Introduction
Hepatocellular carcinoma (HCC) is one of most
frequently occurring neoplasms worldwide (1) and a major cause of cancer-associated
mortality owing to its high potential of invasion and metastasis.
Surgery, including liver resection and transplantation, remains the
most effective treatment for HCC. However, high recurrence or
metastatic rate after surgery hinders further improvements in HCC
survival (2). Molecules involved in
HCC recurrence and metastasis are potential factors predicting
patient outcome, and personalized treatment depends mostly on the
increased use of diagnostic markers for treatment allocation
(3,4). Therefore, the identificaiton of an
effective biomarker is pivotal in screening cancer, assessing
general population risk, evaluating treatment-response and
monitoring recurrence.
Interleukin 9 (IL-9) cytokine is a 14-kDa
glycoprotein comprising 144 amino acids (5). The receptor of IL-9 (IL-9R) is a
member of hematopoietin consisting of two subunits, the IL-9
specific α-chain receptor and a member of the γ-chain-receptor
family commonly shared with IL-2, IL-4, IL-15 and IL-17 (6–8). IL-9
was considered to be a T-lymphocyte growth factor and a cytokine
produced by the Th2 subset (6,9). As
indicated by the T-cell growth function of IL-9, IL-9R is expressed
in T cells and effector T cells (10). Addtionally, IL-9R is highly
expressed in the Th2 and Th17 cell subsets (8). However, in asthma patients, IL-9R was
reported to be expressed in mast and polymorphonuclear cells
(7,8). The secretion of IL-9 was observed in a
variety of immune cells, including Th2, Th9, Th17 and Treg cells,
NKT cells and mast cells (5).
Activation of IL-9R results in phosphorylation of Jak1 in the α
chain and Jak3 in the γ chain, which leads to the downstream
activation of STAT3, MAPK and PI3K pathways (8,11,12).
IL-9 and its receptor have been investigated in
bronchial asthma (13), autoimmune
diseases (14), parasitic
infections (15) and atopic
dermatitis (16). In the mouse
inflammatory models, IL-9 showed proinflammatory activity and
blocking IL-9 inhibited airway remodeling in chronic lung
inflammation (17). In chronic
lymphocytic leukemia (CLL), IL-9R overexpression inhibited
apoptosis significantly (18).
Aberrant expression of IL-9R was also observed in several types of
human leukemia (19).
HCC is a well-known inflammation-related cancer
(20). However, the expression of
IL-9R and its biological role in HCC tumorigenesis and metastasis
remain largely unknown. In the present study, we identified
aberrant IL-9R expression in HCC and investigated its biological
role in HCC cells as well as the clinical significance in 329 HCC
patients undergoing resection. A high expression of IL-9R in HCC
correlated with tumor aggressiveness and served as an independent
prognostic factor for HCC patients.
Materials and methods
Patient samples
We randomly selected 12 HCC tissue samples for
RT-qPCR analysis from the patients who underwent liver resection at
the Zhongshan Hospital, Fudan University (Shanghai, China). Samples
were collected within 15 min of hepatectomy, delivered in liquid
nitrogen and stored at −80°C. Tumor specimens used in the tissue
microarray analysis were consecutively selected from 329 HCC
patients, who underwent liver resection in the Liver Cancer
Institute, Zhongshan Hospital, Fudan University in 2006. After
fixed in 4% formaldehyde for 24 h, tissues of HCC patients were
used for construction of paraffin-embedded blocks as described
previously (21). The
clinicopathological characteristics of the patients are shown in
Table I.
 | Table ICorrelation between IL-9R expression
and clinicopathological characteristics in HCC patients. |
Table I
Correlation between IL-9R expression
and clinicopathological characteristics in HCC patients.
