Introduction
Gastric cancer is one of the leading causes of
cancer-associated mortality and the fourth most common cancer
worldwide. Patients with gastric cancer have a poor prognosis and
the 5-year survival rate is only ~20% (1). Despite the growth of gastric carcinoma
being inhibited by chemotherapy, the side effects and toxicity are
so high that it becomes intolerable for the majority of patients
(2). Therefore, alternative
therapeutic drugs are being sought for the treatment of gastric
cancer.
Flavonoids are family of polyphenolic compounds that
occur naturally in plants. Flavonoids are noted for their
biological activities, including antioxidative, anti-inflammatory,
anti-allergic and anti-carcinogenic properties. Tangeretin
[5,6,7,8-tetramethoxy-2-(4-methoxyphenyl)-4H−1-benzopyran-4-one]
is a natural O-polymethoxylated flavonoid commonly occurring in
fruits of the Citrus genus. Polymethoxylated flavonoids are
known to inhibit tumor cell viability more effectively compared
with free hydroxylated flavonoids (3,4). It
has been identified that tangeretin possesses a number of
biological activities such as anti-proliferative, anti-invasive,
anti-metastatic and antioxidative properties (5). Tangeretin has been identified to
inhibit the viability of breast cancer and colon cancer, and human
leukemic cell lines (6,7). Previous study has demonstrated that
tangeretin induces apoptosis in AGS gastric cancer cells (8). However, to the best of our knowledge,
cellular protein alterations in response to tangeretin in AGS
gastric cancer cells have not yet been investigated.
Proteomic techniques are promising tools for
identifying differentially expressed proteins and they are also
able to screen for novel target proteins. Differential proteomics
is an important area of proteomics that involves the comparison and
identification of proteins that are expressed by a whole genome or
in a complex mixture (9). Previous
studies have identified that a quantitative proteomic profile
reveals markedly abundant differentially expressed proteins that
may serve as novel biomarkers on cancer cells that may be targeted
using phytonutrients (10,11). The aim of the present study was to
identify novel biomarkers for gastric cancer. Despite it having
been revealed that tangeretin induces apoptosis in AGS gastric
cancer cells (8), to the best of
our knowledge, the proteomic profile of tangeretin-induced cell
death in AGS cells has not yet been reported.
The aim of the present study was to identify the
differentially expressed proteins between tangeretin-treated or
untreated AGS cancer cells using a proteomics method. Key
functional proteins involved in the major signaling network were
identified that revealed the various cellular proteins associated
with the regulatory mechanism of cell viability and cell death,
which may serve as predictable biomarkers for therapeutic
targets.
Materials and methods
Chemicals and reagents
RPMI-1640 medium, fetal bovine serum (FBS) and
antibiotics (streptomycin/penicillin) were purchased from Gibco;
Thermo Fisher Scientific, Inc. (Waltham, MA, USA). Materials and
chemicals used for electrophoresis were obtained from Bio-Rad
Laboratories, Inc. (Hercules, CA, USA). Anti-phosphoinositide
4-kinase (PI4K; 230 kDa; cat. no. 4902) and β-actin (45 kDa; cat.
no. 4970) were purchased from Cell Signaling Technology, Inc.
(Danvers, MA, USA), anti-mitogen-activated protein kinase 4 (MAPK4;
65 kDa; PA5-14185) was purchased from Thermo Fisher Scientific,
Inc., anti-protein kinase Cε (PKCε; 90 and 85 kDa; cat. no. 06991)
was purchased from Merck & Co., Inc. (Whitehouse Station, NJ,
USA) and anti-poly(ADP-ribose) polymerase 14 (PARP14; 171 kDa; cat.
no. HPA012063) was purchased from Sigma-Aldrich; Merck KGaA
(Darmstadt, Germany). All other chemicals were purchased from
Amresco, LLC (Solon, OH, USA) and Sigma-Aldrich; Merck KGaA. The
chemicals used were commercially available and of the highest
grade.
Cell culture and treatment
The human AGS gastric cancer cell line was obtained
from the Korean Cell Line Bank (Seoul, Korea). The cells were
maintained in RPMI-1640 medium supplemented with 10% heat
inactivated FBS and 1% penicillin/streptomycin at 37°C in a 5%
CO2 incubator. Cells were treated with vehicle alone [1%
dimethylsulfoxide (DMSO)] or 50, 75, 100 and 150 µM tangeretin
dissolved in 1% DMSO.
