Introduction
Colorectal cancer (CRC) is the third most common
type of tumor worldwide and the fourth leading cause of death.
Approximately 1.2 million new cases and 600,000 deaths due to CRC
are reported annually (1). An
important factor affecting the prognosis of CRC patients is the
stage of the disease at the time of diagnosis. The 5-year survival
rate has been reported as 90.1% for patients with in situ
carcinoma, 69.2% for patients with regional lymph node metastasis,
and 11.7% for patients with distant metastases, among all patients
diagnosed with CRC in the US between 2001 and 2007 (2). Therefore, identifying specific and
effective CRC tumor markers is of great significance for early
diagnosis and improved prognosis.
Long non-coding RNA (lncRNA), the focus of this
research, does not encode protein but is involved in gene
regulation at multiple levels. In recent years, the function of
lncRNAs in cancer has gradually been elucidated by researchers on
account of new methods that have emerged, such as genome-wide
association studies, lncRNA chip-based screening, RIP-RNA
sequencing, and gene overexpression and knockdown assays. There is
growing evidence that lncRNAs play an important role in the
carcinogenesis, invasion and metastasis of cancers of various
tissues (3–5). In many different tumors, lncRNAs play
a role either as a tumor suppressor or as a carcinogenic agent
(6–8). Study of CRC-related lncRNAs has found
that they can regulate the formation and development of CRC by
stimulating or inhibiting different cellular processes, including
tumor cell proliferation, apoptosis, differentiation, invasion and
metastasis (9,10).
Growth arrest-specific transcript 5 (GAS5) was
initially identified by Schneider and others by screening the tumor
suppressor gene library to focus on those genes involved in cell
growth arrest. GAS5 is approximately 630 bases long and is located
at 1q25 (11). Researchers have
proposed that the introns of GAS5 are responsible for its important
biological functions. Kino et al found that the amount of
mature GAS5 was notably higher in serum-starvation and
growth-inhibition conditions caused by lack of growth factors. They
also found that GAS5 could compete with glucocorticoid response
element (GRE) DNA to bind with glucocorticoid receptor (GR) DNA and
form a structural domain, causing inhibition of the target genes
mediated by glucocorticoid (12).
It is becoming increasingly clear that GAS5 acts as a tumor
suppressor and is downregulated in certain tumor tissues, including
those of breast, kidney and prostate cancers, as well as in
non-small cell lung carcinoma (13–16).
It has also been reported that GAS5 can induce growth arrest and
apoptosis in prostate and breast cancer cells (16). Furthermore, genetic aberration of
GAS5 has been found in many types of tumor, including melanoma,
breast cancer and prostate cancer, but its functional significance
remains to be fully elucidated (15). GAS5 exhibits down-regulated
expression in colon cancer, and this is likely connected to the
poor prognosis of patients (17–19).
Meanwhile, GAS5 probably affects the development of colon cancer as
a potential cancer-suppressor gene, although its biological
functions and mechanism of action in CRC remain ambiguous.
With the discovery of a large number of lncRNAs and
the gradual identification of their functions, understanding the
mechanisms of lncRNAs and their target molecules has become a focus
of research. lncRNAs are able to interact with miRNAs as well as
proteins. Sequence analysis has shown that lncRNAs and miRNAs have
complementary target sequences. lncRNAs can make miRNAs out of
mRNAs by binding the miRNAs. These types of lncRNAs are known as
ceRNAs (competing endogenous RNAs), and can reduce the biological
effects of multiple miRNAs, resulting in released suppression of
the miRNA target genes and therefore increased expression levels
(20). Conversely, it has been
reported that GAS5 is suppressed by miR-21 through a potential
binding site for miR-21 on GAS5; this negative correlation between
miR-21 and GAS5 was identified in breast cancer samples (21). Meanwhile, GAS5 in endometrial cancer
cells can bind with miR-103, regulating the expression of the
downstream target gene PTEN (22).
This suggests that the binding of GAS5 with different miRNAs has
different functions.
In this study, we reported an interaction between
GAS5 and miR-182-5p, which regulated CRC cell growth and apoptosis
through the regulation of FOXO3a. Our findings provide a novel
understanding of the role of GAS5 in CRC progression and of the
underlying mechanism involved.
Materials and methods
Patients and tissue samples
A total of 95 pairs of surgically resected CRC
specimens and adjacent normal tissues were collected at the
Department of Gastrointestinal Surgery, People's Hospital of
Tongling City. All tissue samples were obtained with written
informed consent in accordance with the requirements of the
Research Ethics Committee at the People's Hospital of Tongling
City, and all the experimental protocols were approved by the
Research Ethics Committee of the People's Hospital of Tongling
City. All of the methods performed in this study were in accordance
with the approved guidelines.