| Variables | IL-9R high | IL-9R low | P-value |
|---|
| Age, years
(≤52/>52) | 37/47 | 125/120 | 0.270 |
| Gender
(male/female) | 73/11 | 205/40 | 0.480 |
| Hepatitis history
(yes/no) | 74/10 | 218/27 | 0.825 |
| Liver cirrhosis
(yes/no) | 71/13 | 199/46 | 0.496 |
| AFP (≤20/>20
ng/ml) | 36/48 | 92/153 | 0.389 |
| ALT (≤75/>75
U/l) | 67/17 | 210/35 | 0.197 |
| γ-GT (≤54/>54
U/l) | 25/59 | 52/193 | 0.111 |
| Tumor size
(<5/≥5 cm) | 34/50 | 138/107 | 0.012 |
| Tumor number | | | |
|
(Single/multiple) | 66/18 | 209/36 | 0.150 |
| Tumor capsule
(yes/no) | 51/33 | 128/117 | 0.179 |
| Tumor
differentiation | | | |
| (I-II/III-IV) | 62/22 | 176/69 | 0.727 |
| Vascular invasion
(yes/no) | 28/56 | 77/168 | 0.747 |
| TNM stage
(I/II-III) | 39/45 | 148/97 | 0.018 |
| BCLC stage
(0-A/B-C) | 37/47 | 105/140 | 0.849 |
Detailed patient inclusion and exclusion criteria
have been previously described (21). The tumor stage was assessed
according to the Barcelona Clinic Liver Cancer (BCLC) staging
system (22) and the
tumor-node-metastasis (TNM) system of the International Union
against Cancer (UICC, 7th edition). Tumor differentiation was
graded using the Edmondson grading system. Detailed procedures for
postoperative surveillance and treatment modalities have been
previously described (23). The
present study was approved by the Research Ethics Committee of
Zhongshan Hospital (Shanghai, China). Written consent was provided
by the patients. Overall survival (OS) was defined as the date of
surgery and death or the last observation recorded. Relapse-free
survival (RFS) was defined as the interval between resection and
tumor recurrence, death or the last follow up. The data were
censored for surviving patients and patients without signs of
recurrence.
Cell lines
Six human HCC cell lines were used in the present
study: MHCC97H, MHCC97L (Liver Cancer Institute, Fudan University,
Shanghai, China), PLC/PRF/5 (Japanese Cancer Research Bank, Tokyo,
Japan), HepG2, Hep3B (American Type Culture Collection, Manassas,
VA, USA) and the immortalized human normal L-02 liver cell line
(Cell Bank, Chinese Academy of Sciences, China). The cell lines
were routinely maintained in DMEM or RPMI-1640 supplemented with
10% heat-inactivated fetal bovine serum, 100 U/ml penicillin and
100 mg/ml streptomycin at 37°C in a humidified incubator under 5%
CO2.
RNA isolation and RT-qPCR
Total RNA was extracted from cell lines and frozen
tumor specimens using TRIzol reagent (Invitrogen, Carlsbad, CA,
USA) according to the manufacturer’s instructions. mRNA expression
of IL-9 and IL-9R in HCC cell lines and tumor tissues was detected
by RT-qPCR using an ABI7900HT (Applied Biosystems, Foster, CA,
USA). RT-qPCR was performed using a SYBR PrimeScript RT-PCR kit
(Takara Bio Inc., Otsu, Shiga, Japan) according to the
manufacturer’s instructions. The primer sequences used were: IL-9
forward, 5′-CTCTGCCCTGCTCCTGTGCT-3′ and reverse,
5′-GCCTGCCGTGGTTTGGTT-3′; IL-9R forward, 5′-TCACCATCACTTTCCACCAC-3′
and reverse, 5′-CCTCTACCACATCATCCTCC-3′; GAPDH forward,
5′-CGGAGTCAACGGATTTGGTCGTAT-3′ and reverse,
5′-AGCCTTCTCCATGGTGGTGAAGAC-3′. Relative mRNA levels were detected
based on the Ct values, and corrected to GAPDH expression,
according to the 2−ΔCt method. The experiments were
performed in triplicate.
Western blot analysis
The protein level expression of IL-9R and signaling
molecules was detected by western blot analysis. Briefly, the cells
were washed twice with ice-cold PBS and frozen tumor specimens were
ground under liquid nitrogen. Total protein was extracted in lysis
buffer for 45 min on ice. Equal amounts of protein were separated
by 10% SDS-PAGE and transferred to a PVDF membrane (Millipore,
Billerica, MA, USA) using a mini trans-blot apparatus (Bio-Rad
Laboratories, CA, USA). The membrane was blocked with PBSwith 0.05%
Tween-20 containing 5% non-fat dry milk for 1 h and incubated for 2
h at room temperature with monoclonal/polyclonal mouse/rabbit
anti-human antibodies including IL9R (Abcam, Cambridge, MA, USA),
VEGF, p-JNK, JNK, p-STAT3, STAT3, p-STAT5, STAT5, p38, p-p38,
p-ERK, ERK, MMP9 (Cell Signaling, Danvers, MA, USA) and GADPH
(Sigma-Aldrich, St. Louis, MO, USA). Each experiment was repeated
at least three times.
