicates that
the upregulation of miR-218 inhibits the invasive ability of glioma
cells in vitro.
Upregulation of miR-218 reduces Robo1
expression via the inactivation of Slit2-Robo1 signaling
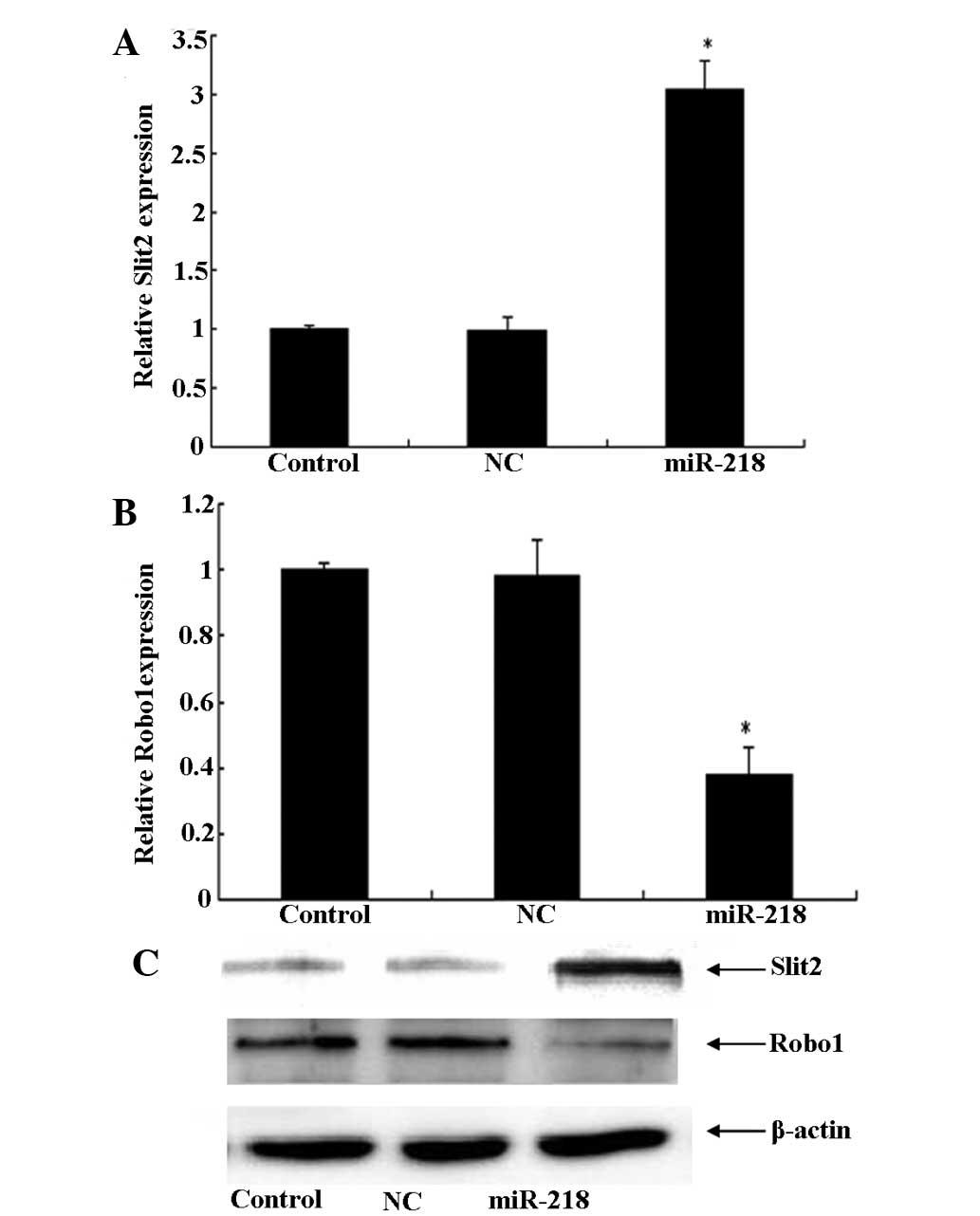
Development of invasiveness by malignant glioma
cells involves multiple genetic alterations in signaling pathways.
