Introduction
Gastric cancer is the second most common malignancy
and one of the principal causes of cancer-related mortality
worldwide (1), and remains a
prevalent disease worldwide with poor prognosis (2). The crude mortality rate of gastric
cancer was 10.2 per 100,000 worldwide in 2012 (3). The top four common cancers diagnosed
in China are lung, stomach, liver, and esophageal cancer. They
account for 57% of cancers diagnosed in China, compared with 18% in
the United States (4). This means
that China is one of the countries with the highest gastric cancer
morbidity and mortality (5,6). The incidence of gastrointestinal
carcinomas is rapidly increasing in China, with more than 404,565
newly diagnosed stomach cancers, and ~287,851 deaths from gastric
cancer in 2010 (7). Similarly, more
than 423,500 stomach cancers were newly diagnosed, and ~298,500
patients died from gastric cancer in 2012 (6). There is no national screening program,
so early detection of gastric cancer relies only on opportunistic
screening (8). Early diagnostic and
screening methods for gastric cancer are limited at present, most
of them being invasive procedures, which include biopsy, endoscopy,
enhanced CT and upper gastrointestinal angiography, or serology
tests (2). To indentify a
non-invasive means of early diagnosis or screening for gastric
cancer is crucial.
The bacterial community in the oral cavity is one of
the most complex mixtures of bacteria known. More than 700
bacterial species have been identified from the oral cavity using
ribosomal RNA (rRNA) gene-based techniques (9,10). The
oral cavity is the entrance to the digestive tract, which is often
regarded as the ‘inner outside’ (11). The gastrointestinal tract is
anatomically continuous and harbors ~1×1014
microorganisms, which is more than the ~6×1013 cells
that constitute the entire human body. As mentioned above, of the
various sites in the body, the oral cavity is one of the most
densely populated sites (12). The
oral microbiota is stable and in harmony with the host unless
disturbed by medication, disease, low pH, or significant changes in
diet (13). As the essential part
of the digestive system, the stomach will receive these oral
microbes, flushed by the saliva during the physical process of
swallowing (14). The oral bacteria
can also travel to the stomach with the intake of food, while some
gastric cancer patients have gastroesophageal reflux disease (GERD)
which might affect some bacteria in the oral cavity environment
(15). It has also been reported
that inflammatory bowel disease and periodontitis share several
factors in their etiology and pathogenesis (16).
With the progress in metagenomic research, many
diseases have been associated with the changes in the endogenous
microbial components of the human body. Like periodontal disease,
the pathogenesis of gastric cancer is closely related to bacterial
infection. Until recently, only a few reports have compared the
oral microbiome between individuals with gastric cancer and the
healthy population. Saliva and dental plaque are two relevant
samples in the study of oral microorganisms, which are clinically
easy to obtain and are non-invasive to the patient. This is a
straightforward and easy way to perform a massive epidemiological
survey. We hypothesized that the oral microbiome has a different
population and phenotype in gastric cancer individuals compared
with that in healthy people. To test this hypothesis, we conducted
an exploratory hospital-based case-control study. In the present
study, high-throughput sequencing was chosen as a tool for analysis
of the microbial populations obtained from both oral salivary and
subgingival plaque samples from subjects with gastric cancer and a
control population. The findings of the present study may indicate
that oral microbial detection has the potential to be an early
diagnostic tool and a means of screening for gastric cancer, in the
future.
Materials and methods
Ethics statement
All participants provided written informed consent
for this institutionally-approved study. Ethics approval for this
study was obtained from the Human Ethics Research Committee of the
Beijing Stomatological Hospital, Capital Medical University,
China.
Participants
A total of 50 participants were enrolled in the
present study. The median age of the population was 44 years (range
35–78), with 52% of the participants being female. Subjects had an
average of 21 teeth (range 6–28). All were referred for an upper
endoscopy at the Gastrointestinal Clinic in the National Clinical
Research Center for Digestive Diseases of Beijing Friendship
Hospital. Briefly, after a standardized endoscopic procedure and
histopathological evaluation, the individuals who were diagnosed
with gastric cancer were selected as the study cases (n=37). The
controls (n=13) were free of any gastric cancerous lesions, as
confirmed by biopsy (Table I). No
significant differences were observed between the two groups
regarding demographic, socioeconomic, or lifestyle characteristics.
Individuals who had received antibiotics or used steroids or other
immunomodulating drugs within 4 weeks were excluded from the study.
Women who were pregnant were also excluded from the study.
 | Table I.Height and weight of the case and
control groups (mean ± SD), N=50. |
Table I.
Height and weight of the case and
control groups (mean ± SD), N=50.
| Variable | Case (37) | Control (13) | P-value |
|---|
| Height (cm) | 163.9±8.5 | 164.6±9.1 | 0.812 |
| Weight (kg) |
65.0±8.5 |
64.9±12.4 | 0.973 |
Sample collection and preparation
Sample collection and preparation were carried out
as previously published (17).
Briefly, the subgingival plaque samples were collected from the 1st
molar, or most posterior tooth in each quadrant available, plus
from two additional teeth with the deepest periodontal pockets. The
patient who had six teeth, plaque samples were collected from all
the teeth. The collected plaque mass was transferred into a 2-ml
prelabled centrifuge tube with Tris-EDTA buffer (TE buffer) (10 mM
Tris-Cl, pH 8.0, 1 mM EDTA), and the wet weight of the plaque
samples was measured. Subjects were asked to chew a piece of
paraffin wax, and then ~1 ml of expectorated whole saliva was
collected in a sterile tube with TE buffer. Both salivary and
plaque samples were vortex mixed thoroughly, kept on ice,
transferred to the laboratory and stored at −70°C until DNA
extraction.