Cell viability assay
Cell viability was determined using an MTT assay.
AGS cells were seeded at a density of 1×105 cells/well
in 12-well plates. Following overnight incubation at 37°C in a 5%
CO2 incubator, cells were treated with 0, 50, 75, 100
and 150 µM tangeretin. The MTT assay was performed after 24 h of
incubation. To each well, 100 µl 0.5% (w/v) MTT dissolved in 1X PBS
was added prior to incubation at 37°C for 3 h. The medium was
aspirated and the formazan contained in the cell was solubilized in
500 µl DMSO. After 15 min of shaking, the absorbance at 540 nm was
determined using a microplate reader. Cell viability was expressed
as a percentage relative to that of controls (untreated cells),
which was set at 100%.
Protein extraction proteins were
extracted from AGS cells treated with vehicle or 100 µM tangeretin
for 24 h
In brief, trypsinized cells were dissolved in lysis
buffer containing 7 M urea, 2 M thiourea and 4% (w/v)
3-[(3-cholamidopropyl)dimethylammonio] propane-1-sulfonic acid
(CHAPS). The lysates were centrifuged at 1,000 × g for 15 min at
4°C, and the collected supernatant was stored at −70°C until
analysis. Proteins were precipitated with an equal volume (1:1) of
20% (v/v) trichloroacetic acid and dissolved in 7 M urea, 2 M
thiourea and 4% (w/v) CHAPS, 0.5% (v/v) immobilized pH gradient
(IPG) buffer and 1% dithiothreitol (DTT). Protein concentration was
then determined using a Non-Interfering™ Protein Assay kit
(G-Biosciences, St. Louis, MO, USA), according to the
manufacturer's protocol.
Two-dimensional gel electrophoresis
(2DE)
IPG strips (18 cm), pH 3–10, were rehydrated in a
rehydration buffer containing 7 M urea, 2 M thiourea, 4% (w/v)
CHAPS and 0.002% bromophenol blue. For the first dimension, 800 µg
protein was focused using the Ettan IPG Phor II isoelectric
focusing (IEF) system (GE Healthcare, Chicago, IL, USA) at 50 V for
1 h, followed by 200 V for 1 h, 400 V for 30 min, 500 V for 30 min,
4,000 V for 1 h for hold, 4,000 V for 1 h for gradient, 10,000 V
for 1 h, 10,000 V for 13 h and 50 V for 3 h. The focused strips
were equilibrated twice for 15 min each, first with 10 mg/ml DTT
and then with 40 mg/ml iodoacetamide (IAA) prepared in
equilibration buffer containing 50 mM Tris/HCl (pH 8.8), 6 M urea,
30% (v/v) glycerol, 2% (w/v) SDS and 0.002% (w/v) bromophenol blue.
The focused proteins were then separated in the second dimension by
SDS-PAGE (12% linear gradient) with a constant current of 15 mA/gel
at 20°C until the dye reached the bottom of the gel.
Gel spot detection and in-gel
digestion
Silver staining was performed for protein spot
visualization. Three independent gels were stained in triplicate.
Scanned gel images were acquired using a GS-800 scanner (Bio-Rad
Laboratories, Inc.) and imported into Progenesis SameSpots software
(version 4.1; Nonlinear Dynamics, Ltd., Newcastle upon Tyne, UK)
for differential spot expression analysis using automatic matching
alignment of the detected protein spots. Spots differing
significantly (P<0.05 and P<0.1) in their intensities with a
fold-change ≥1.5 were used for further analysis. Selected protein
spots were excised manually from the two-dimensional
electrophoresis (2DE) gel and protein digestion was performed
according to a silver stain gel extraction protocol (12) with a slight modification. Briefly,
the excised gel pieces were washed with deionized water prior to
destaining with 30 mM potassium ferricyanide for 10 min until the
silver stain disappeared followed by washing three times with
deionized water for 5 min each and dehydration in 100 µl
acetonitrile for 10 min. Dehydrated pieces were dried in a
lyophilizer (SFDSM06; Samwon Freezing Engineering Co., Busan,
Korea), the gel pieces were rehydrated in 100 µl 100 mM
NH4HCO3 continuing the reduction (10 mM DTT)
and alkylation (100 mM IAA) process at room temperature for 45 min.
