Introduction
As one of the most common malignant tumors,
hepatocellular carcinoma (HCC) has been identified as the third
primary cause of tumor-induced death in the word (1). Despite great advancements in the
diagnosis and treatment of HCC, the rates of distant metastasis and
local recurrence after surgical resection remain high resulting in
a poor long-term patient prognosis (2–4). Thus,
it is extremely critical to uncover the potential mechanisms
underlying HCC progression.
MicroRNAs (miRNAs), a type of endogenous, small and
non-coding RNA, are involved in tumor initiation, development and
progression via binding with the 3′-untranslated region (UTR) of
target genes, which results in the translational inhibition or
degradation of the target mRNAs (5,6).
Accumulating data have revealed that aberrant miRNAs are involved
in HCC initiation, development and progression, which could
represent potential diagnostic, therapeutic and prognostic markers
(7). Recent studies have
demonstrated that miR-577 is dysregulated in cancers (8,9).
miR-577 was found to modulate the Wnt signaling pathway to inhibit
glioblastoma tumor growth (10). Yu
et al reported that, in gastric cancer, E2F transcription
factor 3 works as a direct downstream target of miR-577 (11). Moreover, in colorectal cancer,
miR-577 suppressed tumor growth and enhanced chemosensitivity
(12). In addition, in research
concerning pediatric diabetes, miR-577 was identified as an
inhibitor to pancreatic β-cell function and survival, which
targeted fibroblast growth factor 21 (13). These studies suggest that miR-577 is
a cancer-related gene. However, the expression and the specific
mechanism of miR-577 in HCC remain to be uncovered.
Epithelial-to-mesenchymal transition (EMT) has been
confirmed to be critical in tumor metastasis, including HCC
(14–16). EMT results in decreased expression
of epithelial marker (E-cadherin), while the expression of
mesenchymal markers (N-cadherin and vimentin) are enhanced. EMT
enhances the migratory and invasive properties of HCC cells,
thereby contributing to HCC metastasis (17). However, in HCC, whether miR-577
regulates the process of EMT in tumor cells has been rarely
investigated.
Here, we found that decreased expression of miR-577
was closely related to poor clinicopathological features and HCC
patient survival. Functionally, miR-577 suppressed the migration
and invasion of HCC cells by directly targeting HOXA1.
Additionally, miR-577 inhibited the process of EMT in HCC cells. In
conclusion, these findings demonstrated that miR-577 suppressed HCC
cell migration, invasion and EMT, and miR-577 may act as a possible
valuable target for molecular-targeted therapy of HCC.
Materials and methods
Tissue samples
HCC tissues and adjacent non-tumor tissues were
obtained from patients diagnosed with HCC at the Department of
Hepatobiliary Surgery, The First Affiliated Hospital of Xi'an
Jiaotong University (Xi'an, China) from January 2007 to December
2009. The HCC patients did not receive any adjuvant therapy before
surgery, such as chemotherapy or radiotherapy. The fresh tissues
were stored in liquid nitrogen. All of the patients provided
written informed consent. Xi'an Jiaotong University Ethics
Committee approved the research on the basis of the Declaration of
Helsinki.
Cell culture and transfection
The normal hepatic cell line (LO2) and five human
HCC cell lines (Huh7, MHCC-97L, MHCC-97H, SMMC-7721 sand Hep3B)
were obtained from the Chinese Academy of Sciences (Shanghai,
China). Complete Dulbecco's modified Eagle's medium (DMEM)
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with
1% penicillin-streptomycin (Thermo Fisher Scientific, Inc.) and 10%
FBS (Gibco; Thermo Fisher Scientific, Inc.) was applied to culture
the cells. Cells were then placed in a humidified atmosphere (37°C,
5% CO2).
Lipofectamine 2000 reagent (Invitrogen Life
Technologies; Thermo Fisher Scientific, Inc.) was applied to
conduct cell transfection on the basis of the product specification
at a final concentration of 100 pmol according to previous studies
(9,10). The mimic control (miR-control;
CmiR0001-MR04), miR-577 mimics (miR-577, HmiR0074-MR04), inhibitor
control (anti-miR-NC; CmiR-AN0001-AM02) and miR-577 inhibitors
(anti-miR-577; HmiR-AN0678-AM02) were purchased from Genecopoeia
(Guangzhou, China). HOXA1 clones and HOXA1 siRNAs were obtained
from Sangon Biotech Co., Ltd. (Shanghai, China). The cells were
transfected with the siRNAs and clones above using Lipofectamine
2000 according to the manufacturer's instructions. After 48 h of
transfection, cells were used for the following experiments.
Quantitative real-time PCR (qPCR)
TRIzol (Thermo Fisher Scientific, Inc.) was employed
to extract total RNA from the tissues and cells according to the
product manual. Then, TIANScript RT Kit (Tiangen Biotech, Beijing,
China) was used to synthesize cDNAs. TaqMan Human MiRNA Assay Kit
(Genecopoeia, Guangzhou, China) and the SYBR Premix Ex Taq™ Kit
(Takara Bio Inc.,, Shiga, Japan) were used to conduct PCR
amplifications for miR-577 and HOXA1 mRNA. The detection was
performed in the ABI 7300 system (Applied Biosystems, Foster City,
CA, USA). snRNA U6 qPCR Primer (HmiRQP9001), hsa-miR-577 primer
(HmiRQP0678), GAPDH (HQP006940) and HOXA1 primer (HQP008966) were
purchased from Genecopoeia (Guangzhou, China).