High-throughput sequencing of 16S rRNA
gene amplicons
To evaluate the bacterial diversity, high-throughput
sequencing of the 16S rRNA was performed. Bacterial genomic DNA of
the plaque or saliva was isolated. Then, the hypervariable V4
region of the 16S rRNA gene was amplified using the following
primers (18): 515F,
GTGCCAGCMGCCGCGGTAA and 806R, GGACTACHVGGGTWTCTAAT. Sequencing
libraries were generated using the NEBNext® Ultra™ DNA
Library Prep Kit for Illumina (New England BioLabs, Inc., Ipswich,
MA, USA) following the manufacturer's recommendations, and the
index codes were added. Libraries were sequenced on an Illumina
MiSeq, and 250 bp paired-end reads were generated (Novogene
Bioinformatics Technology Co., Ltd., Beijing, China). The
sequencing data were processed with data filtering and operational
taxonomic unit (OTU) assigning (Annoroad Gene Technology Co., Ltd.,
Beijing, China). Reads with uncorrectable bar codes were discarded.
Sequences were assigned to OTUs and then classified taxonomically
using the Greengenes 16S rRNA reference database. The relative
abundance of genus distribution was generated by QIIME v1.8
software. Venn diagram and species abundance clusters were also
generated (Annoroad Gene Technology Co., Ltd.). The score system
for the screening was designed and calculated in Microsoft Excel.
Origin 9 was used to generate the statistical figures.
Results
Overlap of bacteria within the study
groups
We used high-throughput sequencing to examine the
total bacterial profile of the saliva and plaque samples from 50
subjects, including 37 individuals with gastric cancer and 13
controls. The samples were divided into four groups: saliva from
controls (CS); plaque from controls (CP); saliva from gastric
cancer patients (GS); plaque from gastric cancer patients (GP). For
each sample, 13,116-16,000 OTUs (average 14,869) were detected. We
analyzed the OTUs overlapping between saliva and pooled plaque of
case and control groups to assess the similarities and differences
in bacterial diversity (Fig. 1).
The Chao1 and Shannon indices of each sample were close to
saturation, and the sparse curves of each sample almost reached the
plateau, indicating that the experimental data could be used for
subsequent analysis. The results showed that 712 OTUs were detected
in all of the four groups. There were 1,227 OTUs detected in both
the saliva and pooled plaque of the subjects with gastric cancer
(groups GS and GP) compared with 783 OTUs in the CS and CP groups.
We also found that the OTUs that only appeared in the CS or CP