SiRNA-mediated IL-9R silencing
An Annexin V-FITC apoptosis detection kit was used
to determine IL-9R knockdown-mediated effects on cell apoptosis.
The target siRNA sequence for IL9R used was:
5′-CCTCTCCAGCGATGTTCTT-3′. MHCC97H and MHCC97L were used in the
siRNA analysis and transfection of siRNA was carried out using
Lipofectamine 2000 (Invitrogen) according to the manufacturer’s
instructions.
Cell proliferation, cycle, invasion and
apoptosis assays
Cell proliferation was analyzed using a Cell
Counting kit-8 (CCK-8) assay kit (Dojindo Corp, Japan). The Tecan
Infinite 200 microplate reader (Tecan Inc., Mäennedorf,
Switzerland) was used to detect absorbance at 450 nm.
Transwell (8-µm pore size; Millipore) coated
with Matrigel (BD Biosciences, San Jose, CA, USA) was used to
evaluate cell invasion. Cells (1×105) were added to the
upper chamber and after 48-h incubation, the cells remaining in the
upper chamber were removed using cotton swabs. The cells that had
migrated or invaded were fixed and stained in a dye solution
containing 0.1% crystal violet and 20% methanol. The cells were
counted in five random fields at a magnification of ×100. The
experiments were performed in triplicate.
Cell cycle and apoptosis were analyzed using flow
cytometry. Briefly, the cells were collected, stained and fixed in
ice-cold 70% ethanol for FACS analysis (Beckman-Coulter, Brea, CA,
USA) using MultiCycle AV for Windows 5.0 (Phoenix Flow Systems, San
Diego, CA, USA). Apoptosis was measured using apoptosis detection
kit (BD Biosciences).
Tissue microarray and
immunohistochemistry
Tissue microarray blocks were constructed as
previously described (23).
Briefly, the HCC tissues were reviewed by two histopathologists and
representative tumor areas free from necrotic and hemorrhagic
materials were premarked in the paraffin blocks. Two core biopsies
of 1 mm in diameter were obtained from the tumor and peritumor
region of the donor blocks and transferred to the recipient
paraffin block at defined array positions. Four different tissue
microarray blocks were then constructed. Immunohistochemistry was
performed according to a previously described two-step protocol
(Novocastra, Newcastle, UK) (23).
The primary antibody used in immunohistochemistry was rabbit
polyclonal anti-human IL-9R (Abcam, Cambridge, UK).
Immunohistochemical staining was assessed by two
independent pathologists without knowledge of patient
characteristics. For IL-9R staining, we determined the percentage
of cells with a positive score in the cytosol or membrane in the
whole biopsy cylinder. The scoring procedure was as follows: the
staining intensity was first scored (0, negative; 1, weak; 2,
moderate; 3, high) and the percentage of positive cells was scored
(0, 0% positive cells; 1, 1–10% positive cells; 2, 11–50% positive
cells; 3, >50% positive cells). The final score was calculated
by multiplying the staining intensity score by the percentage of
positive cells. Samples were defined as negative when the final
scores were 0–3 and positive when 4–9.
Statistical analysis
Statistical analysis was performed using SPSS 18.0
for windows (SPSS, IBM, NY, USA). Survival analysis was performed
by the Kaplan-Meier method and the log-rank test. Univariate and
multivariate analyses were based on the Cox proportional hazards
regression model. The Chi-square test, Fisher’s exact probability
and Student’s t-test were used for comparison between groups. The
experimental data were presented as means ± SD. A two-tailed
P<0.05 was considered statistically significant.