Cell line culture and
transfection
Four human CRC cell lines (HCT-116, HT-29, SW480 and
LoVo) and the normal colon epithelial cell line NCM460 were
purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA). Cells were cultured in Gibco™ RPMI-1640 medium
(Thermo Fisher Scientific, Inc., Waltham, MA, USA), supplemented
with 10% fetal bovine serum (FBS; HyClone Laboratories; GE
Healthcare Life Sciences, China) at 37°C in a 5% CO2
incubator. The full-length GAS5 sequence lacking the poly-A tail
was synthesized and sub-cloned into a pcDNA3.1 vector (Invitrogen;
Thermo Fisher Scientific, Inc.). Small interfering RNAs (siRNAs)
against GAS5 (si-GAS5), control siRNA (si-NC), miR-182-5p mimics
(miR-182-5p) and control mimics (miR-NC) were synthesized by
GenePharma Co., Ltd. (Shanghai, China). The target sequence for
GAS5 was 5′-CUUGCCUGGACCAGCUUAAdTdT-3′. The plasmids, siRNAs and
miRNA mimics were transfected into cells separately using
Invitrogen™ Lipofectamine 2000 reagent (Thermo Fisher Scientific,
Inc.), in accordance with the manufacturer's instructions. At 24,
48 and 72 h after transfection, the transfected cells were
harvested and processed for further analysis.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells and tissue
specimens using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's instructions. RNA
samples were reverse transcribed into cDNA using a PrimeScript™ II
1st Strand Synthesis kit (Takara Biotechnology, Co., Ltd., Dalian,
China). qPCR was performed using SYBR Premix Ex Taq II (Takara
Biotechnology) in 96-well optical plates and monitored with a Roche
Cobas® z480 system (Roche, Welwyn Garden City, UK). The
reaction conditions were as follows: 95°C for 10 sec, followed by
40 cycles of 95°C for 5 sec, 60°C for 15 sec and 72°C for 30 sec.
The relative gene expression of GAS5 was normalized to that of
β-actin and calculated using the 2−ΔΔCq method. A TaqMan
MicroRNA Assay Kit (Applied Biosystems; Thermo Fisher Scientific)
was used for miR-182-5p detection, and the expression level of
miR-182-5p was normalized using the 2−ΔΔCq method
relative to U6 snRNA expression. The sequences of specific primers
used in this study were as follows: GAS5 forward,
5′-CTTCTGGGCTCAAGTGATCCT-3′ and reverse,
5′-TTGTGCCATGAGACTCCATCAG-3′; β-actin forward, CTCCATCCTGGCTCGCTGT
and reverse, GCTGTCACCTTCACCGTTCC; miR-182-5p real-time primer,
5′-GGCTTTGGCAATGGTAGAACTCA-3′; and U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′. All assays were performed in
triplicate.
Dual-Luciferase reporter assay
The binding sites between GAS5 and miR-182-5p were
predicted using DIANA tools (http://carolina.imis.athena-innovation.gr/diana_tools/web).
GAS5 fragments containing the predicted wild-type (WT) or mutant
(MUT) miR-182-5p binding sites were generated and inserted into the
luciferase reporter vector psi-CHECK-2 (Promega, Shanghai, China).
HT-29 cells were placed on a 24-well plate and grown to 80%
confluence. Cells were then co-transfected with 100 ng miR-182-5p
mimics or miR-NC, 50 ng of GAS5-WT or GAS5-MUT plasmid, and 5 ng of
pRL-CMV containing Renilla luciferase using Lipofectamine
2000. At 48 h after transfection, luciferase activities were
detected using a Dual-Luciferase® reporter assay system
(Promega). The luciferase activity of each group was normalized to
the Renilla luciferase activity.
Cell proliferation assay
Cells were seeded into 96-well plates at
2×103 cells/well and cultured overnight. After
transfection for 24, 48 or 72 h, cell viability was analyzed with a
Cell Counting Kit-8 (CCK-8; Dojindo Laboratories, Rockville, MD,
USA). CCK-8 reagent was added to each well, and the cells were
incubated at 37°C for 1–4 h according to the manufacturer's
protocol. The absorbance value of each group was
spectrophotometrically determined at a wavelength of 450 nm. Each
group was assayed in a 96-well plate in triplicate.
Western blot analysis
Total protein from transfected cells was extracted
in cell lysis buffer (Pierce; Thermo Fisher Scientific, Inc.).
Protein concentrations were measured using the Micro BCA Protein
Assay kit (Enzyme, Nanjing, China). Proteins were separated by 10%
SDS-PAGE and transferred to polyvinylidene fluoride (PVDF)
membranes (EMD Millipore, Billerica, MA, USA). Target protein was
probed with primary antibodies overnight at 4°C. The primary
antibodies used were rabbit anti-FOXO3a (dilution 1:1,000; cat. no.
3938; Cell Signaling Technology, Danvers, MA, USA) and rabbit
anti-β-actin (dilution 1:2,000; cat. no. 20536-1-AP; Proteintech,
Rosemont, IL, USA). Expression of FOXO3a was quantified using
β-actin as the loading control. Then, the membranes were incubated
with secondary antibody (dilution 1:2,000; cat. no. 8889; Cell
Signaling Technology) for 2 h at room temperature. Protein bands
were visualized using an ECL kit (Tanon Science and Technology Co.,
Ltd., Shanghai, China) according to the manufacturer's
instructions.
Apoptosis analysis
Cells were transfected in 6-well plates for 48 h,
and then washed and resuspended in phosphate-buffered saline (PBS)
twice. Flow cytometric analysis was carried out to evaluate
apoptosis using an Annexin V-FITC/PI Apoptosis Detection kit
(MultiSciences Biotech, Hangzhou, China). Cells were double-stained
with Annexin V-FITC (50 µg/ml) and propidium iodide (PI) (10 µg/ml)
in the dark for 15 min at room temperature before they were
subjected to flow cytometric analysis (FACScan; BD Biosciences, San
Diego, CA, USA).
Statistical analysis
Experimental results are presented as the mean ±
standard deviation. The association of GAS5 expression with
clinical characteristics was analyzed using a Chi-square test.
Comparisons between two groups were conducted using the two-tailed
Student's t-test or the Chi-square test (SPSS 18.0; SPSS, Inc.,
Chicago, IL, USA). The comparison of multiple groups was analyzed
by ANOVA with Holm-Sidak's or Dunnett's multiple comparisons test
as indicated in the Figure legends (GraphPad Prism 6.0; GraphPad
Software Inc., La Jolla, CA, USA). Differences were considered
statistically significant when P<0.05.