group but not in the GS or GP group were much fewer than those that
only appeared in the GS or GP group but not in the CS or CP group
(59 compared with 407). This indicated that the oral bacteria were
more complex in the patients with gastric cancer.
Bacterial profile of the saliva and
plaque samples in the study population
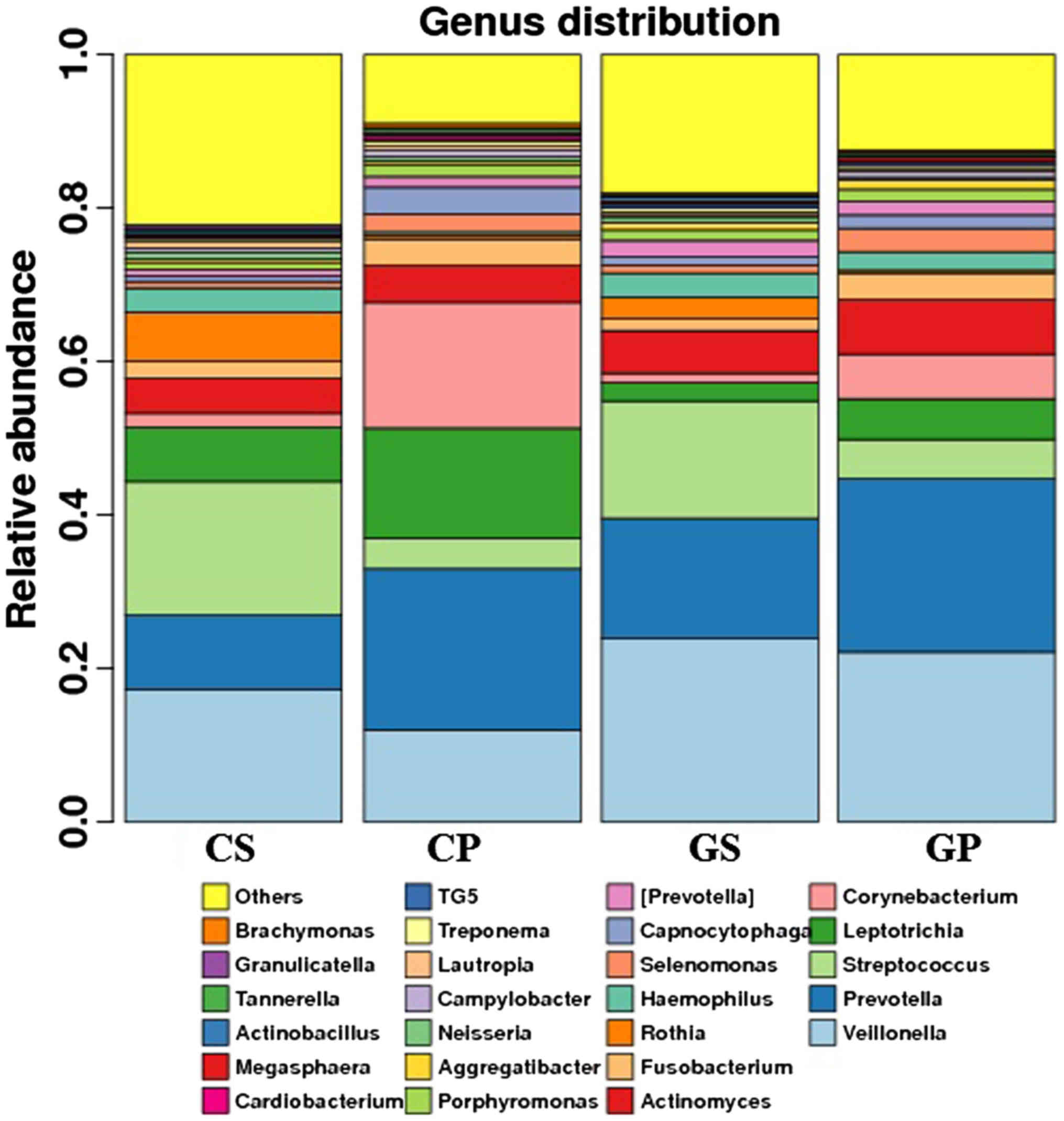
The bacterial distribution within the different
groups was analyzed. The results are shown at the genus level
(Fig. 2). Veillonella,
Prevotella, Streptococcus, Leptotrichia, Corynebacterium and
Actinomyces were the top 10 OTUs detected in all four
groups. We compared the percentages of the top 25 bacteria in the
four groups with some interesting findings. Some of the top 25
bacteria (Veillonella, Prevotella, Aggregatibacter, and
Megasphaera) increased, while others (Leptotrichia,
Rothia, Capnocytophaga, Campylobacter, Tannerella and
Granulicatella) decreased in either the saliva or the plaque
of the gastric cancer group. The difference reached statistical
significance (Table II). It is
worth mentioning that the incidence of Leptotrichia was
lower in both the GS and GP groups compared with the CS and CP
groups. The difference reached a statistically significant level
(P<0.05). Aggregatibacter was twice as prevalent in both
the GS and GP groups compared with the CS and CP groups
(P<0.01).
 | Table II.Percentage of the different bacteria
in study groups. |
Table II.
Percentage of the different bacteria
in study groups.
|
| Saliva | Plaque |
|---|
|
|
|
|
|---|
| Genus | Control (%) | Gastric cancer
(%) | P-value | Control (%) | Gastric cancer
(%) | P-value |
|---|
|
Veillonella | 17 | 24 | 0.28 | 12 | 22 | 0.0073b |
|
Prevotella | 9.7 | 16 | 0.042a | 22 | 23 | 0.86 |
|
Leptotrichia | 7 | 2.4 | 0.045a | 14 | 5.3 |
0.00099b |
| Rothia | 6.5 | 2.8 | 0.0066b | 0.41 | 0.36 | 0.67 |
|
[Prevotella] | 0.85 | 2.2 | 0.023a | 1.4 | 1.8 | 0.48 |
|
Capnocytophaga | 0.74 | 1 | 0.48 | 3.5 | 1.7 | 0.0069b |
|
Aggregatibacter | 0.41 | 1 | 0.0049b | 0.44 | 1.3 | 0.023a |
|
Campylobacter | 0.53 | 0.27 | 0.0052b | 0.84 | 0.7 | 0.63 |
|
Megasphaera | 0.091 | 0.32 | 0.0098b | 0.28 | 0.48 | 0.097 |
|
Tannerella | 0.14 | 0.13 | 0.82 | 0.51 | 0.33 | 0.042a |
|
Granulicatella | 0.53 | 0.27 | 0.024a | 0.11 | 0.13 | 0.57 |
Relative abundance of oral
bacteria
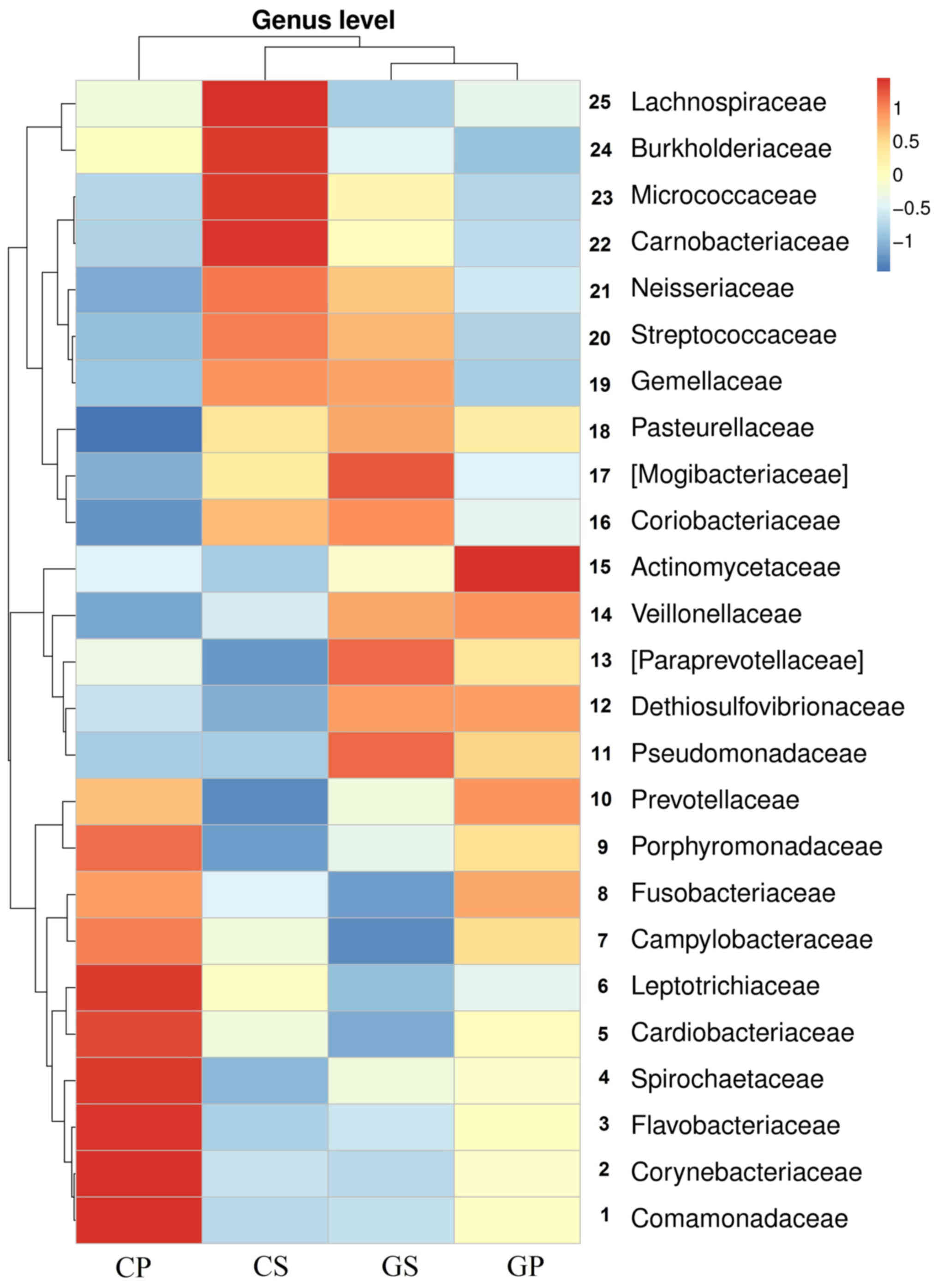
In order to discover the differences in oral
bacterial distribution between gastric cancer patients and healthy
control patients, we clustered the abundance of the top 25
distributed bacteria at the genus level of all groups of samples.