Western blot analysis
Proteins were isolated with RIPA buffer and then
separated on 10% SDS-PAGE gels. After proteins were transferred to
PVDF membranes, the membranes were blocked using 5% non-fat
milk/TBST (Tris-buffered saline Tween-20). Subsequently, the
primary antibodies rabbit anti-HOXA1 (1:1,000; cat. no. ab37563;
Abcam, Cambridge, UK), mouse anti-E-cadherin (cat. no. 14472; Cell
Signaling Technology, Inc. Danvers, MA, USA), rabbit
anti-N-cadherin (cat. no. 13116; Cell Signaling Technology, Inc.),
rabbit anti-vimentin (cat. no. 5741; Cell Signaling Technology,
Inc.) were used to incubate the membranes at 4°C overnight.
Secondary antibodies (anti-rabbit cat. no. 7074 and anti-mouse cat.
no. 7076; Cell Signaling Technology, Inc.) were employed and the
ECL reagent (Beyotime Institute of Biotechnology, Haimen, China)
was applied for detection.
Luciferase reporter assay
The bioinformation public database TargetScan and
miRanda was used. Cells were seeded in triplicate in a 24-well
plate and pGL3-HOXA1 was co-transfected into HCC cells with the
TK-Renilla plasmid as control signals using Lipofectamine 2000.
Moreover, vectors with the wild-type HOXA1 3′-UTR or mutant HOXA1
3′-UTR constructed by Sangon Biotech (Shanghai, China) and relevant
mir-577 or anti-miR-577 vectors were co-transfected into HCC cells.
After 48 h, the luciferase activity was measured by a
Dual-Luciferase Reporter Assay system (E1910; Promega, Madison, WI,
USA). Three independent experiments were performed and the data are
presented as the mean ± SD.
MTT assays
Cell viability was detected by the 3-(4,5-dimethyl
thiazol-2-yl)2,5-diphenyltetrazolium bromide (MTT; Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) assay. Detailed protocol of the
experiment was described in previous studies (18,19).
Transwell assays
Migratory and invasion abilities of the cells were
detected with Matrigel-uncoated and -coated Transwell inserts
(8-µm; EMD Millipore, Billerica, MA, USA). The detailed experiment
was performed similar to previous studies (20,21).
In vivo metastasis assay
Male BALB/c nude mice (4–6 weeks of age) (Centre of
Laboratory Animals, The Medical College of Xi'an Jiaotong
University, Xi'an, China) were randomized into two groups (n=5). We
subsequently injected the stably overexpressing miR-577 cells,
MHCC-97H-miR-577, and MHCC-97H-miR-control cells (1×106)
into the tail veins for the establishment of a pulmonary metastatic
model. Mice were sacrificed by cervical dislocation under
anesthesia with ether 3 weeks post injection and examined
microscopically (Axioskop 2 plus; Carl Zeiss Co., Ltd., Jena,
Germany) by hematoxylin and eosin (H&E) staining for the
development of lung metastatic foci. Animals were housed in cages
maintained in the pathogen-free (SPF) conditions. All in
vivo protocols were approved by the Institutional Animal Care
and Use Committee of Xi'an Jiaotong University.
Statistical analysis
SPSS 20.0 (IBM Corp., Armonk, NY, USA) and GraphPad
Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA) were
employed in this study. All data are denoted as the mean ± SD.
Statistical methods, such as one-way ANOVA, Student t-test,
Kaplan-Meier method, Pearson's correlation analysis and the
log-rank test were applied. A result with P<0.05 was regarded as
having a statistically significant difference.
Results
miR-577 is decreased in HCC tissues
and cell lines
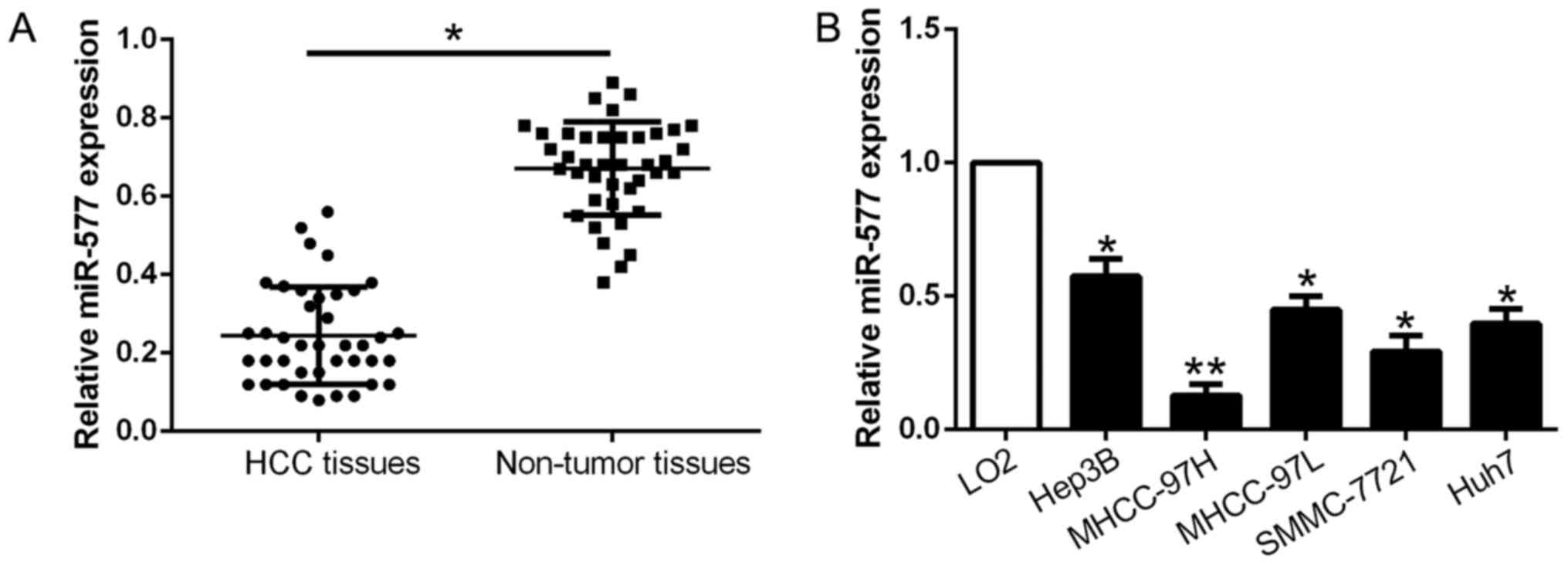
qPCR was conducted to explore miR-577 expression in
40 pairs of tumor tissues and corresponding adjacent non-tumor
tissues. As exhibited in Fig. 1A,
miR-577 was markedly downregulated in the HCC tissues when compared
with that noted in the adjacent non-tumor tissues (P<0.05,
Fig. 1A). Consistently, miR-577
expression was obviously lower in the HCC cell lines compared to
the normal liver cell LO2 (P<0.05, Fig. 1B). The above results revealed that
miR-577 expression was downregulated in HCC and may play a crucial
role in HCC development.