Results
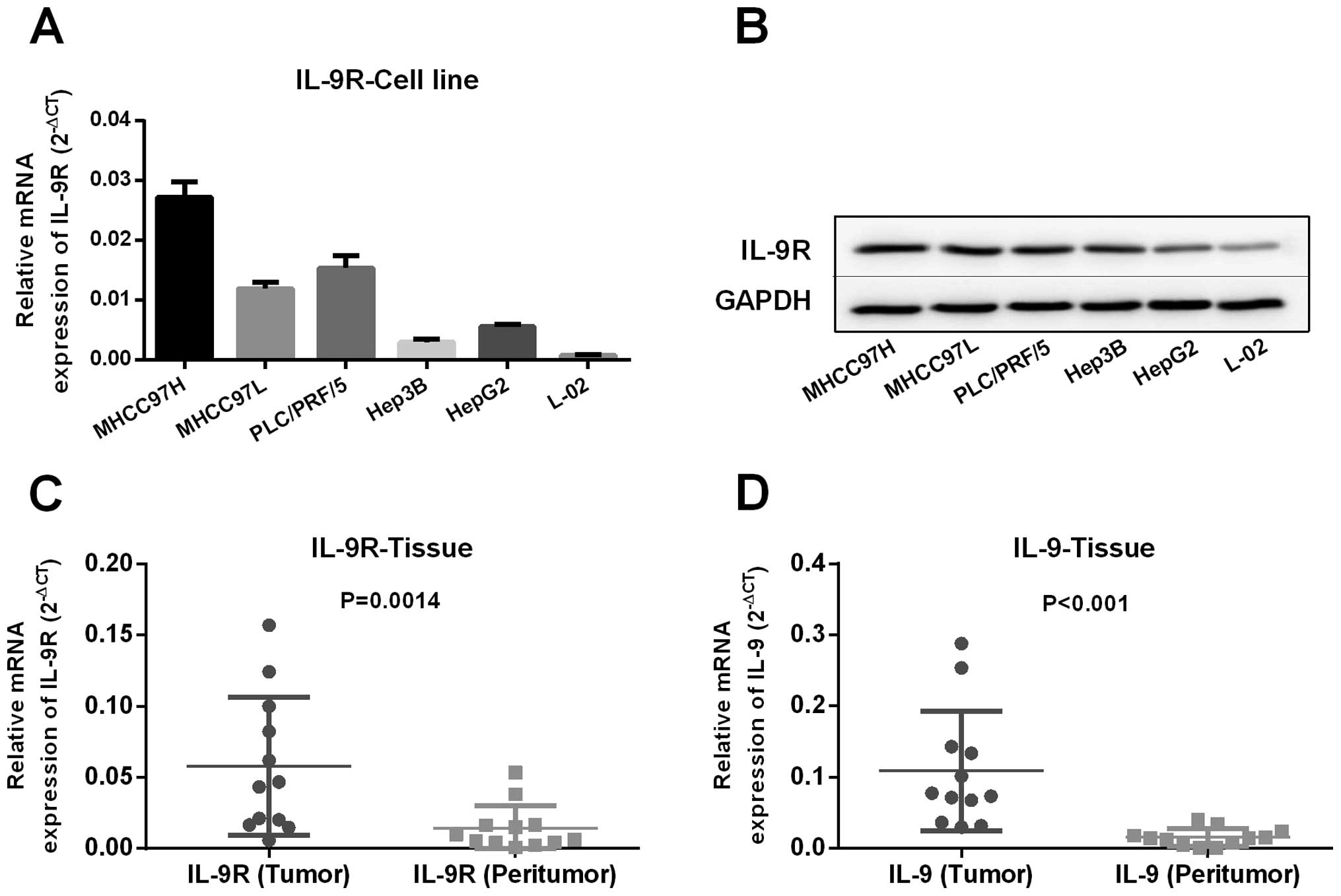
IL-9R is constitutively expressed in HCC
cells and tissues
IL-9R has been reported to be expressed in T
cell-derived subsets, especially Th2 and Th17 cells (19). Expression of IL-9R was significantly
higher in T-cell lymphoma cells than that in normal thymocytes,
leading to aberrant downstream STAT3 and STAT5 phosphorylation
(24). In the present study, we
used western blot and RT-qPCR analyses to determine whether IL-9R
was expressed in HCC cells and tissues. RT-qPCR showed that IL-9R
was constitutively expressed in HCC cell lines and that the
expression level correlated with HCC cell line metastatic ability
(Fig. 1A). Additionally, we found
the ligand for IL-9R, such as IL-9, was expressed in HCC cell lines
at the mRNA level (data not shown). Western blot analysis validated
RT-qPCR results and again indicated that IL-9R was expressed on HCC
cells (Fig. 1B). Furthermore,
expression level of IL-9R and IL-9 was significantly higher in
tumor tissues compared to peritumor tissues by means of RT-qPCR
analysis (Fig. 1C and D,
P<0.001).