The results of the heatmap are shown in Fig. 3. The gastric cancer salivary and
plaque groups were clustered together, demonstrating the
differences between gastric cancer patients and the healthy control
population in the abundance of the top 25 bacterial genera in the
oral cavity. The results showed that, according to the different
groups, the top 25 genera were clustered into three parts. Genera
1–10 appeared more often in the CP group. There were five genera
(genus 11–15) which showed an abnormal increase in both saliva and
plaque samples of gastric cancer patients, namely:
Pseudomonadaceae, Dethiosulfovibrionaceae, Paraprevotellaceae,
Veillonellaceae and Actinomycetaceae. Genera 16–25
appeared more often in the CS group.
Relative abundance and correlation of
oral pathogen distribution
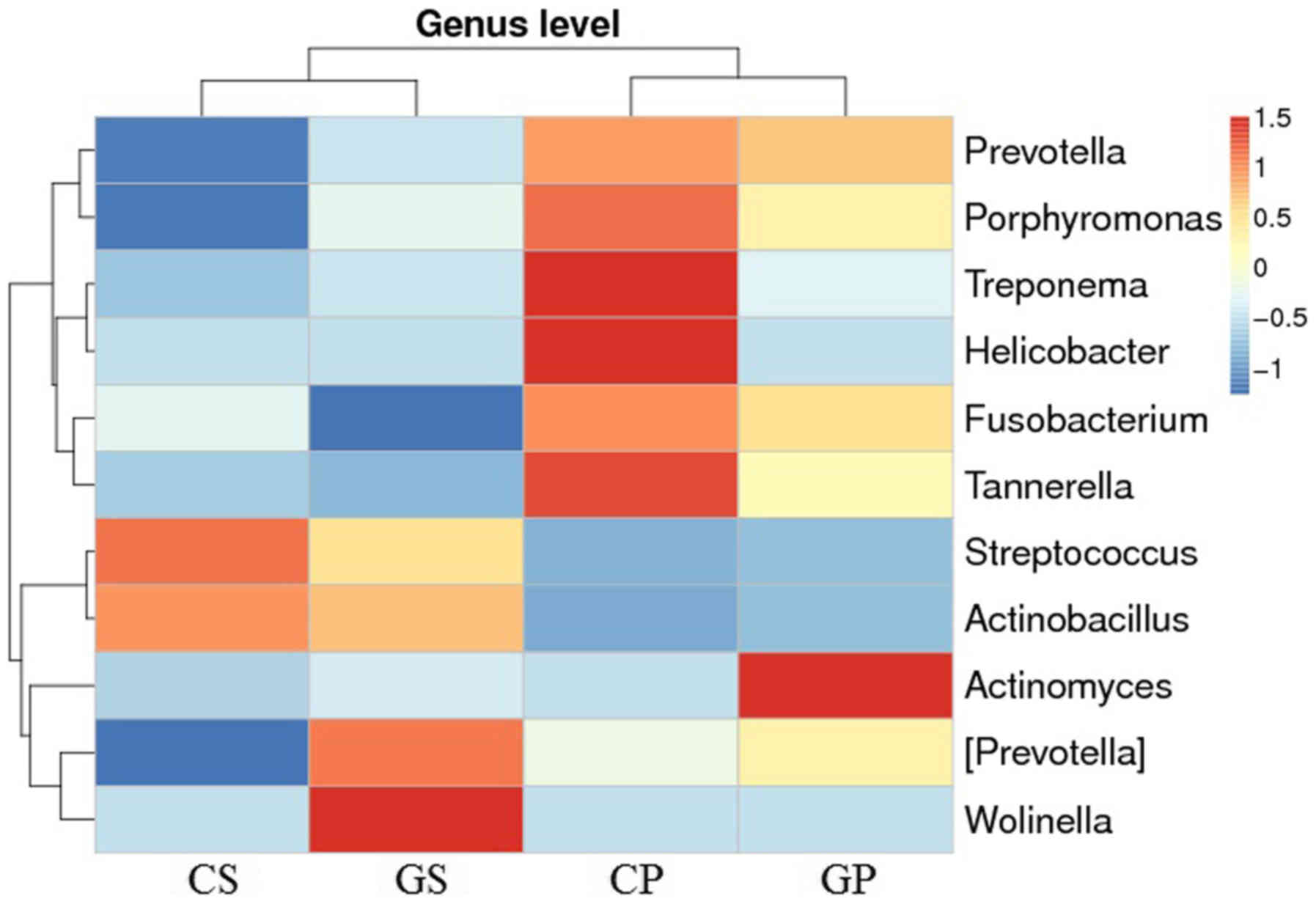
In order to discover the distribution of dental
caries and periodontal pathogens in the oral cavity of gastric
cancer patients and the healthy population, we selected 11
varieties of main caries and periodontal pathogenic bacteria for
analysis (Fig. 4). At the genus
level, the selected oral pathogens were clustered in two groups (or
branches). At the group level, the CS and GS groups and the CP and
GP groups were relatively clustered together. More Prevotella,
Porphyromonas, Fusobacterium and Tannerella appeared in
plaque samples compared with the saliva samples. The six bacteria
(Prevotella, Porphyromonas, Fusobacterium, Tannerella,
Streptococcus and Actinobacillus) were more
representative of the differences between the saliva and plaque
samples and not the differences between the gastric cancer patients
and the control population. Comparing Fig. 4 with Fig. 3, we noted that several dental caries
and periodontal pathogens were also present in the top 25
distributed bacterial orders. Among the 11 oral pathogens, only two
periodontal pathogens were found more commonly in patients with
gastric cancer, including more Wolinella in the saliva of
patients with gastric cancer, and more Actinomyces appeared
in the plaque of patients with gastric cancer. The relationship
between gastric cancer and periodontal disease has been noted and
probably associated with abnormal increases in these two types of
bacteria.
A potential way of screening gastric
cancer by oral microbiome detection
Since there are indeed some characteristics of the
oral microbiome in gastric cancer individuals, in the present
study, a scoring system was designed to screen gastric cancers.
Eleven out of the top 25 distributed bacterial genera were chosen