Numerous studies have reported that the Slit2-Robo1 signaling
channels can inhibit glioma invasion and migration. However, the
specific roles of Slit2/Robo1 in cancer cell invasion have not yet
been completely elucidated in vivo. The present study also
observed, using qPCR and western blot analysis, that treatment with
the miR-218 mimic for 24 h significantly upregulated the expression
of Slit2, which was followed by a decrease in Robo1 (Fig. 3). These results indicate that
miR-218 reduced the expression of Robo1 via the inactivation of
Slit2-Robo1 signaling.
Robo1 is a functional downstream target
of miR-218
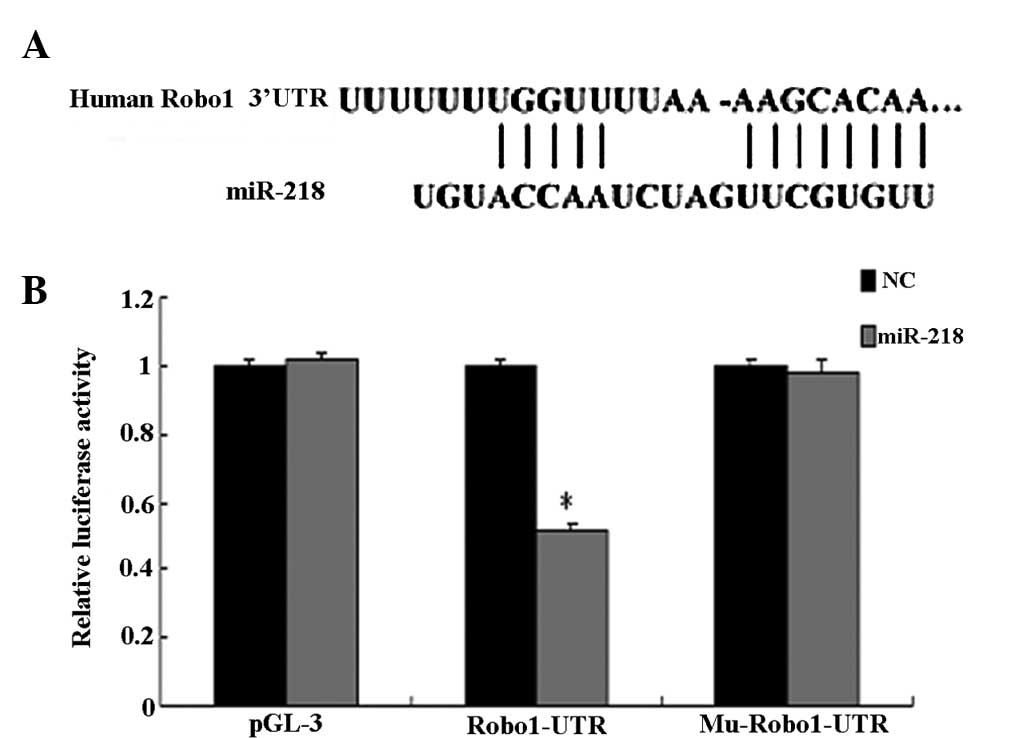
A luciferase reporter assay further confirmed the
direct interaction between miR-218 and the 3′-UTR of Robo1 mRNA
(Fig. 4). The luciferase activity
for the wild-type 3′-UTR of Robo1 was significantly inhibited by
co-transfection with miR-218 mimics compared with constructs
containing mutated 3′-UTRs. The present study demonstrated that
Robo1 is a direct target of miR-218.