Clinical significance of miR-577 in
HCC
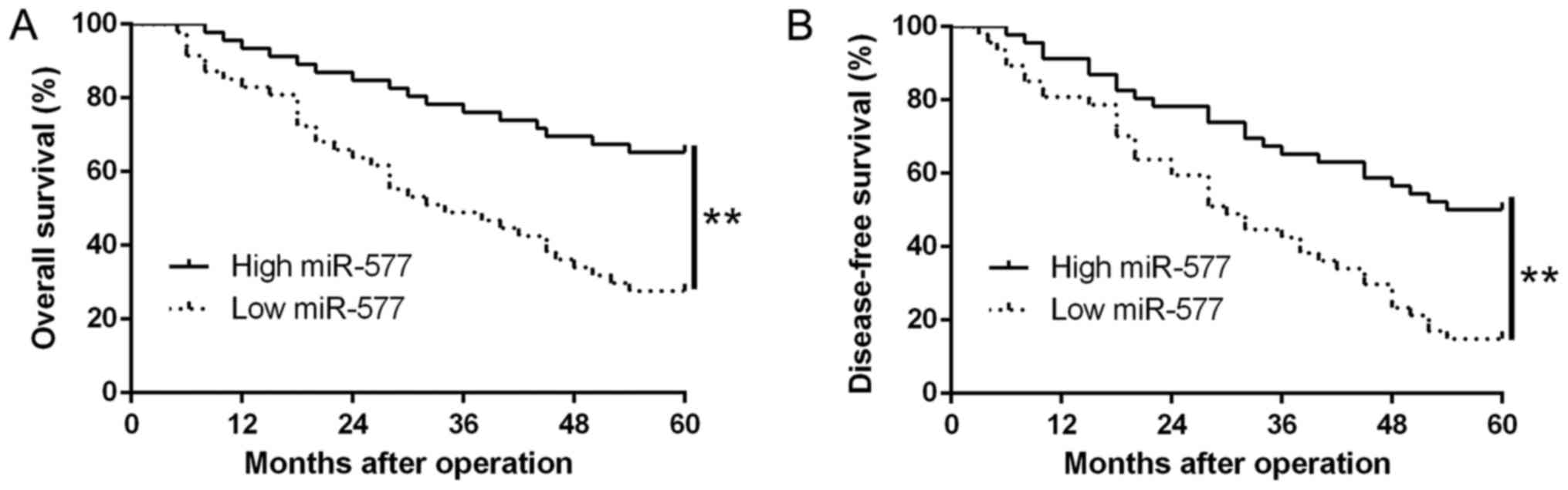
Ninety-three patients were assigned into two
subgroups (high/low miR-577 group), based on the median value of
miR-577 expression in HCC tissues. We found that low miR-577
expression was notably related to venous invasion (P=0.007,
Table I) as well as advanced
tumor-node-metastasis (TNM) stage (P=0.018, Table I). Moreover, results from the
Kaplan-Meier analysis revealed that patients with low miR-577
expression possessed worse overall survival (OS) (P=0.0001,
Fig. 2A) and disease-free survival
(DFS) (P=0.0001, Fig. 2B). Thus,
the above data suggest that miR-577 could be used to predict the
outcome of HCC patients.
 | Table I.Correlation between the
clinicopathological features and miR-577 expression in the HCC
cases (n=93). |
Table I.
Correlation between the
clinicopathological features and miR-577 expression in the HCC
cases (n=93).