Following simultaneous drying and rehydrating, and vacuum drying,
the gel pieces were trypsinized with 20 ng/µl trypsin (Promega
Corporation, Madison, WI, USA) on ice. After 45 min, 10–20 µl 50 mM
NH4HCO3 was added followed by overnight
digestion at 37°C. These peptide mixtures were extracted for
subsequent steps of matrix-assisted laser-desorption ionization
(MALDI) spot targeting.
MALDI-time-of-flight (TOF)-mass
spectrometry (MS) and tandem MS (MS/MS) analysis
The aforementioned pooled extracts were dried in a
lyophilizer and the extracts were redissolved in 1 µl extraction
buffer (50 µl acetonitrile, 20 µl trifluoroacetic acid and 930 µl
distilled water) and 1 µl matrix solution
(α-acyano-4-hydroxycinnamic acid) and targeted onto a MALDI-TOF
plate. Following drying the samples completely onto the targeting
plate, MALDI-TOF-MS was performed using a Voyager-DE STR mass
spectrometer (Applied Biosystems; Thermo Fisher Scientific, Inc.)
equipped with delay ion extraction. Mass spectra were acquired over
a mass range between 800 and 3,000 Da. The peptide mass peak list
was processed using DataExplorer software (version 4.8; Applied
Biosystems; Thermo Fisher Scientific, Inc.) to search the protein
against the SwissProt database (www.ebi.ac.uk/uniprot) using the Mascot-Peptide Mass
Fingerprint program (www.matrixscience.com). The following parameters were
used for database searches: Taxonomy, Homo sapiens (human);
cleavage specificity, trypsin with one missed cleavage allowed;
peptide tolerance of 100 p.p.m. for the fragment ions; and allowed
modifications, cysteine carbamidomethyl (fixed) and oxidation of
methionine (variable). The MOWSE scores (>56) and species were
considered to identify the correct protein from the Mascot results
list.
Bioinformatics analysis
Functional genome ontology of the identified
proteins was performed using Protein ANalysis THrough Evolutionary
Relationships (PANTHER; version 11.1; pantherdb.org)
database. PANTHER uses GO-Slim which is a subset of Gene Ontology
(GO). Proteins were further annotated using Database for
Annotation, Visualization and Integrated Discovery (DAVID) gene
bioinformatics resource for enrichment analysis and association
(version 6.8; david.ncifcrf.gov). Expression Analysis Systematic
Explorer (EASE) was used for the biological interpretation of the
genes derived from the proteomics profile. Protein interactions
were identified using Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING; version 10) database (13). Markov Cluster Algorithm (MCL) is
used for clustering the proteins that were displayed in the
network.
Western blot analysis
Briefly, AGS cells treated with vehicle or 100 µM
tangeretin for 24 h and were lysed overnight with lysis buffer
(radioimmunoprecipitation assay buffer) containing phosphatase
inhibitor cocktail along with protease inhibitor and EDTA (Thermo
Fisher Scientific, Inc.). The extracted proteins were then
centrifuged at 1,000 × g for 30 min at 4°C to remove debris.
Amounts of 20 µg proteins, determined using the Bradford assay,
were resolved by SDS-PAGE (8–12% gel) and subsequently transferred
onto a polyvinylidene difluoride membrane (Immobilon-P, 0.45 µm;
EMD Millipore, Billerica, MA, USA) using a TE 77 Semi-Dry Transfer
Unit (GE Healthcare Life Sciences, Little Chalfont, UK). The
membranes were blocked with 5% bovine serum albumin (BioShop Canada
Inc., Burlington, Canada) in Tris-buffered saline containing 1%
Tween-20 (TBS-T, pH 7.4) or 1X Phospho blocking solution (TransLab
Biosciences, Daejon, Korea) at room temperature for 1 h. Blots were
probed with 1:500 (anti-PI4K and anti-PKCε) or 1:1,000 (anti-MAPK4
and anti-PARP14) dilutions of the respective primary antibodies at
4°C for overnight. Following washing five times with TBS-T, the
membranes were incubated with horseradish peroxidase-linked
anti-rabbit IgG (cat. no. 7074; Cell Signaling Technology, Inc.)
secondary antibodies diluted 1:1,000 (for detection of PI4K and
PKCε) or 1:2,000 (for detection of MAPK4, PARP14 and β-actin) at
room temperature for 3 h. The immunoblots were visualized using an
enhanced chemiluminescence kit and western blotting detection
reagents (GE Healthcare Life Sciences). Each protein band was
quantified densitometrically using ImageJ software (version 1;
imagej.nih.gov/ij; National Institutes of Health,
Bethesda, MD, USA) following normalization to β-actin
expression.