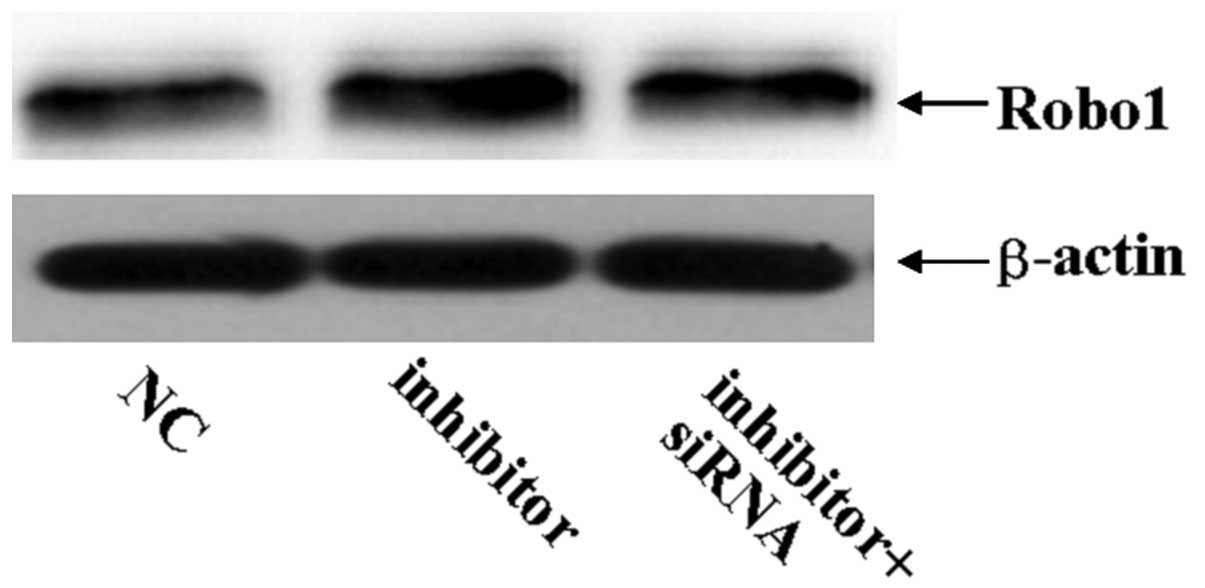
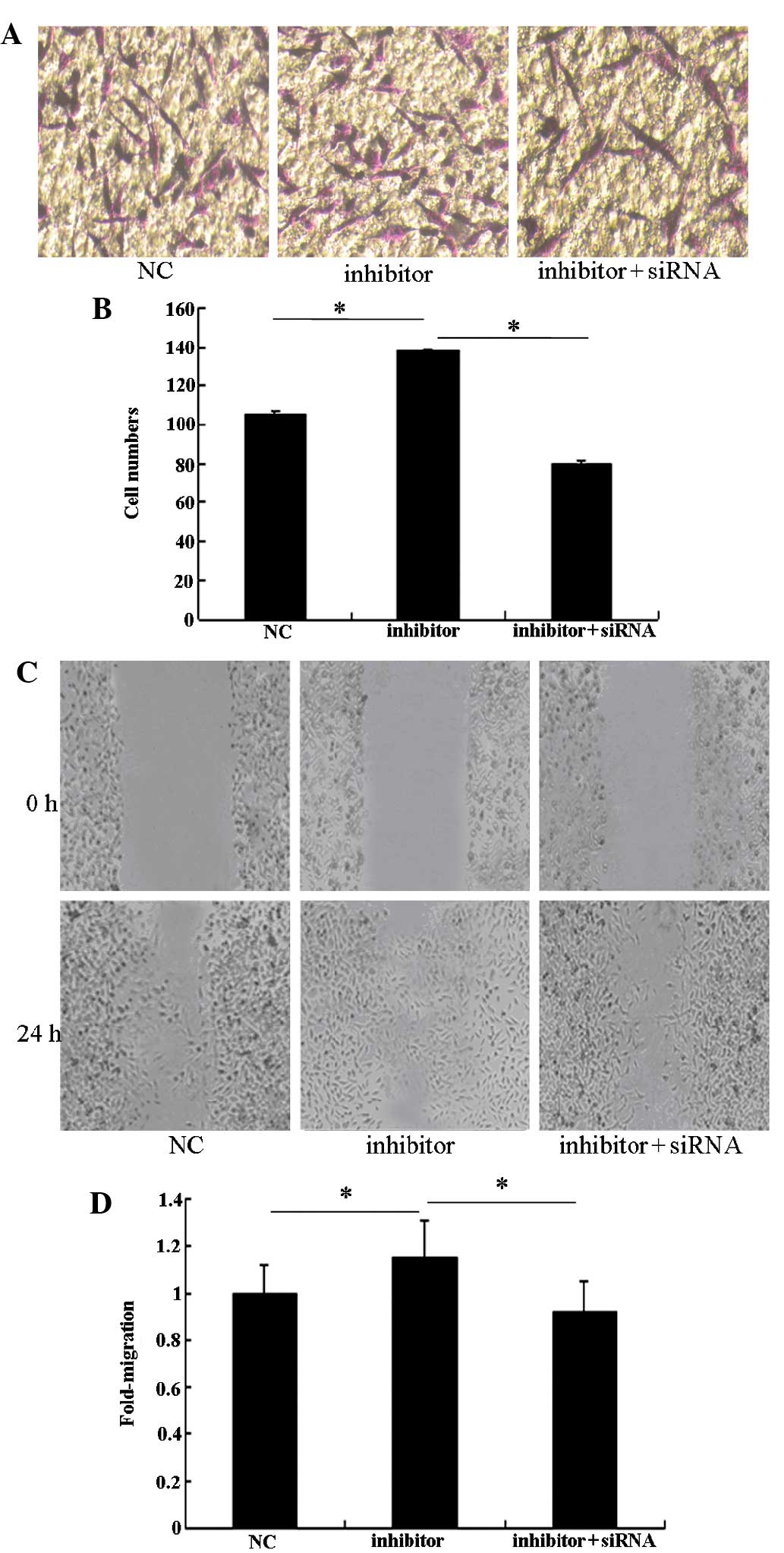
Robo1 siRNA can imitate the role of
miR-218 in U87 glioma cells
To assess the role of Robo1 in the miR-218-dependent
inhibition of cell migration and invasion, miR-218 inhibitor was
transfected into U87 cells treated with Robo1 siRNA. As expected,
Robo1 protein expression was significantly reduced by the specific
Robo1 siRNA (Fig. 5). The enhanced
invasive ability of miR-218 inhibitor-transfected U87 cells
declined when Robo1 siRNA was co-transfected with the miR-218
inhibitor (Fig. 6). These results
indicate that Robo1 is essential for miR-218-dependent cell
migration and invasion.
Discussion
Malignant gliomas are diffuse tumors that are
extremely invasive and usually multifocal. Glioma possesses a
dismal prognosis, with a median survival of ~16 months (18). The ability for single tumor cell
infiltration, which involves the extension of tendrils of the tumor
several centimeters away from the main tumor mass, is one of the
major obstacles for the effective treatment of gliomas, as this
infiltration results in incomplete surgical removal and contributes
to the high frequency of tumor recurrence (19). Despite the increasing quantity of
evidence that demonstrates the promotion of glioma cell
infiltration into the brain parenchyma by various stimuli, the
mechanisms underlying the dysregulation of cell motility during
tumor invasion by the insidious glioma cells have yet to be
elucidated (20). As a result,
exploring novel treatment methods is an urgent clinical challenge