|
|
| Expression
level |
|
|---|
|
|
|
|
|
|---|
| Clinical
parameters | Cases (N) |
miR-577high (n=46) |
miR-577low (n=47) | P-value |
|---|
| Age (years) |
|
|
| 0.536 |
| <65
years | 27 | 12 | 15 |
|
| ≥65
years | 66 | 34 | 32 |
|
| Sex |
|
|
| 0.838 |
|
Male | 74 | 37 | 37 |
|
|
Female | 19 | 9 | 10 |
|
| Tumor size
(cm) |
|
|
| 0.423 |
|
<5 | 78 | 40 | 38 |
|
| ≥5 | 15 | 6 | 9 |
|
| Tumor number |
|
|
| 0.392 |
|
Solitary | 80 | 41 | 39 |
|
|
Multiple | 13 | 5 | 8 |
|
| Edmondson |
|
|
| 0.166 |
|
I+II | 32 | 19 | 13 |
|
|
III+IV | 61 | 27 | 34 |
|
| TNM stage |
|
|
| 0.018a |
|
I+II | 76 | 42 | 34 |
|
|
III+IV | 17 | 4 | 13 |
|
| Venous
invasion |
|
|
| 0.007a |
|
Present | 16 | 3 | 13 |
|
|
Absent | 77 | 43 | 34 |
|
| AFP (ng/ml) |
|
|
| 0.667 |
|
<400 | 22 | 10 | 12 |
|
|
≥400 | 71 | 36 | 35 |
|
| HBsAg |
|
|
| 0.751 |
|
Positive | 84 | 42 | 42 |
|
|
Negative | 9 | 4 | 5 |
|
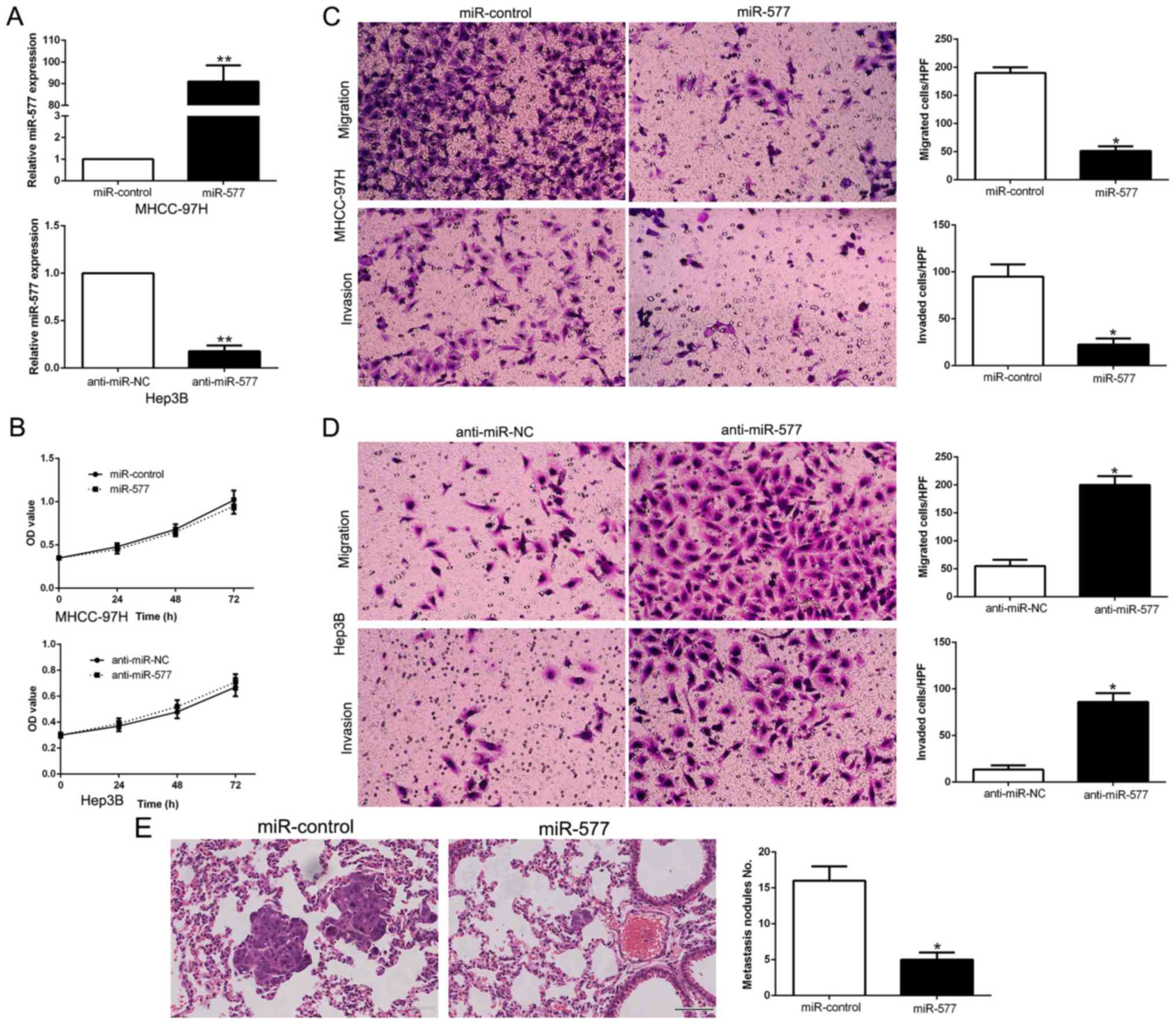
miR-577 suppresses the migration and
invasion of HCC cells
miR-577 levels were manipulated by stable
transfection with miR-577 mimics into MHCC-97H cells whose
expression of miR-577 was the lowest, while miR-577 inhibitors were
transfected into Hep3B cells which had the highest miR-577
expression (P<0.05, Fig. 3A).
MTT assays revealed that the changes in miR-577 expression did not
have any significant influence on HCC cell growth compared to the
control groups (Fig. 3B). Then,
data from Transwell assays confirmed that overexpression of miR-577
notably suppressed the migration and invasion abilities of the
MHCC-97H cells (P<0.05, respectively, Fig. 3C), while silencing of miR-577
expression had the contrary effects on Hep3B cells (P<0.05,
respectively, Fig. 3D). To confirm
the in vitro functional effects of miR-577 on HCC, we
performed in vivo metastatic experiments to examine whether
miR-577 could inhibit the metastasis of HCC cells in vivo.
We subsequently injected the stably overexpressing miR-577 cells,
MHCC-97H-miR-577 and MHCC-97H-miR-control cells into the lateral
veins of the nude mice. The results showed that injection of the
miR-577 overexpressing cells resulted in fewer and smaller foci in
the lungs of the nude mice through microscopic evaluation (5 vs. 16
nodules per lung in MHCC-97H-miR-577 and miR-control cells,
respectively; P<0.01, Fig. 3E).
Thus, we demonstrated that miR-577 exerts an anti-metastatic effect
in HCC cells in vitro and in vivo.