Results
GAS5 is downregulated in CRC tissues
and cell lines
The expression levels of the lncRNA GAS5 in 95
paired samples (CRC specimens and corresponding adjacent non-tumor
tissues) were examined using real-time RT-qPCR. GAS5 expression was
significantly lower in tumor tissues than that noted in the
adjacent tissues (Fig. 1A). We next
investigated the levels of GAS5 expression in different CRC cell
lines (HCT-116, HT-29, SW480 and LoVo) and in the normal colon
epithelial cell line NCM460. GAS5 was expressed at a significantly
low level in HCT-116, HT-29, SW480 and LoVo cells in comparison to
its level in NCM460 cells; this difference was especially evident
in HT-29 and SW480 cells (Fig. 1B).
Therefore, HT-29 and SW480 cells were used to establish
GAS5-overexpressing cell lines with pcDNA3.1-GAS5 plasmids to
investigate the function of GAS5 in the development of CRC. The
transfection efficiency was verified by RT-qPCR (Fig. 1C). The median GAS5 expression was
used as the cut-off value. When the expression of GAS5 was greater
than the median, it was defined as high expression; otherwise it
was defined as low expression. As indicated in Table I, low expression of GAS was
significantly associated with lymph node metastasis and advanced
clinical stage in breast cancer. Therefore, downregulation of GAS5
is associated with the malignant progression of breast cancer.
 | Table I.Association between GAS5 expression
and clinicopathological characteristics of the colorectal cancer
cases. |
Table I.
Association between GAS5 expression
and clinicopathological characteristics of the colorectal cancer
cases.
|
| GAS5 levels |
|---|
|
|
|
|---|
|
Characteristics | Low (n=47) | High (n=48) | P-value |
|---|
| Age, years |
|
| 0.615 |
|
<55 | 19 | 17 |
|
|
≥55 | 28 | 31 |
|
| Sex |
|
| 0.576 |
|
Male | 31 | 29 |
|
|
Female | 16 | 19 |
|
| Depth of
invasion |
|
T1-T2 | 13 | 21 | 0.102 |
|
T3-T4 | 34 | 27 |
|
| Tumor size
(cm) |
| ≤4 | 21 | 28 | 0.183 |
|
>4 | 26 | 20 |
|
| Lymphatic
metastasis |
| N0 | 14 | 25 | 0.027a |
| N1 or
above | 33 | 23 |
|
| TNM stage |
|
I–II | 14 | 25 | 0.027a |
|
III–IV | 33 | 23 |
|
GAS5 overexpression suppresses CRC
cell proliferation and promotes apoptosis
Next, we investigated the association of GAS5
expression with CRC cell proliferation and apoptosis. HT-29 and
SW480 cells were transfected with pcDNA3.1-NC or pcDNA3.1-GAS5.
Cellular proliferation was evaluated by CCK-8 assay. The results
revealed that overexpression of GAS5 significantly attenuated the
proliferation rate of both cell lines over time compared with the
control group rate (Fig. 2A and B).
Flow cytometric analysis was subsequently used to detect the rate
of cellular apoptosis. We found that the cellular apoptosis rate
was increased significantly in response to GAS5 overexpression
compared with the rate in the pcDNA3.1-NC control group (Fig. 2C and D). Taken together, these data
indicate that GAS5 overexpression suppresses proliferation and
promotes apoptosis in CRC cells.
GAS5 negatively regulates the
expression of miR-182-5p in CRC cells
To further investigate the mechanism by which GAS5
regulates CRC cell proliferation and apoptosis, we studied the
relationship between miR-182-5p and GAS5. We used a bioinformatics
online software program (DIANA tools) to predict the potential
miRNAs that interact with GAS5. As expected, we found that
miR-182-5p contains a complementary nucleotide sequence for GAS5
(Fig. 3A). We measured the
expression level of miR-182-5p in CRC specimens and examined the
potential correlation between the RNA expression levels of GAS5 and
miR-182-5p. We observed a negative correlation between GAS5 and
miR-182-5p levels (Fig. 3B). As we
described, GAS5 negatively regulated miR-182-5p, yet, GAS5
regulated miR-182-5p through its function as an miRNA sponger.
Thus, we constructed a GAS5 fragment, which contained the
miR-182-5p-binding site into luciferase reporter vector, and
checked the mimic miR-182-5p effects on luciferase activity. This
is a common and standard process as reported previously (23,24).
The luciferase activity of the reporter containing GAS5-WT was
reduced in cells transfected with miR-182-5, while GAS5-MUT
provided resistance to miR-182-5p-induced luciferase reporter
repression (Fig. 3C and D). To
determine the effect of GAS5 on the expression of miR-182-5p, the
CRC cell line HT-29 was transfected with pcDNA3.1-GAS5 and SW480
cells were treated with si-GAS5. Expression levels of miR-182-5p
were detected by RT-qPCR in the GAS5-knockdown SW480 cells and
GAS5-overexpressing HT-29 cells. The knockdown of GAS5 markedly
increased the expression of miR-182-5p in the SW480 cells (Fig. 3E), while GAS5 upregulation
significantly decreased miR-182-5p expression in the HT-29 cells
(Fig. 3F), compared with expression
in the corresponding controls. Taken together, these results imply
that GAS5 functions as a sponge, which negatively regulates the
expression of miR-182-5p in CRC cells.