Statistical analysis
Results are expressed as the mean ± standard
deviation of a minimum three replicates in independent experiments.
The data were analyzed using one-way analysis of variance followed
by a Newman-Keuls post hoc test using GraphPad Prism (version 5;
GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Effect of tangeretin on AGS cell
viability
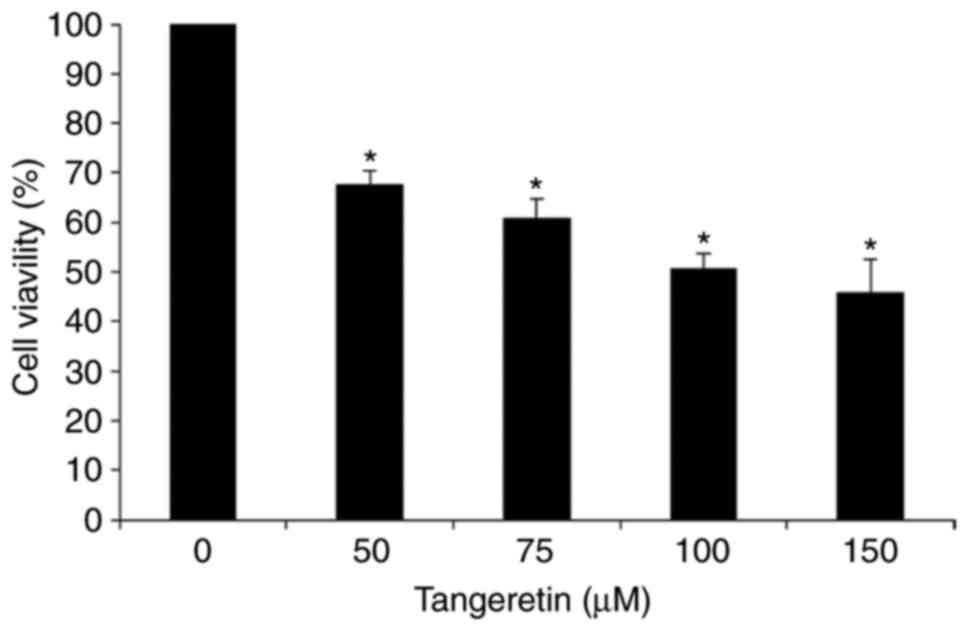
In order to assess the effect of tangeretin on the
viability of AGS cells, an MTT assay was performed. It was observed
that tangeretin treatment decreased the cell viability of AGS cells
dose-dependently with an IC50 value of 100 µM (Fig. 1). This result suggests that
tangeretin induced significant inhibition and cell death in AGS
cells, and the 100 µM concentration of tangeretin was selected for
further experiments.
Proteomic analysis of AGS cells in
response to tangeretin treatment
To analyze the underlying molecular mechanism of
tangeretin-induced AGS cell death, 800 µg total proteins were
separated by IEF on 18 cm IPG strips in the first dimension and
resolved by 2DE followed by silver staining for visualization. A
total of 300 protein spots were identified, with 36 spots differing
significantly in their intensities with a fold change ≥1.5
(Fig. 2). These 36 differentially
expressed proteins spots were selected for further analysis using
MALDI-TOF-MS. As MALDI-TOF-MS detects fewer peptides, these 36
differentially expressed spots were analyzed further using the
MASCOT search engine. Owing to post-translational modification or
proteolytic cleavage, a number of proteins may be detected from one
spot or the same protein may be detected from different spots. Of
the 38 spots, 16 significantly differentially expressed proteins
were successfully identified using the MASCOT search engine, and
the SwissProt database revealed two upregulated proteins and 14
downregulated proteins (Table
I).
 | Table I.Differentially expressed proteins
identified by matrix-assisted laser-desorption
ionization-time-of-flight-mass spectrometry in AGS cells treated
with tangeretin. |
Table I.