for neuroscientists.
Previous studies have revealed that miR-218
expression is often downregulated in several human cancers,
including gastric cancer, lung squamous cell carcinoma, malignant
astrocytomas and medulloblastomas, which indicates that miR-218 may
function as a tumor suppressor (21–23).
Studies have identified that the ectopic expression of miR-218
contributes to the inhibition of proliferation, invasion and
migration in glioma cells, as well as the induction of apoptosis by
downregulating the gene that was directly targeted by miR-218
(12–15). Firstly, miR-218 inhibits the
expression of the target gene inhibitor of nuclear factor κB
(NF-κB) kinase β, and in a dose-dependent manner, inhibits the
expression of NF-κB, whilst reducing the expression of matrix
metalloproteinase (MMP) 9 and inhibiting the invasion and migration
ability of glioma cells (12).
Secondly, epidermal growth factor receptor-coamplified and
overexpressed protein (ECOP) has been identified as a functional
downstream target gene of miR-218 that can regulate NF-κB
transcription activity and is associated with the apoptotic
response. Overexpression of miR-218 can restrain the activity of
NF-κB through ECOP, thus inducing glioma cell apoptosis and
inhibiting the activity, proliferation and tumorigenicity of glioma
cells (13). Thirdly, the
expression of lymphoid enhancer-binding factor 1 (LEF1) and MMP-9
in the high grade glioma group is extremely high, while the
expression in the low-grade glioma group is extremely low, and is
negatively correlated with the expression of miR-218.