GAS5 overexpression suppresses CRC
cell proliferation and promotes apoptosis by inhibiting
miR-182-5p
Cell proliferation and apoptosis assays were
performed on HT-29 cells transfected with miR-182-5p mimics or
miR-NC. miR-182-5p overexpression caused a significant increase in
cell viability at 48 and 72 h (Fig.
4A), and consequently reduced the proportion of apoptotic cells
(Fig. 4B). To further evaluate the
effect of miR-182-5p regulation by GAS5 on CRC progression, HT-29
cells were transfected with pcDNA3.1-GAS5, pcDNA3.1-NC,
pcDNA.1-GAS5+miR-182-5p or pcDNA3.1-GAS5+miR-NC. CCK-8 assay
results indicated that the reduction in cell proliferation induced
by GAS5 overexpression was abrogated by miR-182-5p upregulation
(Fig. 4C). Furthermore, flow
cytometric analysis indicated that pcDNA3.1-GAS5 transfection led
to a significant increase in apoptotic rates; while miR-182-5p
overexpression obviously attenuated the GAS5-induced apoptosis in
HT-29 cells (Fig. 4D). Taken
together, all of these data suggest that GAS5 overexpression
inhibits cell proliferation and promotes apoptosis by targeting
miR-182-5p in CRC cells.
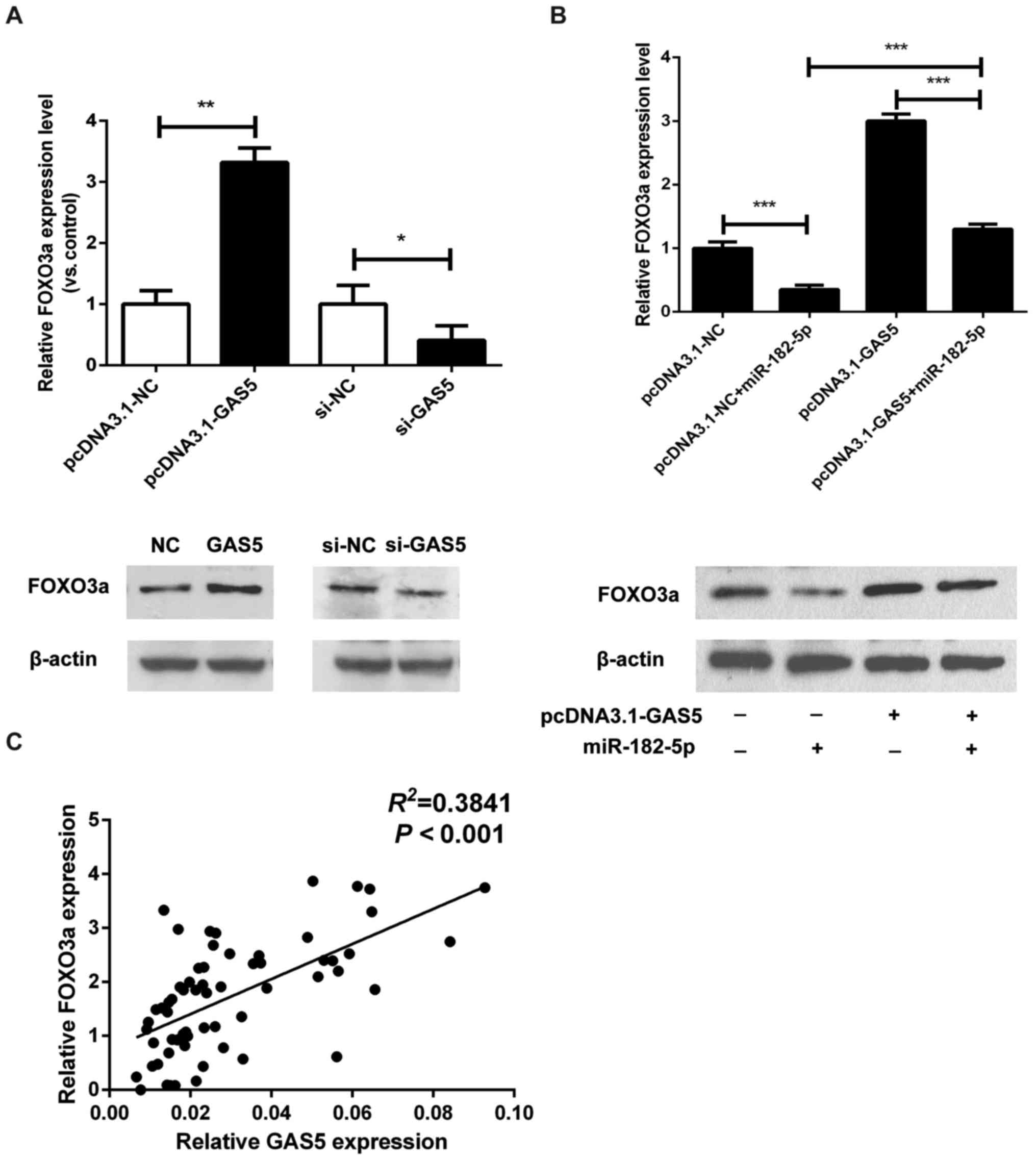
GAS5 inhibits the function of
miR-182-5p in CRC cells by regulating FOXO3a
It has been reported that FOXO3a, a pro-apoptotic
transcription factor and direct target of the PI3K-AKT signaling
pathway, may be the direct target of miR-182-5p (25). Therefore, we explored the potential
correlation between FOXO3a and GAS5 expression. As shown in
Fig. 5A, the mRNA and protein
levels of FOXO3a were increased in HT-29 cells transfected with
pcDNA.1-GAS5, and were decreased in GAS5-knockdown SW480 cells. To
further investigate the expression correlation between FOXO3a and
the GAS5/miR-182-5p axis, we examined the regulation of FOXO3a by
miR-182-5p and GAS5. The expression of FOXO3a was significantly
downregulated by miR-182-5p overexpression (Fig. 5B). More notably, miR-182-5p mimics
reversed the increase in expression of FOXO3a that was induced by
GAS5 overexpression (Fig. 5B).