IL-9R knockdown mediated by siRNA
significantly inhibits cell proliferation
IL-9 stimulates the proliferation of human T-ALL,
Hodgkin lymphoma and T-cell leukemic cells (19). To examine the biological function of
IL-9R in HCC cells, MHCC97H and MHCC97L were selected for
transfection with siRNA duplexes against IL-9R. Successful
knockdown in MHCC97H and MHCC97L was confirmed by RT-qPCR (Fig. 2A, P<0.001) and western blot
analysis (Fig. 2B). IL-9R
downregulation mediated by siRNA significantly inhibited MHCC97H
and MHCC97L cell proliferation following transfection at 72 h
(Fig. 2C and D, P<0.001). An
Annexin V-FITC apoptosis detection kit was used to examine the
IL-9R knockdown-mediated effects on cell apoptosis. After IL-9R
silencing, the percentage of cells in S phase markedly decreased,
which was consistent with our proliferation analysis showing that
inhibition of IL-9R attenuated HCC cell proliferation.
IL-9R downregulation significantly
reduces invasiveness and promoted apoptosis of HCC cells
IL-9 is a versatile cytokine with various functions
in different types of cells. Previous study of IL-9R in leukemia
has suggested that overexpression of IL-9R inhibited leukemic cell
apoptosis (18). In the present
study, we used a Transwell assay and Annexin V-FITC apoptosis
detection kit to determine the biological effect of IL-9R on HCC
cells. In the Transwell experiment, MHCC97H and MHCC97L cells
invading through the membrane decreased significantly after IL-9R
silencing by siRNA (Fig. 3A and B,
P<0.01). Furthermore, in the apoptosis experiment, the
percentage of apoptotic cells increased significantly in the
IL-9R-silenced groups compared to the control (Fig. 3C and D, P<0.01).
Molecular mechanism involved in IL-9R
signaling pathway
IL-9 mediated signal transduction activated
molecular members of STAT, MAPK and PI3K pathways (8). In the present study, we analyzed the
expression of VEGF, p-p38, p-JNK, p-STAT3, p-STAT5, STAT5, p-ERK
and MMP9, which were previously reported to be potentially involved
in the activity of the IL-9/IL9R axis (8,11,25).
After the significant downregulation of IL-9R, western blot
analysis indicated that the expression of VEGF, p-p38, p-STAT3 and
MMP9 obviously decreased (Fig. 3E and
F). However, no difference was found in the expression of
p-JNK, p-STAT5 and p-ERK (data not shown) following IL-9R
downregulation.
High IL-9R expression significantly
correlates with reduced survival and early recurrence
Tissue microarrays containing 329 patients were used
to evaluate the prognostic value of IL-9R in HCC. Patients with a
high (score between 0 and 3, n=84) or low (score between 4 and 9,
n=245) IL-9R expression were divided into two groups. Comparisons
were determined between various clinicopathologic characteristics
and IL-9R expression (Table I). The
correlation analysis showed that a high IL-9R expression
significantly correlated with larger tumor (P=0.012) and advanced
TNM stage (P=0.018, Table I).
Immunohistochemical staining of IL-9R in HCC cells exhibited a
focal or scattered pattern, with different intensity and percentage
of staining tumor cells (Fig.
4A).
For the entire cohort (n=329), the OS and RFS rates
at 3, 5 and 7 years of age were 69.60, 61.10 and 56.23, and 72.34,
66.26, and 59.27%, respectively. In the univariate analysis, liver
cirrhosis, γ-GT, tumor size, tumor differentiation, vascular
invasion, TNM stage and BCLC stage were shown to be prognostic
factors for OS and RFS (Table II).
Additionally, the univariate analysis showed IL-9R expression was a
significant prognostic factor for OS (P=0.003) and RFS (P=0.007,
Table II). The multivariate
analysis was used to determine whether IL-9R was an independent
factor for HCC. Significant factors in the univariate analysis were
selected for multivariate analysis. IL-9R was shown to be an
independent prognostic factor for OS (P=0.005) and RFS (P=0.030,
Table II). Patients with a high
IL-9R expression had significantly decreased OS (P=0.002, Fig. 4B) and RFS (P=0.018, Fig. 4C). HCC patients with high IL-9R
expression were 1.66-fold more likely to succumb [95% CI,
1.17–2.36; P=0.005] and 1.5-fold more likely to experience
recurrence (95%CI, 1.04–2.17; P=0.03) than the low IL-9R expression
patients (Table II).
 | Table IIUnivariate and multivariate analyses
of factors associated with survival and recurrence. |
Table II
Univariate and multivariate analyses
of factors associated with survival and recurrence.