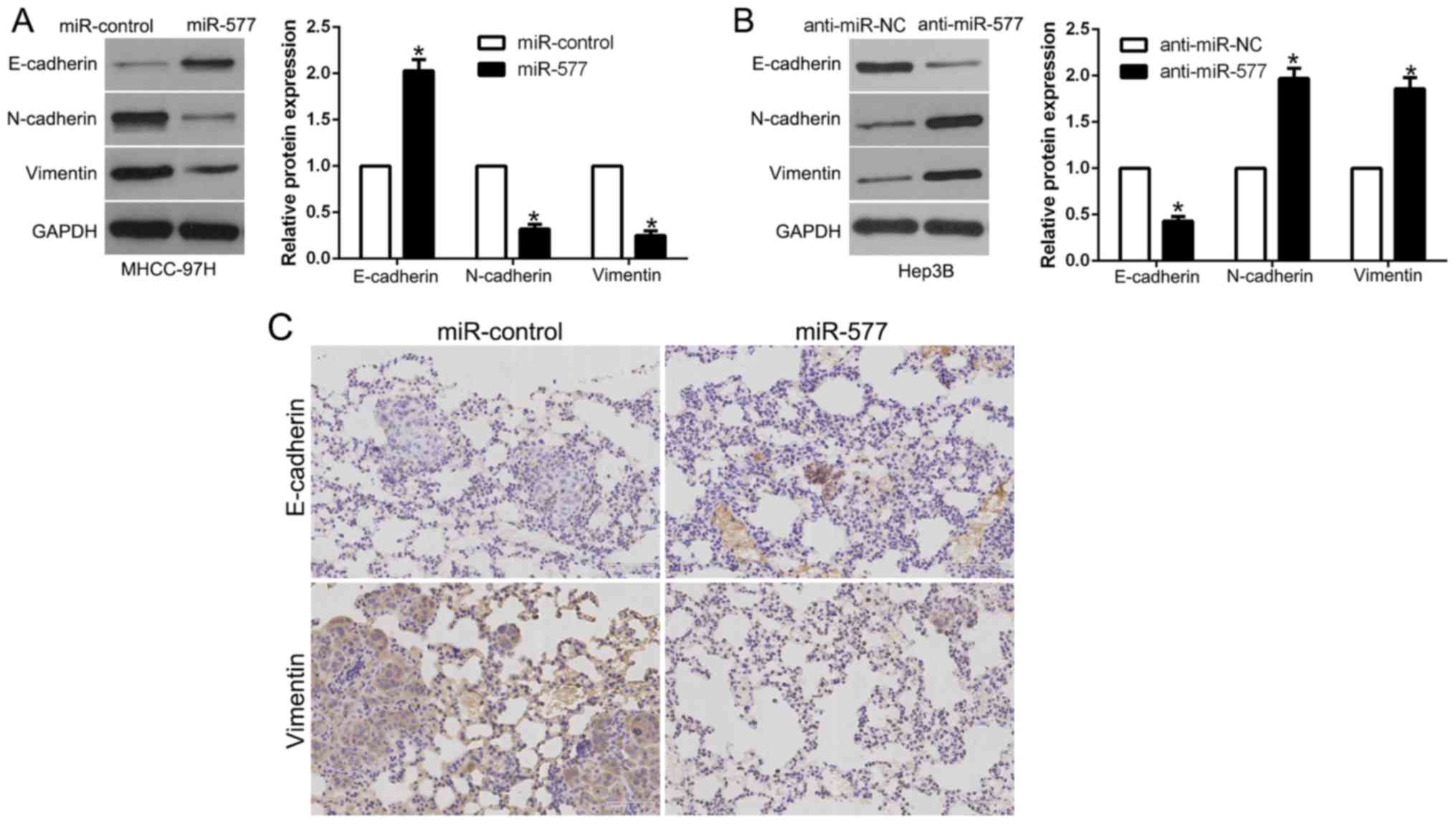
miR-577 inhibits the EMT process of
HCC cells
In order to explore the association between miR-577
and EMT, western blot analysis was performed. The results revealed
that, in MHCC-97H cells, overexpression of miR-577 induced
E-cadherin and suppressed N-cadherin and vimentin (P<0.05,
Fig. 4A). However, in Hep3B cells,
miR-577 knockdown showed the opposite effects (P<0.05, Fig. 4B). Moreover, we examined the
metastatic phenotype of these cells and found that lung sections of
the mice injected with the miR-577-overexpressing cells in fact
showed increased E-cadherin expression and conversely decreased
vimentin expression (Fig. 4C).
Thus, our results revealed that miR-577 acts as an inhibitor of the
EMT process in HCC cells.
miR-577 directly targets HOXA1 in HCC
cells
Two public databases (miRanda and TargetScan) were
employed to predict the potential target of miR-577 in HCC cells.
Then we focused on HOXA1 and speculated HOXA1 was a candidate
target, whose 3′-UTR could bind to miR-577 (Fig. 5A). Additionally, HOXA1 has been
identified as an oncogene in HCC by repressing migration and
invasion of HCC cells (22). To
investigate whether miR-577 could interact with the 3′-UTR of
HOXA1, luciferase assays were conducted. The results indicated that
miR-577 negatively regulated luciferase activity of wt 3′-UTR of
HOXA1 (P<0.05, Fig. 5B).
However, the results could not be found in the miR-577 mutant
groups (Fig. 5B). Moreover, we
performed luciferase assays and found that miR-577 overexpression
significantly decreased the luciferase activity of wild-type (wt)
HOXA1 3′-UTR while had no influence on that of the mutant (mt)
HOXA1 3′-UTR (P<0.05, Fig. 5B).
In contrary, miR-577 knockdown increased the luciferase activity of
wt HOXA1 3′-UTR (P<0.05, Fig.