for further analysis according to their significant distribution
between the control group and the gastric cancer group:
Veillonella, Prevotella, Leptotrichia, Rothia,
[Prevotella], Capnocytophaga, Aggregatibacter,
Campylobacter, Megasphaera, Tannerella, and
Granulicatella. [Prevotella] is a candidate genus
proposed by the greengenes curators and has not been found in NCBI.
Normalization was carried out using the following formula: NV = (C
- Cmean)/Cmean, where NV, normalization
value; C, the content of the genus in the individual sample;
Cmean, the average content of the genus in all the same
type of sample.
A normalized value >0 was defined as positive,
anything below was defined as negative. The percentage of NV >0
individuals in each genus of different groups is shown in Table III, as well as the ratios of the
gastric cancer group to the control group. For genera with a ratio
>2, which suggests it is enriched in the gastric cancer group,
we defined it as an ‘active’ genus; for genera with a ratio
<0.5, which suggests it is enriched in the healthy control
group, we defined it as a ‘passive’ genus which is marked in
Table III (e.g.,.
Prevotella in saliva is an ‘active’ genus,
Leptotrichia in both saliva and plaque is a ‘passive’
genus).
 | Table III.Percentage of genuses with normalized
value >0. |
Table III.
Percentage of genuses with normalized
value >0.
|
| Saliva | Plaque |
|---|
|
|
|
|
|---|
| Genus | Control (%) | Gastric cancer
(%) | Ratio G/C | Score | Control (%) | Gastric cancer
(%) | Ratio G/C | Score |
|---|
|
Veillonella | 23 | 38 | 1.6 | 0 | 15 | 49 | 3.2 | +1 |
|
Prevotella | 15 | 46 | 3.0 | +1 | 31 | 38 | 1.2 | 0 |
|
Leptotrichia | 54 | 19 | 0.35 | −1 | 69 | 27 |
0.39 | −1 |
| Rothia | 69 | 16 | 0.23 | −1 | 54 | 41 |
0.75 | 0 |
|
[Prevotella] | 7.7 | 35 | 4.6 | +1 | 23 | 46 | 2.0 | 0 |
|
Capnocytophaga | 23 | 32 | 1.4 | 0 | 69 | 27 |
0.39 | −1 |
|
Aggregatibacter | 0 | 30 | ∞ | +1 | 7.7 | 27 | 3.5 | +1 |
|
Campylobacter | 77 | 30 | 0.39 | −1 | 23 | 32 | 1.4 | 0 |
|
Megasphaera | 0 | 24 | ∞ | +1 | 31 | 30 |
0.97 | 0 |
|
Tannerella | 46 | 35 | 0.76 | 0 | 77 | 22 |
0.28 | −1 |
|
Granulicatella | 77 | 16 | 0.21 | −1 | 31 | 27 |
0.88 | 0 |
Our scoring system is based on the NV of ‘active’
genera and ‘passive’ genera in each individual. When the NV of an
‘active’ genus was >0, we give individuals a score +1; when the
NV of a ‘passive’ genus is >0, we give individual a score −1;
otherwise the individual was given a score of 0 (e.g.,
Leptotrichia in both saliva and plaque is a ‘passive’ genus.