| Variables |
Univariate
P-value | Overall survival
Multivariate
|
Univariate
P-value | Relapse-free
survival Multivariate
|
|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age, year (>52
vs. ≤52) | 0.788 | NA | | 0.580 | NA | |
| Gender (male vs.
female) | 0.104 | NA | | 0.172 | NA | |
| HBsAg (positive vs.
negative) | 0.378 | NA | | 0.306 | NA | |
| Liver cirrhosis
(yes vs. no) | 0.012 | 1.39
(1.11-1.75) | 0.004 | 0.002 | 1.97
(1.19-3.30) | 0.009 |
| HCC family history
(yes vs. no) | 0.375 | NA | | 0.974 | NA | |
| AFP, ng/ml (>20
vs. ≤20) | 0.141 | NA | | 0.931 | NA | |
| ALT, U/L (>75
vs. ≤75) | 0.427 | NA | | 0.281 | NA | |
| γ-GT, U/L (>54
vs. ≤54) | 0.008 | 1.59
(1.02-2.48) | 0.040 | 0.245 | NA | |
| Tumor size (>5
vs. ≤5) | <0.001 | 2.21
(1.57-3.11) | <0.001 | <0.001 | 1.97
(1.19-3.30) | <0.001 |
| Tumor number
(multiple vs. single) | 0.093 | NA | | 0.163 | NA | |
| Tumor capsule (yes
vs. no) | 0.105 | NA | | 0.826 | NA | |
| Tumor
differentiation (III-IV vs. I-II) | 0.009 | NS | | 0.022 | NS | |
| Vascular invasion
(yes vs. no) | 0.001 | 1.57
(1.15-2.20) | 0.010 | 0.025 | NS | |
| TNM stage (II/III
vs. I) | <0.001 | NA | | 0.013 | NA | |
| BCLC stage (B/C vs.
0/A) | <0.001 | NA | | 0.021 | NA | |
| IL-9R (high vs.
low) | 0.003 | 1.66
(1.17-2.36) | 0.005 | 0.007 | 1.50
(1.04-2.17) | 0.030 |
Discussion
IL-9 is a versatile cytokine that functions in
immune and inflammatory responses as well as in growth-promoting
and anti-apoptotic activities suggesting its multifunctional role
in tumorigenesis (19). In mast
cells, IL-9 acted as a growth factor while in human B cells, IL-9
promoted the production of IgE and IgG through IL-4 signaling
(5). The abnormal expression of
IL-9R significantly promotes T-ALL cells and inhibits apoptosis in
chronic leukemia (5,25).
In the present study, we found that IL-9R was
constitutively expressed in the HCC cell lines and tissues of HCC
patients. mRNA and protein expression results in the HCC cell lines
showed that a high expression of IL-9R correlated with a higher
metastatic potential of the cell line. Previous findings showed
that there was an abnormal feedback loop of IL-9 and IL-9R in
various human leukemias, including T-cell leukemia,
megakaryoblastic leukemia and Hodgkin lymphomas (19,26,27).
The RT-qPCR detection experiment in this study showed that IL-9R
was overexpressed in HCC tumor tissues and the mRNA expression
level of ligand IL-9 was significantly higher in tumors than that
in peritumors, which indicated a potential feedback loop existed in
HCC and it seems to be an active mechanism for HCC
tumorigenesis.
IL-9R was found to be constitutively expressed in
HCC tumor cells. We then used proliferation, cell cycle, apoptosis
and Transwell assays to determine the biological effect of IL-9R on
HCC cells. Downregulation of IL-9R by siRNA in MHCC97H and MHCC97L
significantly inhibited cell proliferation compared to the control
groups. The percentages of cells in the S-phase were markedly lower
than that in the control groups. Results of the two experiments
indicated that overexpression of IL-9R in HCC cells promoted cell
proliferation. This finding was consistent with previous findings
showing that IL-9 promoted the proliferation of T-ALL cells
(26), Hodgkin lymphoma cells
(28) and natural killer/T-cell
lymphoma cells (19,29). It has been shown that IL-9 inhibited
apoptosis of the human CLL cell line MEC-1 (18). In the present study, we showed that
IL-9R downregulation significantly promoted cell apoptosis and
inhibited invasive potential of HCC cell lines. Our in vivo
analysis simultaneously demonstrated that a high IL-9R expression
significantly correlated with larger tumor volume and advanced TNM
stages.