5B) but did not affect the luciferase activity of mt HOXA1
3′-UTR constructs. Furthermore, results from qPCR and western blot
assays revealed that both HOXA1 mRNA and protein expression were
negatively regulated by miR-577 in the HCC cells (P<0.05,
respectively, Fig. 5C and D). Thus,
we conclude that miR-577 directly targets HOXA1 in HCC cells.
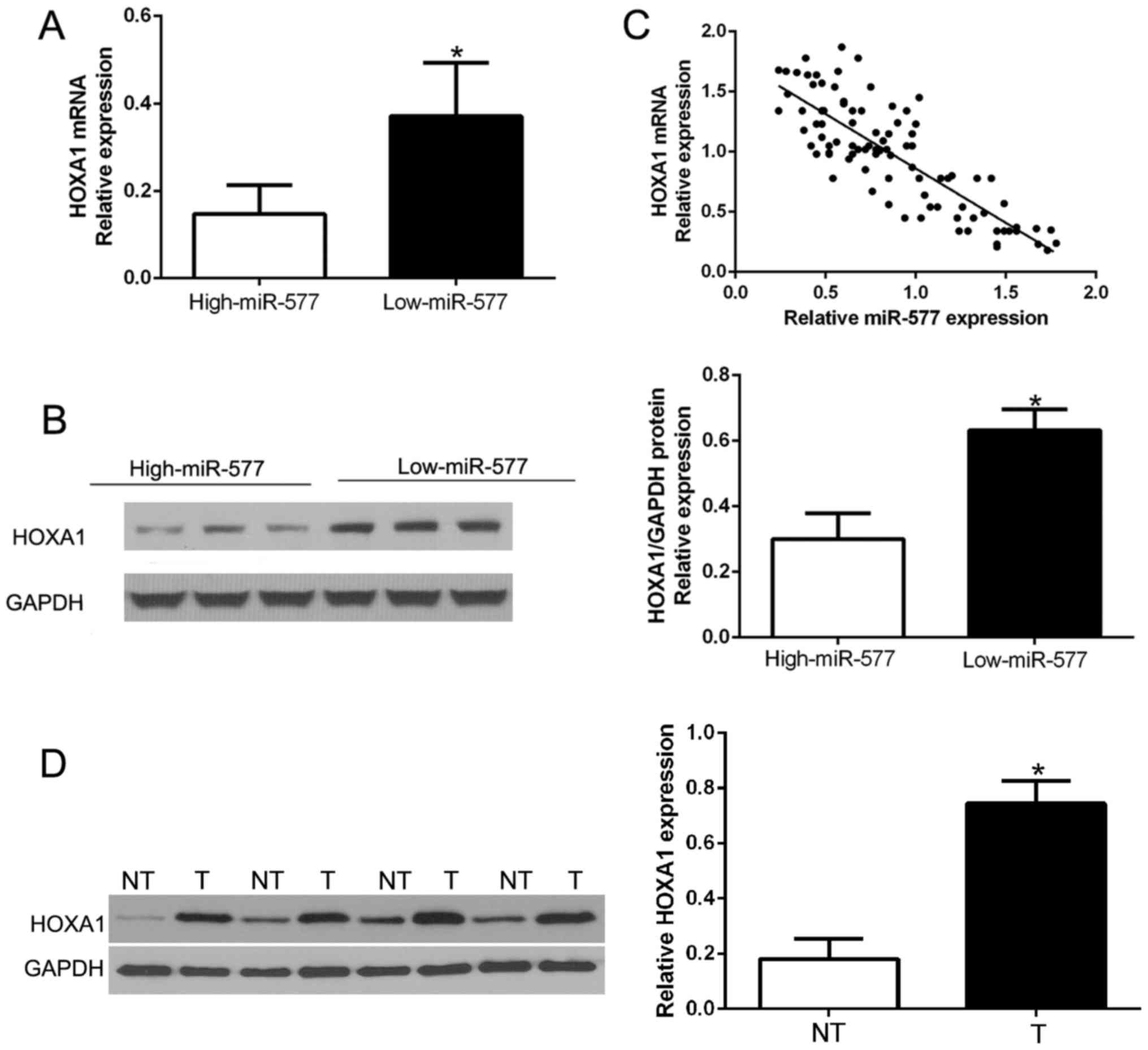
miR-577 is inversely correlated with
the expression of HOXA1 in HCC tissues
Next, we attempted to determine the correlation of
miR-577 and HOXA1 in HCC. The expression levels of HOXA1 mRNA and
protein in HCC tissues with different miR-577 expression were
detected. As expected, tissues with high miR-577 had obviously
lower HOXA1 mRNA and protein expression compared to the tissues
with low miR-577 (P<0.05, Fig. 6A
and B). Furthermore, there existed a negative correlation
between HOXA1 mRNA and miR-577 in the HCC tissues
(R2=0.6866, P<0.001, Fig.
6C). In addition, we performed western blot analysis to confirm
that HOXA1 was overexpressed in HCC tissues compared to that in the
corresponding adjacent non-tumor tissues (P<0.05, Fig. 6D). These results confirmed that
HOXA1 acts as a downstream target of miR-577 in HCC, and HOXA1 is
negatively regulated by miR-577 in HCC.
Restoration of HOXA1 reverses the
biological effects of miR-577 on HCC cells
To determine whether HOXA1 abrogates the effects of
miR-577, we restored HOXA1 expression in miR-577-overexpressing
MHCC-97H cells (P<0.05, Fig.