Furthermore, in CRC tissues, GAS5 expression was found to be
positively association with the expression of FOXO3a (Fig. 5C). These data indicate that GAS5
overexpression inhibits the function of miR-182-5p in CRC cells by
regulating FOXO3a.
Discussion
The discovery of ceRNAs was made during the study of
miRNA regulation. As it was reported that the lncRNA IPS1 altered
the protein level of PHO2 in plants by inhibiting miR-399 activity
(26), lncRNA PVT1 was found to
regulate HIF-1α expression through sponge miRNA-199a5p in lung
cancer cells (27), lncRNA SNHG1
functions as a ceRNA to antagonize the effect of miR-145a-5p on the
downregulation of NUAK1 (28).
These early studies demonstrated that the process of miRNA
targeting may be reciprocal rather than unidirectional. It has been
found that multiple lncRNAs can act as ceRNAs and play a role in
regulating downstream targets by base pairing with and inactivating
miRNAs. For instance, the lncRNA HULC is highly expressed in HCC
cells and is implicated in tumor cell proliferation (29); and the lncRNA Loc285194 can bind to
miR-211 by complementary pairing to downregulate the expression of
miR-211 and exert an anti-oncogenic function (8). In this study, we found that expression
levels of the lncRNA GAS5 were significantly downregulated in CRC
tissues compared with that noted in the corresponding adjacent
tissues. Further functional and mechanistic studies revealed that
GAS5 exerts apoptosis-promoting effects by acting as a ceRNA of
miR-18-5p.
To date, many studies have reported the biological
roles of GAS5 in multiple cancers and its underlying molecular
mechanisms. Li et al found that GAS5 was significantly
decreased in ovarian cancer, which promoted cell proliferation,
migration and invasion partly by regulating cyclin D1, p21 and
apoptosis protease activating factor 1 (APAF1) expression,
suggesting that lower GAS5 expression may indicate a poor prognosis
in ovarian cancer (30). In
colorectal cancer (CRC), overexpressed GAS5 was reported to inhibit
cell proliferation both in vitro and vivo, and the
expression level of GAS5 was significantly associated with
susceptibility to and the progression of CRC (18,19).
Other reports have revealed that GAS5 acts as an important
regulator of the genesis and development of CRC by influencing
inflammatory cytokines via the NF-κB and Erk1/2 pathways (31). Here, we confirmed that GAS5
expression was lower in both CRC tissues and cell lines, and
downregulation of GAS5 was found to be associated with CRC stage
and lymphatic metastasis as reported previously (18,19).
We studied the effect of GAS5 on cell proliferation and apoptosis
using GAS5-overexpressing HT-29 and SW480 cells. We found that the
overexpression of GAS5 suppressed proliferation and promoted
apoptosis in the CRC cells. Other research has shown that GAS5
plays an important role in the process of cell proliferation
(13). To evaluate the underlying
mechanism in our study, the expression levels of miR-182-5p were
detected by RT-qPCR in GAS5-knockdown SW480 and GAS5-overexpressing
HT-29 cells; furthermore, upon analysis of CRC tissues, we found
that GAS5 expression was negatively correlated with miR-182-5p
expression. Additionally, bioinformatics analysis and a dual
luciferase activity assay indicated that GAS5 could directly
interact with miR-182-5p and negatively regulate its expression in
CRC cells. These findings suggest that the interaction between GAS5
and miR-182-5p may be involved in the regulation of CRC
progression. Thus, GAS5 might play a crucial role in inhibiting the
proliferation and promoting the apoptosis of CRC cells by
regulating miR-182-5p.
In recent years, miRNAs, which have been identified
to act as both oncogenes and cancer-suppressor genes, have been
widely implicated in the occurrence and development of CRC as well
as in tumor pathological processes including metastasis (32–34).
Furthermore, miRNAs may also serve as prognostic target molecules
for patients with CRC. A study by Tazawa et al revealed that
high expression of miR-34a could inhibit the proliferation of CRC
cells by negatively regulating the E2F1 signaling pathway; E2F1 may
affect the cell cycle by promoting cellular transition from the G1
phase to the S phase (34). Based
on comparison and analysis of miR-195 expression between CRC and
normal tissues, Liu et al noted downregulation of miR-195 in
cancer tissues, and their further experiments demonstrated that
miR-195 could function as a tumor-suppressor gene to induce the
apoptosis of tumor cells (33).
Therefore, we may conclude that miRNAs play an important role in
the occurrence and development of CRC. As a member of the miR-183
family, also composed of miR-183 and miR-96, miR-182-5p exhibits a
highly conserved coding sequence across animal species (35,36).