Activation of IL-9R leads to the downstream
activation of STAT3, MAPK and PI3K pathway (8,12,25).
Additionally, the function of JNK and p38 MAPK family members had a
pivotal role in affecting tumor cell proliferation, differentiation
and migration (30). Consistent
with the importance of these facts, our western blot analysis
indicated that after silencing IL-9R, the expression of VEGF,
p-p38, p-STAT3 and MMP9 was significantly decreased. Involvement of
the four molecules may be the underlying mechanism of the
biological effects of IL-9R in HCC cells. By contrast, p-JNK,
p-STAT5 or p-ERK did not show any significant changes. Future
studies are needed to elaborate the detailed signaling pathways and
the exact mechanism involved.
Apart from the tumor-promoting potential of IL-9R,
we found that IL-9R was an independent predictor for HCC patients.
A cohort of 329 HCC patients indicated that the OS and RFS in the
high IL-9R expression group was significantly lower than that in
the low expression patients. Patients with a high IL-9R expression
were predisposed to succumbing to the disease (1.66; 95% CI,
1.17–2.36; P=0.005) and relapse (1.50; 95% CI, 1.04–2.17;
P=0.03).
In conclusion, to the best of our knowledge, we
report for the first time the biological and tumor-promoting role
of IL-9R in HCC. IL-9R was constitutively expressed in HCC cells,
promoted cell proliferation and invasive ability, inhibited
apoptosis and acted as an independent predictor for the survival of
HCC. Therefore, together with our findings, IL-9R may be used as a
potential biomarker in HCC therapy.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Niu ZS, Niu XJ and Wang M: Management of
hepatocellular carcinoma: Predictive value of immunohistochemical
markers for postoperative survival. World J Hepatol. 7:7–27. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hanash SM, Baik CS and Kallioniemi O:
Emerging molecular biomarkers - blood-based strategies to detect
and monitor cancer. Nat Rev Clin Oncol. 8:142–150. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stassen M, Schmitt E and Bopp T: From
interleukin-9 to T helper 9 cells. Ann N Y Acad Sci. 1247:56–68.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Renauld JC, Druez C, Kermouni A, Houssiau
F, Uyttenhove C, Van Roost E and Van Snick J: Expression cloning of
the murine and human interleukin 9 receptor cDNAs. Proc Natl Acad
Sci USA. 89:5690–5694. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Noelle RJ and Nowak EC: Cellular sources
and immune functions of interleukin-9. Nat Rev Immunol. 10:683–687.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Goswami R and Kaplan MH: A brief history
of IL-9. J Immunol. 186:3283–3288. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Demoulin JB and Renauld JC: Interleukin 9
and its receptor: An overview of structure and function. Int Rev
Immunol. 16:345–364. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Druez C, Coulie P, Uyttenhove C and Van
Snick J: Functional and biochemical characterization of mouse
P40/IL-9 receptors. J Immunol. 145:2494–2499. 1990.PubMed/NCBI
|
|
11
|
Demoulin JB, Uyttenhove C, Van Roost E,
DeLestré B, Donckers D, Van Snick J and Renauld JC: A single
tyrosine of the interleukin-9 (IL-9) receptor is required for STAT
activation, antiapoptotic activity, and growth regulation by IL-9.
Mol Cell Biol. 16:4710–4716. 1996.PubMed/NCBI
|
|
12
|
Demoulin JB, Louahed J, Dumoutier L,
Stevens M and Renauld JC: MAP kinase activation by interleukin-9 in
lymphoid and mast cell lines. Oncogene. 22:1763–1770. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yamasaki A, Saleh A, Koussih L, Muro S,
Halayko AJ and Gounni AS: IL-9 induces CCL11 expression via STAT3
signalling in human airway smooth muscle cells. PLoS One.
5:e91782010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jäger A, Dardalhon V, Sobel RA, Bettelli E
and Kuchroo VK: Th1, Th17, and Th9 effector cells induce
experimental autoimmune encephalomyelitis with different
pathological phenotypes. J Immunol. 183:7169–7177. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Forbes EE, Groschwitz K, Abonia JP, Brandt
EB, Cohen E, Blanchard C, Ahrens R, Seidu L, McKenzie A, Strait R,
et al: IL-9- and mast cell-mediated intestinal permeability
predisposes to oral antigen hypersensitivity. J Exp Med.