Differentially expressed proteins
identified by matrix-assisted laser-desorption
ionization-time-of-flight-mass spectrometry in AGS cells treated
with tangeretin.
| Spot no. | Protein | UniProt | Theoretical mass
(Mr) | pI | Protein sequence
coverage, % | Matched
peptides | Score | Regulation | Function
(www.uniprot.org) |
|---|
| 15 | Mitogen-activated
protein kinase 4 | MK04_HUMAN | 66393 | 5.25 | 69 | 38 | 58 | ↓ | Protein kinase
intracellular signaling |
| 15 | Microtubule-actin
cross-linking factor 1, isoforms 1/2/3/5 | MACF1_HUMAN | 843033 | 5.28 | 41 | 353 | 90 | ↓ | Cell proliferation
and migration |
| 17 | LIM domain-only
protein 7 | LMO7_HUMAN | 194002 | 8.34 | 61 | 121 | 76 | ↓ | Self-renewal, cell
cycle regulation and metastasis |
| 18 | Apolipoprotein
B-100 | APOB_HUMAN | 516651 | 6.58 | 47 | 233 | 98 | ↑ | Cellular binding
and internalization of low-density lipoprotein particles by the
apolipoprotein B/E receptor |
| 20 | Myosin 13 | MYH13_HUMAN | 224605 | 5.54 | 68 | 164 | 57 | ↓ |
| 21 | T-complex protein 1
subunit θ | TCPQ_HUMAN | 60153 | 5.42 | 71 | 54 | 65 | ↓ | Cell
proliferation |
| 22 |
Microtubule-associated protein 6 | MAP6_HUMAN | 86680 | 9.2 | 69 | 69 | 61 | ↓ |
Calmodulin-binding |
| 23 | Cytosolic
carboxypeptidase 3 | CBPC3_HUMAN | 117135 | 8.98 | 65 | 80 | 61 | ↓ |
Metallocarboxypeptidase that mediates
tubulin deglu tamylation |
| 25 | Chromobox protein
homolog 8 | CBX8_HUMAN | 43483 | 9.92 | 70 | 33 | 58 | ↓ | Cell proliferation,
acts as a tran scriptional repressor |
| 26 | Protein kinase Cε
type | KPCE_HUMAN | 84989 | 6.73 | 61 | 57 | 66 | ↓ | Tumor promoter |
| 28 |
Serrotransferrin | TRFE_HUMAN | 79294 | 6.81 | 62 | 55 | 57 | ↓ | Stimulates cell
proliferation |
| 28 | Phosphoinositide
4-kinase α | PI4KA_HUMAN | 239244 | 6.64 | 47 | 134 | 72 | ↓ | Regulates receptor
tyrosine kinase signaling, implicated in chemoresistance, tumor
angio genesis and metastasis |
| 28 |
Chromodomain-helicase-DNA-binding protein
3 | CHD3_HUMAN | 227989 | 6.92 | 48 | 132 | 76 | ↓ | Transcription,
proliferation, and DNA damage repair |
| 33 |
Microtubule-associated
serine/threonine-protein kinase 4 | MAST4_HUMAN | 286426 | 8.85 | 50 | 162 | 64 | ↑ | ATP binding,