Most previous studies suggest that miR-182-5p is highly expressed
in various types of human cancer, including prostate, breast,
bladder, liver, colon, cervical and ovarian cancers and glioma, and
indicate an oncogenic role of miR-182-5p in tumor progression
(37–42). In patients with early-diagnosed
colon cancer, expression of miR-182-5p and miR-21-3p has been
reported to be significantly upregulated (43). Our data are consistent with previous
studies and, furthermore, confirm that miR-182-5p overexpression
significantly promotes CRC cell proliferation while suppressing
apoptosis. In the present study, to further evaluate the effect of
miR-182-5p regulation by GAS5 on CRC progression, HT-29 cells were
transfected with pcDNA3.1-GAS5, pcDNA3.1-NC,
pcDNA.1-GAS5+miR-182-5p or pcDNA3.1-GAS5+miR-NC. CCK-8 and flow
cytometric analyses showed that GAS5 overexpression obviously
inhibited cell proliferation and induced apoptosis in CRC cells,
while miR-182-5p overexpression markedly abolished these effects,
suggesting that GAS5 served as a tumor suppressor by inhibiting
miR-182-5p expression in CRC cells. With increasing studies on the
relationship between cancers and miR-182-5p-mediated regulation in
recent years, it has been found that miR-182-5p can promote the
proliferation of kidney cancer cells by activating the AKT/FOXO3a
signaling pathway (44). miR-182-5p
is also capable of regulating FOXO1 and FOXO3 genes and
contributing to the metastasis of breast cancer and melanoma
(35). Furthermore, by acting on
the downstream target gene FOXO1 in liver cancer, miR-183, miR96
and miR-182 activate the Wnt/β-catenin pathway, thereby
accelerating cell migration and invasion (45). Other reports have also detected
elevated expression of miR-182 in hepatocellular carcinoma and
found that this may promote liver tumor metastasis by regulating
the MTSS1 gene (46).
FOXO3a has a pivotal role in both oncogenesis and
tumor suppression (47). Loss of
FOXO3a has been observed in various cancers, and its cellular
localization and phosphorylation status are considered to be
prognostic factors for breast, prostate, bladder and ovarian
cancers (48–52). It has been reported that FOXO3a, a
pro-apoptotic transcription factor and direct target of the
PI3K-AKT signaling pathway, might be a direct target of miR-182-5p
(25,44). In this study, we analyzed the
expression level of FOXO3a and found that the expression level of
GAS5 was positively association with FOXO3a in CRC tissues. In
addition, we found that FOXO3a expression was increased in
GAS5-overexpressing CRC cells and decreased in GAS5-knockdown SW480
cells. Furthermore, miR-182-5p abrogated the upregulation of FOXO3a
induced by GAS5. Thus, we suggested that GAS5 overexpression
inhibited the function of miR-182-5p in CRC cells by regulating
FOXO3a expression. This adds to previous research findings that the
ectopic expression of miR-182-5p has a tumor-promoting effect in
melanoma, lung cancer and stomach cancer by targeting MITF, BCL2,
cyclin D2, RGS17 and CREB1 (53–55).
Overall, this study demonstrated that GAS5 was
downregulated in CRC tissues and cell lines. GAS5 overexpression in
CRC cells suppressed cell proliferation and promoted apoptosis by
inhibiting miR-182-5p. We also found that GAS5 inhibits the
function of miR-182-5p in CRC cells by regulating FOXO3a. In
conclusion, the GAS5/miR-182-5p/FOXO3a axis might play a key role
in CRC growth and serve as a target for potential therapeutic
applications.
Acknowledgements
The authors thank Dr Wei Zhao for his technical
support.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
KWC and WMZ conceived and designed the study. KWC,
ZGZ, GW and JW performed the experiments. WMZ and KWC wrote the
paper. KWC, ZGZ, GW and JW reviewed and edited the manuscript. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
All tissue samples were obtained with written
informed consent in accordance with the requirements of the
Research Ethics Committee at the People's Hospital of Tongling
City, and all the experimental protocols were approved by the
Research Ethics Committee of the People's Hospital of Tongling
City. All of the methods performed in this study were in accordance
with the approved guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, DeSantis C, Virgo K, Stein K,
Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al:
Cancer treatment and survivorship statistics, 2012. CA Cancer J
Clin. 62:220–241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huarte M and Rinn JL: Large non-coding
RNAs: Missing links in cancer? Human Mol Genet. 19:R152–R161. 2010.
View Article : Google Scholar
|
|
4
|
Maruyama R and Suzuki H: Long noncoding
RNA involvement in cancer. BMB Rep. 45:604–611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu FJ, Zheng JJ, Dong PH and Fan XM: Long
non-coding RNAs and hepatocellular carcinoma. Mol Clin Oncol.
3:13–17. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Redis RS, Sieuwerts AM, Look MP, Tudoran
O, Ivan C, Spizzo R, Zhang X, de Weerd V, Shimizu M, Ling H, et al:
CCAT2, a novel long non-coding RNA in breast cancer: Expression
study and clinical correlations. Oncotarget. 4:1748–1762. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gutschner T, Hammerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu Q, Huang J, Zhou N, Zhang Z, Zhang A,
Lu Z, Wu F and Mo YY: LncRNA loc285194 is a p53-regulated tumor
suppressor. Nucleic Acids Res. 41:4976–4987. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ye LC, Zhu X, Qiu JJ, Xu J and Wei Y:
Involvement of long non-coding RNA in colorectal cancer: From
benchtop to bedside (Review). Oncol Lett. 9:1039–1045. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Han D, Wang M, Ma N, Xu Y, Jiang Y and Gao
X: Long noncoding RNAs: Novel players in colorectal cancer. Cancer
Lett. 361:13–21. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schneider C, King RM and Philipson L:
Genes specifically expressed at growth arrest of mammalian cells.