205:897–913. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sismanopoulos N, Delivanis DA,
Alysandratos KD, Angelidou A, Vasiadi M, Therianou A and
Theoharides TC: IL-9 induces VEGF secretion from human mast cells
and IL-9/IL-9 receptor genes are overexpressed in atopic
dermatitis. PLoS One. 7:e332712012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kearley J, Erjefalt JS, Andersson C,
Benjamin E, Jones CP, Robichaud A, Pegorier S, Brewah Y, Burwell
TJ, Bjermer L, et al: IL-9 governs allergen-induced mast cell
numbers in the lung and chronic remodeling of the airways. Am J
Respir Crit Care Med. 183:865–875. 2011. View Article : Google Scholar
|
|
18
|
Chen N, Lu K, Li P, Lv X and Wang X:
Overexpression of IL-9 induced by STAT6 activation promotes the
pathogenesis of chronic lymphocytic leukemia. Int J Clin Exp
Pathol. 7:2319–2323. 2014.PubMed/NCBI
|
|
19
|
Shang Y, Kakinuma S, Nishimura M,
Kobayashi Y, Nagata K and Shimada Y: Interleukin-9 receptor gene is
transcriptionally regulated by nucleolin in T-cell lymphoma cells.
Mol Carcinog. 51:619–627. 2012. View
Article : Google Scholar
|
|
20
|
El-Serag HB: Epidemiology of viral
hepatitis and hepatocellular carcinoma. Gastroenterology.
142:1264–1273.e1. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shi JY, Gao Q, Wang ZC, Zhou J, Wang XY,
Min ZH, Shi YH, Shi GM, Ding ZB, Ke AW, et al: Margin-infiltrating
CD20(+) B cells display an atypical memory phenotype and correlate
with favorable prognosis in hepatocellular carcinoma. Clin Cancer
Res. 19:5994–6005. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vitale A, Morales RR, Zanus G, Farinati F,
Burra P, Angeli P, Frigo AC, Del Poggio P, Rapaccini G, Di Nolfo
MA, et al: Italian Liver Cancer group: Barcelona Clinic Liver
Cancer staging and transplant survival benefit for patients with
hepatocellular carcinoma: A multicentre, cohort study. Lancet
Oncol. 12:654–662. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gao Q, Zhao YJ, Wang XY, Qiu SJ, Shi YH,
Sun J, Yi Y, Shi JY, Shi GM, Ding ZB, et al: CXCR6 upregulation
contributes to a proinflammatory tumor microenvironment that drives
metastasis and poor patient outcomes in hepatocellular carcinoma.
Cancer Res. 72:3546–3556. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shang Y, Kakinuma S, Amasaki Y, Nishimura
M, Kobayashi Y and Shimada Y: Aberrant activation of interleukin-9
receptor and downstream Stat3/5 in primary T-cell lymphomas in vivo
in susceptible B6 and resistant C3H mice. In Vivo. 22:713–720.
2008.
|
|
25
|
Chen N and Wang X: Role of IL-9 and STATs
in hematological malignancies (Review). Oncol Lett. 7:602–610.
2014.PubMed/NCBI
|
|
26
|
Barata JT, Keenan TD, Silva A, Nadler LM,
Boussiotis VA and Cardoso AA: Common gamma chain-signaling
cytokines promote proliferation of T-cell acute lymphoblastic
leukemia. Haematologica. 89:1459–1467. 2004.PubMed/NCBI
|
|
27
|
Glimelius I, Edström A, Amini RM, Fischer
M, Nilsson G, Sundström C, Enblad G and Molin D: IL-9 expression
contributes to the cellular composition in Hodgkin lymphoma. Eur J
Haematol. 76:278–283. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gruss HJ, Brach MA, Drexler HG, Bross KJ
and Herrmann F: Interleukin 9 is expressed by primary and cultured
Hodgkin and Reed-Sternberg cells. Cancer Res. 52:1026–1031.
1992.PubMed/NCBI
|
|
29
|
Nagato T, Kobayashi H, Kishibe K, Takahara
M, Ogino T, Ishii H, Oikawa K, Aoki N, Sato K, Kimura S, et al:
Expression of interleukin-9 in nasal natural killer/T-cell lymphoma
cell lines and patients. Clin Cancer Res. 11:8250–8257. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wagner EF and Nebreda AR: Signal
integration by JNK and p38 MAPK pathways in cancer development. Nat
Rev Cancer. 9:537–549. 2009. View
Article : Google Scholar : PubMed/NCBI
|