Overexpression of miR-218 inhibits the Wnt/LEF1 signaling pathways
that lead to a reduction in MMP-9 synthesis, inhibiting tumor
invasion (14). Finally, the
abnormal expression of miR-218 in glioma cells decreased, but there
was an abnormal increase in cyclin-dependent kinase (CDK) 6
expression, with the expression level of the two being negatively
correlated. Overexpression of miR-218 in the glioma cell lines can
inhibit CDK6 expression and glioma cell proliferation and promote
its apoptosis (15).
Tumor development is a complex multi-step process
that includes malignant tumor invasion and metastasis (24). The development of invasiveness in
malignant glioma cells involves several genetic alterations within
signaling pathways. Numerous studies have reported that the
Slit2/Robo1 signaling channels can inhibit glioma invasion and
migration. An in vitro study performed by Mertsch et
al (25) identified, using a
modified Boyden chamber assay, that the Slit2/Robo1 system serves
as a chemorepellent for glioma cells, indicating that glioblastoma
cells migrate away from increased Slit2 concentrations and prompt
Robo1-positive glioma cell invasion along gray matter tracts and
into white matter, including the corpus callosum (25). A previous in vivo study
revealed that ectopic expression of Slit2 in SNB19 cells attenuates
cell migration and invasion (20).
These results indicate that Slit2/Robo1 can inhibit glioma invasion
and migration in vivo and in vitro. Slit2 may also be
a tumor suppressor gene that inhibits the migration and invasion of
tumor cells and this inhibition appears to be mediated by Robo1
(20). Previous studies identified
that miR-218 inhibited invasion and metastasis of gastric cancer by
targeting the Robo1 receptor and suppressed nasopharyngeal cancer
progression through the downregulation of survivin and the
Slit2-Robo1 pathway (16,17).
In the present study, Robo1 was identified as a
notable novel target of miR-218 using the conventional prediction
tool TargetScan (www.targetscan.org). The expression
levels of miR-218, Robo1 and Slit2 were detected in 70 tissue
samples, consisting of normal brain tissue and low- and high-grade
glioma tissues, using RT-qPCR and western blot analysis. It was
found that the expression of miR-218 and Slit2 was always inverse
to that of Robo1. Notably, the mRNA and protein levels of Robo1
were significantly decreased and the mRNA and protein levels of
Slit2 were significantly increased subsequent to the transfection
of miR-218 mimics into U87 cells. Furthermore, it was found that
miR-218 was involved in modulation of the Slit2-Robo1 signaling
pathway and downregulation of Robo1 expression by directly
targeting the Robo1 3′-UTR. In addition, Robo1 siRNA can reduce the
invasive ability of the cells subsequent to its enhancement by the
exogenous expression of the miR-218 inhibitor. Overall, the present
results indicate that miR-218 inhibits the migration and invasion
of glioma cells through the Slit2-Robo1 signaling pathway.
Therefore, the development miR-218 as a biomarker for glioma or as
a potential therapeutic candidate for miRNA replacement therapy is
extremely promising (26,27).
References
|
1
|
Dallol A, Krex D, Hesson L, Eng C, Maher
ER and Latif F: Frequent epigenetic inactivation of the SLIT2 gene
in gliomas. Oncogene. 22:4611–4616. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Louis DN, Ohgaki H, Wiestler OD, et al:
The 2007 WHO classification of tumours of the central nervous
system. Acta Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Grauer OM, Wesseling P and Adema GJ:
Immunotherapy of diffuse gliomas: biological background, current
status and future development. Brain Pathol. 19:674–693. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dontula R, Dinasarapu A, Chetty C, et al:
MicroRNA 203 Modulates Glioma Cell Migration via Robo1/ERK/MMP-9
Signaling. Genes Cancer. 4:285–296. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Long JM and Lahiri DK: Advances in
microRNA experimental approaches to study physiological regulation
of gene products implicated in CNS disorders. Exp Neurol.
235:402–418. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Park S and James CD: ECop
(EGFR-coamplified and overexpressed protein), a novel protein,
regulates NF-kappaB transcriptional activity and associated
apoptotic response in an IkappaBalpha-dependent manner. Oncogene.