For an individual, when its NV of Leptotrichia was >0 in
both saliva and plaque, it was given a score of −2; when its NV of
Leptotrichia was >0 only in the saliva, it was given a
score of −1). The scores of ‘active’ genera and ‘passive’ genera in
each individual were summed, giving their evaluation score for
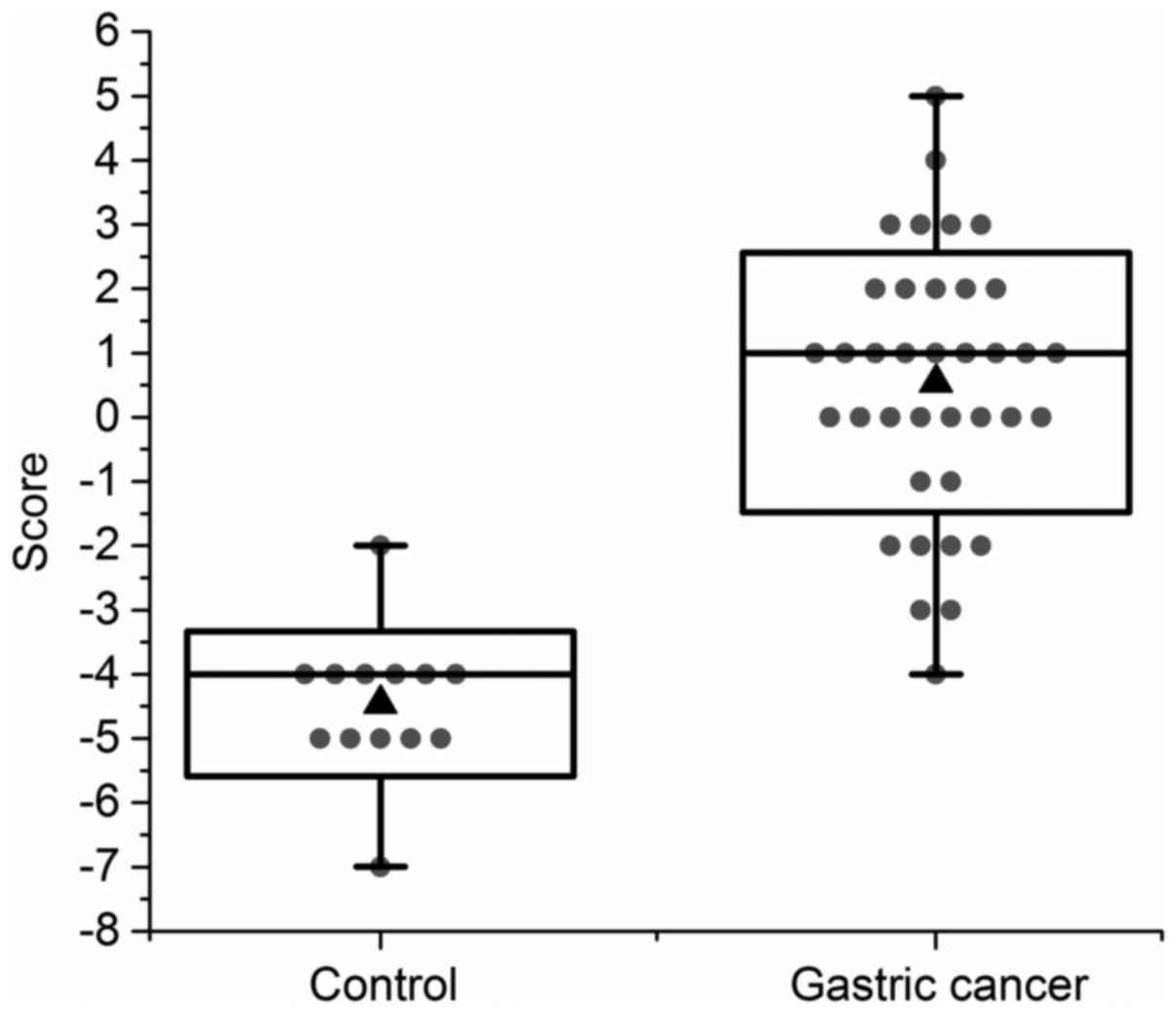
gastric cancer screening. The results are shown in Fig. 5.
The average score for the gastric cancer group was
0.54, and the average score for the control group was −4.46. Using
the Student's t-test, a P-value of 2.30E-13 was calculated, which
suggested a very significant difference between the two groups. If
we defined −2 as the threshold of gastric cancer screening
standard, 36 out of 37 individuals in the gastric cancer group
could be screened as a high-risk population, giving a sensitivity
rate of 97%. One out of the 13 individuals in the control group
could be screened as a high-risk population (Helicobacter
pylori infection was detected in following checkup of the
individual) giving a false-positive rate of 7.7%. Due to the high
sensitivity rate and low false-positive rate, the oral microbiome
detection could be used as an applicable potential method of the
screening for gastric cancer.
Discussion
Gastric cancer is a severe threat to human health. A
non-invasive and easy means of early diagnosis or screening would
positively contribute to reducing the threat of gastric cancer, as
well as reducing the related medical expense. Since the oral cavity
is home to microbial communities, with significant implications for
human health and disease (19), in
the present study, we analyzed the oral microbiome of both the
saliva and subgingival plaque in gastric cancer patients and
healthy controls using 16s rRNA sequencing. Our results indicated a
difference in the biomass, species richness, and species diversity
between gastric cancer and normal human subjects, seen not only in
the saliva but also in the dental plaque. This finding also
supported Momen-Heravi's et al research ‘Periodontal
disease, tooth loss and colorectal cancer risk: Results from the
Nurses’ Health Study (20). This
study pointed out that oral health might biologically increase
systemic inflammation, lead to immune dysregulation, and alter the
gut microbiota, thereby possibly influencing colorectal
carcinogenesis. The findings of the present study suggest that oral
microbial detection has the potential to be an aid to early
diagnosis and a means of screening for gastric cancer in the
future.