Cell. 54:787–793. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kino T, Hurt DE, Ichijo T, Nader N and
Chrousos GP: Noncoding RNA gas5 is a growth arrest- and
starvation-associated repressor of the glucocorticoid receptor. Sci
Signal. 3:ra82010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shi X, Sun M, Liu H, Yao Y, Kong R, Chen F
and Song Y: A critical role for the long non-coding RNA GAS5 in
proliferation and apoptosis in non-small-cell lung cancer. Mol
Carcinog. 54 Suppl 1:E1–E12. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pickard MR, Mourtada-Maarabouni M and
Williams GT: Long non-coding RNA GAS5 regulates apoptosis in
prostate cancer cell lines. Biochim Biophys Acta. 1832:1613–1623.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qiao HP, Gao WS, Huo JX and Yang ZS: Long
non-coding RNA GAS5 functions as a tumor suppressor in renal cell
carcinoma. Asian Pac J Cancer Prev. 14:1077–1082. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mourtada-Maarabouni M, Pickard MR, Hedge
VL, Farzaneh F and Williams GT: GAS5, a non-protein-coding RNA,
controls apoptosis and is downregulated in breast cancer. Oncogene.
28:195–208. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Krell J, Frampton AE, Mirnezami R, Harding
V, De Giorgio A, Alonso Roca L, Cohen P, Ottaviani S, Colombo T,
Jacob J, et al: Growth arrest-specific transcript 5 associated
snoRNA levels are related to p53 expression and DNA damage in
colorectal cancer. PLoS One. 9:e985612014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yin D, He X, Zhang E, Kong R, De W and
Zhang Z: Long noncoding RNA GAS5 affects cell proliferation and
predicts a poor prognosis in patients with colorectal cancer. Med
Oncol. 31:2532014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zheng Y, Song D, Xiao K, Yang C, Ding Y,
Deng W and Tong S: LncRNA GAS5 contributes to lymphatic metastasis
in colorectal cancer. Oncotarget. 7:83727–83734. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C,
Xu M, Wu F and Mo YY: Negative regulation of lncRNA GAS5 by miR-21.
Cell Death Differ. 20:1558–1568. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guo C, Song WQ, Sun P, Jin L and Dai HY:
LncRNA-GAS5 induces PTEN expression through inhibiting miR-103 in
endometrial cancer cells. J Biomed Sci. 22:1002015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM,
Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF and Waye MM: The lncRNA
H19 promotes epithelial to mesenchymal transition by functioning as
miRNA sponges in colorectal cancer. Oncotarget. 6:22513–22525.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY,
Gong W and Quan ZW: Long non-coding RNA CCAT1 promotes gallbladder
cancer development via negative modulation of miRNA-218-5p. Cell
Death Dis. 6:e15832015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Y, Li A, Wu J, He Y, Yu H, Chai R and
Li H: MiR-182-5p protects inner ear hair cells from
cisplatin-induced apoptosis by inhibiting FOXO3a. Cell Death Dis.
7:e23622016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang CY, Shirley N, Genc Y, Shi B and
Langridge P: Phosphate utilization efficiency correlates with
expression of low-affinity phosphate transporters and noncoding
RNA, IPS1, in barley. Plant Physiol. 156:1217–1229. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang C, Han C, Zhang Y and Liu F: LncRNA
PVT1 regulate expression of HIF1α via functioning as
ceRNA for miR199a5p in nonsmall cell lung cancer under hypoxia. Mol
Med Rep. 17:1105–1110. 2018.PubMed/NCBI
|
|
28
|
Lan X and Liu X: LncRNA SNHG1 functions as
a ceRNA to antagonize the effect of miR-145a-5p on the
down-regulation of NUAK1 in nasopharyngeal carcinoma cell. J
Cell Mol Med. Mar 25–2018.(Epub ahead of print). doi:
10.1111/jcmm.13497. View Article : Google Scholar
|
|
29
|
Li J, Wang X, Tang J, Jiang R, Zhang W, Ji
J and Sun B: HULC and Linc00152 act as novel biomarkers in
predicting diagnosis of hepatocellular carcinoma. Cell Physiol
Biochem. 37:687–696. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li J, Huang H, Li Y, Li L, Hou W and You
Z: Decreased expression of long non-coding RNA GAS5 promotes cell
proliferation, migration and invasion, and indicates a poor
prognosis in ovarian cancer. Oncol Rep. 36:3241–3250. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Y, Li Y, Huang S, He K, Zhao M, Lin H,
Li D, Qian J, Zhou C, Chen Y and Huang C: Long non-coding RNA
growth arrest specific transcript 5 acts as a tumour suppressor in
colorectal cancer by inhibiting interleukin-10 and vascular
endothelial growth factor expression. Oncotarget. 8:13690–13702.
2017.PubMed/NCBI
|
|
32
|
Lin M, Chen W, Huang J, Gao H, Ye Y, Song
Z and Shen X: MicroRNA expression profiles in human colorectal
cancers with liver metastases. Oncol Rep. 25:739–747.
2011.PubMed/NCBI
|
|
33
|
Liu L, Chen L, Xu Y, Li R and Du X:
microRNA-195 promotes apoptosis and suppresses tumorigenicity of
human colorectal cancer cells. Biochem Biophys Res Commun.