24:2495–2502. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu B, Hsu PK, Karayiorgou M and Gogos JA:
MicroRNA dysregulation in neuropsychiatric disorders and cognitive
dysfunction. Neurobiol Dis. 46:291–301. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Godlewski J, Nowicki MO, Bronisz A,
Williams S, et al: Targeting of the Bmi-1 oncogene/stem cell
renewal factor by microRNA-128 inhibits glioma proliferation and
self-renewal. Cancer Res. 68:9125–9130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Silber J, Lim DA, Petritsch C, et al:
MiR-124 and miR-137 inhibit proliferation of glioblastoma
multiforme cells and induce differentiation of brain tumor stem
cells. BMC Med. 6:142008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rao SA, Santosh V and Somasundaram K:
Genome-wide expression profiling identifies deregulated miRNAs in
malignant astrocytoma. Mod Pathol. 23:1404–1417. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Song L, Huang Q, Chen K, et al: miR-218
inhibits the invasive ability of glioma cells by direct
downregulation of IKK-β. Biochem Biophys Res Commun. 402:135–140.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xia H, Yan Y, Hu M, et al: MiR-218
sensitizes glioma cells to apoptosis and inhibits tumorigenicity by
regulating ECOP-mediated suppression of NF-κB activity. Neuro
Oncol. 15:413–422. 2013. View Article : Google Scholar :
|
|
14
|
Liu Y, Yan W, Zhang W, et al: MiR-218
reverses high invasiveness of glioblastoma cells by targeting the
oncogenic transcription factor LEF1. Oncol Rep. 28:1013–1021.
2012.PubMed/NCBI
|
|
15
|
Zhang JM, Sun CY, Yu SZ, et al:
Relationship between miR-218 and CDK6 expression and their
biological impact on glioma cell proliferation and apoptosis.
Zhonghua Bing Li Xue Za Zhi. 40:454–459. 2011.(In Chinese).
PubMed/NCBI
|
|
16
|
Tie J, Pan Y, Zhao L, et al: MiR-218
inhibits invasion and metastasis of gastric cancer by targeting the
Robo1 receptor. PLoS Genet. 6:e10008792010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Alajez NM, Lenarduzzi M, Ito E, et al:
MiR-218 suppresses nasopharyngeal cancer progression through
downregulation of survivin and the SLIT2-ROBO1 pathway. Cancer Res.
71:2381–2391. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Stupp R, Mason WP, van den Bent MJ, et al;
European Organisation for Research and Treatment of Cancer Brain
Tumor and Radiotherapy Groups; National Cancer Institute of Canada
Clinical Trials Group. Radiotherapy plus concomitant and adjuvant
temozolomide for glioblastoma. N Engl J Med. 352:987–996. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Furnari FB, Fenton T, Bachoo RM, et al:
Malignant astrocytic glioma: genetics, biology, and paths to
treatment. Genes Dev. 21:2683–2710. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yiin JJ, Hu B, Jarzynka MJ, et al: Slit2
inhibits glioma cell invasion in the brain by suppression of Cdc42
activity. Neuro Oncol. 11:779–789. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Martinez I, Gardiner AS, Board KF, et al:
Human papillomavirus type 16 reduces the expression of microRNA-218
in cervical carcinoma cells. Oncogene. 27:2575–2582. 2008.
View Article : Google Scholar :
|
|
22
|
Petrocca F, Visone R, Onelli MR, et al:
E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest
and apoptosisin gastric cancer. Cancer Cell. 13:272–286. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yanaihara N, Caplen N, Bowman E, et al:
Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu Y, Li WL, Fu L, et al: Slit2/Robo1
signaling in glioma migration and invasion. Neurosci Bull.
26:474–478. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mertsch S, Schmitz N, Jeibmann A, et al:
Slit2 involvement in glioma cell migration is mediated by Robo1
receptor. J Neurooncol. 87:1–7. 2008. View Article : Google Scholar
|
|
26
|
Bader AG, Brown D and Winkler M: The
promise of microRNA replacement therapy. Cancer Res. 70:7027–7030.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Roth P, Wischhusen J, Happold C, et al: A
specific miRNA signature in the peripheral blood of glioblastoma
patients. J Neurochem. 118:449–457. 2011. View Article : Google Scholar : PubMed/NCBI
|