7A). Interestingly, regaining HOXA1 partially abrogated the
inhibitory functions of the ovexpression of miR-577 in regards to
migration, invasion and EMT of MHCC-97H cells (P<0.05, Fig. 7B and E). In contrast, HOXA1
inhibition by a specific siRNA significantly reversed the promotive
effects of miR-577 knockdown in Hep3B cells (P<0.05, Fig. 7C, D and F). In brief, our findings
demonstrated that HOXA1 reversed the anti-metastatic effects of
miR-577 in HCC cells.
Discussion
Cumulative evidence indicates that miRNAs take part
in the progression of human malignancies as either oncogenes or
tumor suppressors (23). Recently,
miR-577 was identified as a new tumor-related miRNA. In esophageal
squamous cell carcinoma, miR-577 was found to regulate cell
proliferation and the cell cycle by targeting TSGA10 (24). Moreover, miR-577 was found to be
involved in non-alcoholic fatty liver disease (25). In the present study, both in HCC
tissues and cell lines, miR-577 was notably underexpressed.
Downregulation of miR-577 was closely related to poor
clinicopathological characteristics of HCC patients. Importantly,
we demonstrated that HCC patients in the low miR-577 group
exhibited an obviously worse 5-year overall survival and
disease-free survival. These results indicate an important role of
miR-577 in HCC development and could be a predictor of patient
survival for HCC patients. Therefore, these data suggest that
reduced miR-577 might be able to serve as a potential biomarker for
HCC, and a prognostic indicator for HCC patients.
Metastasis is one of the main causes of treatment
failure and poor outcome in HCC patients. During the initiation of
metastasis, EMT is a vital step. In this research, by gain- and
loss-of-function experiments, miR-577 was identified to be an
inhibitor of HCC cell migration and invasion in vitro and
in vivo. The same biological effects of miR-577 were also
noted in other cancers (10,11).
Nevertheless, alteration of miR-577 had no effect on HCC cell
proliferation. Moreover, miR-577 suppressed HCC cell EMT process.
Moreover, in lung metastatic tissues, miR-577 overexpression also
inhibited the EMT process. Thus, our findings indicated that
miR-577 suppresses HCC metastasis via influencing EMT.
HOXA1, which is a member of the HOX gene family,
regulates cell differentiation, embryonic development, survival and
migration. Increasing evidence has confirmed that HOXA1 expression
is dysregulated in diverse cancer types (26–29).
In small cell lung cancer, HOXA1 is targeted by miR-100 to regulate
tumor cell growth and chemoresistance (30). In gastric cancer, elevated HOXA1
acts as an oncogene to promote tumor cell proliferation (31). In HCC, overexpression of HOXA1
promotes cell growth, migration and invasion and is closely related
to the poor prognosis of HCC patients (22). We also confirmed that HOXA1 is
overexpressed in HCC compared to that noted in corresponding
adjacent non-tumor tissues, which shows a similar result. These
data confirmed the critical roles of HOXA1 in cancer development.
Here, our data revealed that miR-577 modulated HCC cell metastasis
and the process of EMT by directly interacting with HOXA1. However,
how HOXA1 regulates the EMT process remains unclear. miR-577 was
found to negatively regulate HOXA1 expression in HCC cells and to
change the luciferase activity of 3′-UTR of HOXA1-wt, rather than
3′-UTR of HOXA1-mt. Moreover, restoration of HOXA1 significantly
reversed the effect of miR-577 on HCC cell migration, invasion and
EMT. However, the detailed molecular mechanism of HOXA1 downstream
pathway warrants further investigation. In a word, HOXA1, a
downstream target of miR-577, reversed the inhibitory effects of
miR-577 on HCC cell migration, invasion and EMT process.
In conclusion, aberrant expression of microRNAs
(miRNAs) is closely associated with HCC pathogenesis and
tumorigenicity. Recent studies suggest that miR-577 is a
cancer-related miRNA. In the present study, both in HCC tissues and
cell lines, miR-577 expression was found to be downregulated.
Decreased miR-577 was distinctly related to malignant
clinicopathologic features and worse outcome of HCC patients.
Functionally, miR-577 modulated HCC cell migration, invasion and
EMT. Additionally, miR-577 directly targets HOXA1 to exert its
effects on HCC cells. Taken together, miR-577 could act as a
prognostic tumor biomarker and a potential target for
molecular-targeted therapy of HCC.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the National
Natural Science Foundation of China (nos. 81602566, 81402039 and
81572847), and the Natural Science Basic Research Plan in Shaanxi
Province of China (no. 2016JQ8029).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
QGL and SSH conceived and designed the experiments;
ZKL, YFW, LW, BWY, CG and TS performed the experiments; SSH and ZKL
analyzed the data; KST and SSH contributed
reagents/materials/analysis tools; ZKL and SSH wrote the paper. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
All of the patients provided written informed
consent. Xi'an Jiaotong University Ethics Committee (Xi'an, China)
approved the research on the basis of the Declaration of
Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
EMT
|
epithelial-mesenchymal transition
|
|
HOXA1
|
homeobox A1
|
|
qPCR
|
quantitative real-time polymerase
chain reaction
|
|
UTR
|
untranslated region
|
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Maluccio M and Covey A: Recent progress in
understanding, diagnosing, and treating hepatocellular carcinoma.