400:236–240. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tazawa H, Tsuchiya N, Izumiya M and
Nakagama H: Tumor-suppressive miR-34a induces senescence-like
growth arrest through modulation of the E2F pathway in human colon
cancer cells. Proc Natl Acad Sci USA. 104:15472–15477. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Segura MF, Hanniford D, Menendez S, Reavie
L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A,
Bogunovic D, et al: Aberrant miR-182 expression promotes melanoma
metastasis by repressing FOXO3 and microphthalmia-associated
transcription factor. Proc Natl Acad Sci USA. 106:1814–1819. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cao LL, Xie JW, Lin Y, Zheng CH, Li P,
Wang JB, Lin JX, Lu J, Chen QY and Huang CM: miR-183 inhibits
invasion of gastric cancer by targeting Ezrin. Int J Clin Exp
Pathol. 7:5582–5594. 2014.PubMed/NCBI
|
|
37
|
Tang T, Wong HK, Gu W, Yu MY, To KF, Wang
CC, Wong YF, Cheung TH, Chung TK and Choy KW: MicroRNA-182 plays an
onco-miRNA role in cervical cancer. Gynecol Oncol. 129:199–208.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cekaite L, Rantala JK, Bruun J, Guriby M,
Agesen TH, Danielsen SA, Lind GE, Nesbakken A, Kallioniemi O, Lothe
RA, et al: MiR-9, −31, and −182 deregulation promote proliferation
and tumor cell survival in colon cancer. Neoplasia. 14:868–879.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hirata H, Ueno K, Shahryari V, Tanaka Y,
Tabatabai ZL, Hinoda Y and Dahiya R: Oncogenic miRNA-182-5p targets
Smad4 and RECK in human bladder cancer. PLoS One. 7:e510562012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chiang CH, Hou MF and Hung WC:
Up-regulation of miR-182 by β-catenin in breast cancer
increases tumorigenicity and invasiveness by targeting the matrix
metalloproteinase inhibitor RECK. Biochim Biophys Acta.
1830:3067–3076. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yao J, Xu C, Fang Z, Li Y, Liu H, Wang Y,
Xu C and Sun Y: Androgen receptor regulated microRNA miR-182-5p
promotes prostate cancer progression by targeting the ARRDC3/ITGB4
pathway. Biochem Biophys Res Commun. 474:213–219. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang YQ, Guo RD, Guo RM, Sheng W and Yin
LR: MicroRNA-182 promotes cell growth, invasion, and
chemoresistance by targeting programmed cell death 4 (PDCD4) in
human ovarian carcinomas. J Cell Biochem. 114:1464–1473. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang X, Chen L, Jin H, Wang S, Zhang Y,
Tang X and Tang G: Screening miRNAs for early diagnosis of
colorectal cancer by small RNA deep sequencing and evaluation in a
Chinese patient population. Onco Targets Ther. 9:1159–1166.
2016.PubMed/NCBI
|
|
44
|
Xu X, Wu J, Li S, Hu Z, Xu X, Zhu Y, Liang
Z, Wang X, Lin Y and Mao Y: Downregulation of microRNA-182-5p
contributes to renal cell carcinoma proliferation via activating
the AKT/FOXO3a signaling pathway. Mol Cancer. 13:1092014.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Leung WK, He M, Chan AW, Law PT and Wong
N: Wnt/β-catenin activates MiR-183/96/182 expression in
hepatocellular carcinoma that promotes cell invasion. Cancer Lett.
362:97–105. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang J, Li J, Shen J, Wang C, Yang L and
Zhang X: MicroRNA-182 downregulates metastasis suppressor 1 and
contributes to metastasis of hepatocellular carcinoma. BMC Cancer.
12:2272012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Guo S and Sonenshein GE: Forkhead box
transcription factor FOXO3a regulates estrogen receptor alpha
expression and is repressed by the Her-2/neu/phosphatidylinositol
3-kinase/Akt signaling pathway. Mol Cell Biol. 24:8681–8690. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lu M, Xiang J, Xu F, Wang Y, Yin Y and
Chen D: The expression and significance of pThr32-FOXO3a in human
ovarian cancer. Med Oncol. 29:1258–1264. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yu C, Zhang Z, Liao W, Zhao X, Liu L, Wu
Y, Liu Z, Li Y, Zhong Y, Chen K, et al: The tumor-suppressor gene
Nkx2.8 suppresses bladder cancer proliferation through upregulation
of FOXO3a and inhibition of the MEK/ERK signaling pathway.
Carcinogenesis. 33:678–686. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dubrovska A, Kim S, Salamone RJ, Walker
JR, Maira SM, García-Echeverría C, Schultz PG and Reddy VA: The
role of PTEN/Akt/PI3K signaling in the maintenance and viability of
prostate cancer stem-like cell populations. Proc Natl Acad Sci USA.
106:268–273. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yang L, Xie S, Jamaluddin MS, Altuwaijri
S, Ni J, Kim E, Chen YT, Hu YC, Wang L, Chuang KH, et al: Induction
of androgen receptor expression by phosphatidylinositol
3-kinase/Akt downstream substrate, FOXO3a, and their roles in
apoptosis of LNCaP prostate cancer cells. J Biol Chem.
280:33558–33565. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Habashy HO, Rakha EA, Aleskandarany M,
Ahmed MA, Green AR, Ellis IO and Powe DG: FOXO3a nuclear
localisation is associated with good prognosis in luminal-like
breast cancer. Breast Cancer Res Treat. 129:11–21. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kong WQ, Bai R, Liu T, Cai CL, Liu M, Li X
and Tang H: MicroRNA-182 targets cAMP-responsive element-binding
protein 1 and suppresses cell growth in human gastric
adenocarcinoma. FEBS J. 279:1252–1260. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sun Y, Fang R, Li C, Li L, Li F, Ye X and
Chen H: Hsa-mir-182 suppresses lung tumorigenesis through down
regulation of RGS17 expression in vitro. Biochem Biophys Res
Commun. 396:501–507. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yan D, Dong XD, Chen X, Yao S, Wang L,
Wang J, Wang C, Hu DN, Qu J and Tu L: Role of microRNA-182 in
posterior uveal melanoma: Regulation of tumor development through
MITF, BCL2 and cyclin D2. PLoS One. 7:e409672012. View Article : Google Scholar : PubMed/NCBI
|