Our results showed that the oral bacteria are more
complex in patients with gastric cancer compared with the normal
control population. This may possibly be due to the fact that the
immunity of gastric cancer patients is weaker than that of normal
people, thus it is easier for the aggregation of specific bacteria
to occur. We found a difference in the distribution of bacteria in
the oral cavity between normal and gastric cancer patients, in
which we also found oral pathogens, such as Prevotella and
Aggregatibacter. Analysis of the relative abundance and
correlation of oral pathogen distribution found that the same kind
of samples (group CS and GS, group CP and GP) were relatively
clustered together but not the same groups of subjects. The
heterogeneity of tissue types in the oral cavity, such as teeth,
tongue and mucosa, means that a variety of sites are available for
colonization by oral microorganisms (21). Each site has unique characteristics
and allows those microorganisms best suited to the environment to
inhabit the site. For oral bacteria, saliva and dental plaque are
two entirely different microbial environments, with the liquidity
of the saliva and the saliva lysozyme, which may result in a
difference of the bacteria in the saliva and dental plaque. Thus,
this may lead to differences in the flora structure in the saliva
and plaque that is greater than the differences between the healthy
individuals and those with gastric cancer.
Several studies have suggested a positive
association between tooth loss and the risk of gastric cancer
(22–26). Periodontal disease is one of the
most important causes of tooth loss. The abnormal increases in some
periodontal pathogens in gastric cancer patients in the present
study confirmed our previous research (17,23).
We previously showed that specific oral health conditions and
behaviors, such as gingival bleeding, not flossing teeth and
smoking, were associated with an increased risk of gastric
precancerous lesions (23). Tooth
loss due to periodontal disease may be a surrogate for periodontal
pathogen infection. There is strong evidence that chronic
inflammation is widely responsible for the early stages of disease
progression (27). Periodontal
infection can lead to chronic systemic inflammation, which is an
important risk factor in the development of gastric cancer. Light
and electron microscopy studies, in vitro adhesion,
co-aggregation models and in vitro continuous culture
studies have been helpful in describing the possible changes that
might occur in species composition during biofilm formation
(28). From the microbial
perspective, our results proved the role of periodontal disease in
the process of gastric cancer development.
Helicobacter pylori is the main pathogen
involved in gastric cancer, which can also be detected in the oral
cavity. In the 50 subjects in the present study, H. pylori
was detected only in the plaque samples of one subject of the
healthy control group. The following can be considered. The oral
cavity is not the most suitable living environment for H.
pylori. Additionally, the content of H. pylori in the
oral cavity is lower than the other common oral bacteria. The
sequencing depth (13,116-16,000 OTUs) of this study was not
sufficient to detect H. pylori. The person that was positive
for H. pylori was also confirmed by a breath test,
indicating that our results are reliable. In this study, the
gastric cancer study population is not associated with H.
pylori gastric carcinogenesis.
We collected all the data and designed a scoring
system for screening suspected gastric cancer patients through the
distribution of their oral bacteria. Eleven strains were selected
for the scoring system. The content of Veillonella, Prevotella,
Leptotrichia, Rothia, [Prevotella], Capnocytophaga,
Aggregatibacter, Campylobacter, Megasphaera, Tannerella,
Granulicatella bacteria was compared with the average of the
sample levels in all subjects as criteria for scoring. Using our
screening method, 97.3% of patients with gastric cancer were
identified, in comparison one of 13 control subjects was
identified, giving a false-positive rate of 7.7%. This positive
subject in the scoring system was the individual who had detected
positive for H. pylori. Although the follow-up examination
found that this person did not have stomach cancer, the reason for
the false positive may have been due to the abnormal enrichment of
H. pylori, resulting in an oral distribution of bacteria
similar to that seen in gastric cancer patients. Bacterial species
inhabiting the biofilm formation do not interact passively, but
through specific interactions with other bacterial species. We
suggest for this kind of subjects; physicians can recommend they
undergo conventional upper endoscopy, to achieve the purpose of
early detection, early diagnosis, and early treatment.
Our findings indicate that the changes in the oral
microbiome might be a microbial indicator for gastric cancer and
could be considered more in the screening and diagnostic procedure.
This study was carried out in order to obtain scientific evidence
of the association between oral flora and gastric cancer, and to
provide a scientific basis for prevention of gastric cancer.
Elucidating the composition of bacteria in the oral cavity of
patients with gastric cancer is conducive to the early detection,
early diagnosis and early treatment of gastric cancer. The
population at high-risk of gastric cancer could be identified
during a large-scale epidemiological investigation or by oral
health education when oral saliva and plaque samples would be
tested. This screening system is not limited to the results of
high-throughput sequencing, many detection methods, for example
microaray and RT-PCR, could be combined with our system for early
screening of gastric cancer.
The main limitations of the study included the small
sample size, since only 50 participants were enrolled in the
present study. Although the sample size was small, the
characteristics of the oral microbiome in gastric cancer
individuals still provided valuable indications. Another
shortcoming of this study was the lack of detection rate of H.
pylori, one of the strongest risk factors for gastric cancer.
Thus, a larger study population and additional investigations are
needed to address those issues and further confirm the study
findings.
Acknowledgements
Thanks are due to Fiona Ruge and Jane Lane (CCMRC,
Cardiff University School of Medicine) for the proofreading of this
manuscript.
References
|
1
|
Torre LA, Siegel RL, Ward EM and Jemal A:
Global cancer incidence and mortality rates and trends - an update.