CA Cancer J Clin. 62:394–399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Ann Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tu K, Liu Z, Yao B, Han S and Yang W:
MicroRNA-519a promotes tumor growth by targeting PTEN/PI3K/AKT
signaling in hepatocellular carcinoma. Int J Oncol. 48:965–974.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ji H, Chen M, Greening DW, He W, Rai A,
Zhang W and Simpson RJ: Deep sequencing of RNA from three different
extracellular vesicle (EV) subtypes released from the human LIM1863
colon cancer cell line uncovers distinct miRNA-enrichment
signatures. PloS One. 9:e1103142014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang LY, Li B, Jiang HH, Zhuang LW and Liu
Y: Inhibition effect of miR-577 on hepatocellular carcinoma cell
growth via targeting β-catenin. Asian Pac J Trop Med. 8:923–929.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang W, Shen C, Li C, Yang G, Liu H, Chen
X, Zhu D, Zou H, Zhen Y, Zhang D and Zhao S: miR-577 inhibits
glioblastoma tumor growth via the Wnt signaling pathway. Mol
Carcinog. 55:575–585. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu Z, Zhang W and Deng F: MicroRNA-577
inhibits gastric cancer growth by targeting E2F transcription
factor 3. Oncol Lett. 10:1447–1452. 2015.PubMed/NCBI
|
|
12
|
Jiang H, Ju H, Zhang L, Lu H and Jie K:
microRNA-577 suppresses tumor growth and enhances chemosensitivity
in colorectal cancer. J Biochem Mol Toxicol. 31:2017. View Article : Google Scholar
|
|
13
|
Chen XY, Li GM, Dong Q and Peng H: MiR-577
inhibits pancreatic β-cell function and survival by targeting
fibroblast growth factor 21 (FGF-21) in pediatric diabetes. Genet
Mol Res. 14:15462–15470. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamanaka C, Wada H, Eguchi H, Hatano H,
Gotoh K, Noda T, Yamada D, Asaoka T, Kawamoto K, Nagano H, et al:
Clinical significance of CD13 and epithelial mesenchymal transition
(EMT) markers in hepatocellular carcinoma. Jpn J Clin Oncol.
48:52–60. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vincent CT and Fuxe J: EMT, inflammation
and metastasis. Semin Cancer Biol. 47:168–169. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guarino M, Rubino B and Ballabio G: The
role of epithelial-mesenchymal transition in cancer pathology.
Pathology. 39:305–318. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu Z, Dou C, Yao B, Xu M, Ding L, Wang Y,
Jia Y, Li Q, Zhang H, Tu K, et al: Ftx non coding RNA-derived
miR-545 promotes cell proliferation by targeting RIG-I in
hepatocellular carcinoma. Oncotarget. 7:25350–25365.
2016.PubMed/NCBI
|
|
19
|
Liu Z, Dou C, Jia Y, Li Q, Zheng X, Yao Y,
Liu Q and Song T: RIG-I suppresses the migration and invasion of
hepatocellular carcinoma cells by regulating MMP9. Int J Oncol.
46:1710–1720. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Z, Dou C, Yao B, Xu M, Ding L, Wang Y,
Jia Y, Li Q, Zhang H, Tu K, et al: Methylation-mediated repression
of microRNA-129-2 suppresses cell aggressiveness by inhibiting high
mobility group box 1 in human hepatocellular carcinoma. Oncotarget.
7:36909–36923. 2016.PubMed/NCBI
|
|
21
|
Liu Z, Dou C, Wang Y, Jia Y, Li Q, Zheng
X, Yao Y, Liu Q and Song T: High-mobility group box 1 has a
prognostic role and contributes to epithelial mesenchymal
transition in human hepatocellular carcinoma. Mol Med Rep.
12:5997–6004. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zha TZ, Hu BS, Yu HF, Tan YF, Zhang Y and
Zhang K: Overexpression of HOXA1 correlates with poor prognosis in
patients with hepatocellular carcinoma. Tumour Biol. 33:2125–2134.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dou C, Liu Z, Xu M, Jia Y, Wang Y, Li Q,
Yang W, Zheng X, Tu K and Liu Q: miR-187-3p inhibits the metastasis
and epithelial-mesenchymal transition of hepatocellular carcinoma
by targeting S100A4. Cancer Lett. 381:380–390. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yuan X, He J, Sun F and Gu J: Effects and
interactions of MiR-577 and TSGA10 in regulating esophageal
squamous cell carcinoma. Int J Clin Exp Pathol. 6:2651–2667.
2013.PubMed/NCBI
|
|
25
|
Li Z, Feng S, Zhou L, Liu S and Cheng J:
NS5ATP6 modulates intracellular triglyceride content through FGF21
and independently of SIRT1 and SREBP1. Biochem Biophys Res Commun.
475:133–139. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Q, Zhang X, Li N, Liu Q and Chen D:
miR-30b inhibits cancer cell growth, migration, and invasion by
targeting homeobox A1 in esophageal cancer. Biochem Biophys Res
Commun. 485:506–512. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang H, Liu G, Shen D, Ye H, Huang J, Jiao
L and Sun Y: HOXA1 enhances the cell proliferation, invasion and
metastasis of prostate cancer cells. Oncol Rep. 34:1203–1210. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang X, Li Y, Qi W, Zhang N, Sun M, Huo Q,
Cai C, Lv S and Yang Q: MicroRNA-99a inhibits tumor aggressive
phenotypes through regulating HOXA1 in breast cancer cells.
Oncotarget. 6:32737–32747. 2015.PubMed/NCBI
|
|
29
|
Kraft S, Moore JB, Muzikansky A, Scott KL
and Duncan LM: Differential UBE2C and HOXA1 expression in
melanocytic nevi and melanoma. J Cutan Pathol. 44:843–850. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xiao F, Bai Y, Chen Z, Li Y, Luo L, Huang
J, Yang J, Liao H and Guo L: Downregulation of HOXA1 gene affects
small cell lung cancer cell survival and chemoresistance under the
regulation of miR-100. Eur J Cancer. 50:1541–1554. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yuan C, Zhu X, Han Y, Song C, Liu C, Lu S,
Zhang M, Yu F, Peng Z and Zhou C: Elevated HOXA1 expression
correlates with accelerated tumor cell proliferation and poor
prognosis in gastric cancer partly via cyclin D1. J Exp Clin Cancer
Res. 35:152016. View Article : Google Scholar : PubMed/NCBI
|