Cancer Epidemiol Biomarkers Prev. 25:16–27. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
den Hoed CM and Kuipers EJ: Gastric
cancer: How can we reduce the incidence of this disease? Curr
Gastroenterol Rep. 18:342016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Arnold M, Moore SP, Hassler S,
Ellison-Loschmann L, Forman D and Bray F: The burden of stomach
cancer in indigenous populations: A systematic review and global
assessment. Gut. 63:64–71. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen W, Zheng R, Zeng H and Zhang S: The
incidence and mortality of major cancers in China, 2012. Chin J
Cancer. 35:732016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen W, Zheng R, Zuo T, Zeng H, Zhang S
and He J: National cancer incidence and mortality in China, 2012.
Chin J Cancer Res. 28:1–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen W, Zheng R, Zhang S, Zhao P, Zeng H,
Zou X and He J: Annual report on status of cancer in China, 2010.
Chin J Cancer Res. 26:48–58. 2014.PubMed/NCBI
|
|
8
|
Bu ZJJ: A current view of gastric cancer
in China. Transl Gastrointest Cancer. 2:1–4. 2013.
|
|
9
|
Leys EJ, Griffen AL, Kumar PS and Maiden
MF: Isolation, classification, and identification of oral
microbioorganismsOral Microciology and Immunology. Lamont RJ, Burne
RA, Lantz MS and Leblanc DJ: ASM Press, American Society for
Microbiology; Washington, DC: pp. 73–88. 2006
|
|
10
|
Aas JA, Paster BJ, Stokes LN, Olsen I and
Dewhirst FE: Defining the normal bacterial flora of the oral
cavity. J Clin Microbiol. 43:5721–5732. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Redinbo MR: The microbiota, chemical
symbiosis, and human disease. J Mol Biol. 426:3877–3891. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Takahashi N: Microbial ecosystem in the
oral cavity: Metabolic diversity in an ecological niche and its
relationship with oral diseases. Int Congr Ser. 1284:103–112. 2005.
View Article : Google Scholar
|
|
13
|
Johansson I, Witkowska E, Kaveh B,
Holgerson Lif P and Tanner AC: The microbiome in populations with a
low and high prevalence of caries. J Dent Res. 95:80–86. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Camp SLY, Costalonga M, Zhang Y, Zaia A,
Vajna R, Ross KF and Herzberg MC: Systemic disease and the oral
microbiotaOral Microbiology and Immunology. Richard JLMS, Robert AB
and Donald JL: ASM Press, American Society for Microbiology;
Washington, DC: pp. 361–375. 2006
|
|
15
|
Sheh A and Fox JG: The role of the
gastrointestinal microbiome in Helicobacter pylori
pathogenesis. Gut Microbes. 4:505–531. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Indriolo A, Greco S, Ravelli P and
Fagiuoli S: What can we learn about biofilm/host interactions from
the study of inflammatory bowel disease. J Clin Periodontol. 38
Suppl 11:36–43. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Salazar CR, Sun J, Li Y, Francois F, Corby
P, Perez-Perez G, Dasanayake A, Pei Z and Chen Y: Association
between selected oral pathogens and gastric precancerous lesions.
PLoS One. 8:e516042013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Evans CC, LePard KJ, Kwak JW, Stancukas
MC, Laskowski S, Dougherty J, Moulton L, Glawe A, Wang Y, Leone V,
et al: Exercise prevents weight gain and alters the gut microbiota
in a mouse model of high fat diet-induced obesity. PLoS One.
9:e921932014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bik EM, Long CD, Armitage GC, Loomer P,
Emerson J, Mongodin EF, Nelson KE, Gill SR, Fraser-Liggett CM and
Relman DA: Bacterial diversity in the oral cavity of 10 healthy
individuals. ISME J. 4:962–974. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Momen-Heravi F, Babic A, Tworoger SS,
Zhang L, Wu K, Smith-Warner SA, Ogino S, Chan AT, Meyerhardt J,
Giovannucci E, et al: Periodontal disease, tooth loss and
colorectal cancer risk: Results from the Nurses' Health Study. Int
J Cancer. 140:646–652. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Teles RP, Haffajee AD and Socransky SS:
Microbiological goals of periodontal therapy. Periodontol 2000.
42:180–218. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Seymour RA: Is oral health a risk for
malignant disease? Dent Update. 37(279–280): 282–283.
2010.https://doi.org/10.12968/denu.2010.37.5.279
|
|
23
|
Salazar CR, Francois F, Li Y, Corby P,
Hays R, Leung C, Bedi S, Segers S, Queiroz E, Sun J, et al:
Association between oral health and gastric precancerous lesions.
Carcinogenesis. 33:399–403. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yin XH, Wang YD, Luo H, Zhao K, Huang GL,
Luo SY, Peng JX and Song JK: Association between tooth loss and
gastric cancer: A meta-analysis of observational studies. PLoS One.
11:e01496532016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Boylan MR, Khalili H, Huang ES, Michaud
DS, Izard J, Joshipura KJ and Chan AT: A prospective study of
periodontal disease and risk of gastric and duodenal ulcer in male
health professionals. Clin Transl Gastroenterol. 5:e492014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shakeri R, Malekzadeh R, Etemadi A,
Nasrollahzadeh D, Abedi-Ardekani B, Khoshnia M, Islami F, Pourshams
A, Pawlita M, Boffetta P, et al: Association of tooth loss and oral
hygiene with risk of gastric adenocarcinoma. Cancer Prev Res
(Phila). 6:477–482. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Uzel NG, Teles FR, Teles RP, Song XQ,
Torresyap G, Socransky SS and Haffajee AD: Microbial shifts during
dental biofilm re-development in the absence of oral hygiene in
periodontal health and disease. J Clin Periodontol. 38:612–620.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Y, Ku CY, Xu J, Saxena D and Caufield
PW: Survey of oral microbial diversity using PCR-based denaturing
gradient gel electrophoresis. J Dent Res. 84:559–564. 2005.
View Article : Google Scholar : PubMed/NCBI
|