Mg2+ ion binding, nucleotide binding, protein
binding |
| 34 | DNA topoisomerase
2α | TOP2A_HUMAN | 175017 | 8.82 | 48 | 89 | 72 | ↓ | Cell cycle
progression |
| 36 | Poly(ADP-ribose)
polymerase 14 | PAR14_HUMAN | 204725 | 6.81 | 57 | 121 | 73 | ↓ | Promotes survival
and anti-apoptotic role |
Functional classification of
identified proteins
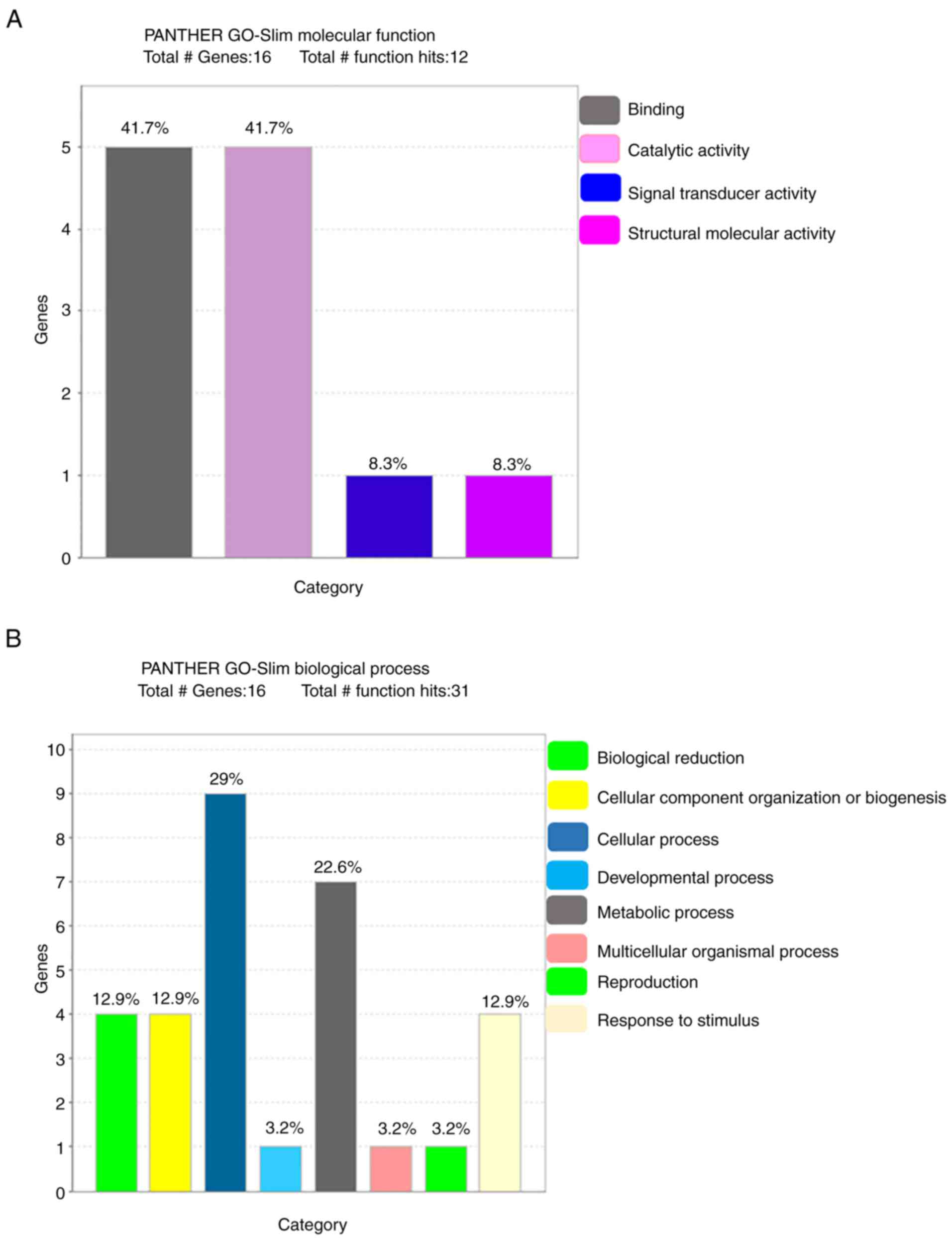
The PANTHER database was used to analyze the 16
identified proteins in terms of molecular function, biological
process, cellular component, protein class and pathway (Fig. 3). The most common molecular
functions were binding protein (41.7%), catalytic activity (41.7%),
signal transducer activity (8.3%) and structural molecular activity
(8.3%). The major biological processes were cellular process (29%),
metabolic process (22.6%), biological regulation (12.9%),
biogenesis (12.9%), stimulus (12.9%), developmental process (3.2%),
multicellular organismal process (3.2%) and reproduction (3.2%).
The cell component carries cell part (46.7%), organelle (26.7%),
macromolecular complex (20.0%) and cell junction (6.7%). Regarding
the protein class, the most common were transferase (26.7%),
transfer/carrier protein (20.0%), calcium-binding protein (13.3%),
hydrolase (13.3%), chaperone (6.7%), enzyme modulator (6.7%),
cytoskeleton protein (6.7%) and receptor (6.7%).
PANTHER classification identified 21 pathways with
signaling mechanisms that are involved in the effect of tangeretin
on AGS cancer cells (Fig. 4). Among
the 16 derived proteins, PKCε (encoded by KPCE) is the major
protein leading the cellular signaling mechanism in the obtained
pathways (Table II).
 | Table II.Signaling pathways and the regulating
genes in tangeretin-treated AGS cells using the Protein ANalysis
THrough Evolutionary Relationships (PANTHER) database. |
Table II.
Signaling pathways and the regulating
genes in tangeretin-treated AGS cells using the Protein ANalysis
THrough Evolutionary Relationships (PANTHER) database.
| No. | Pathway | Symbol of regulated
gene |
|---|
| 1 | 5-Hydroxytryptamine
type 2 receptor-mediated signaling pathway | KPCE |
| 2 | α-adrenergic
receptor signaling pathway | KPCE |
| 3 | Alzheimer's
disease-amyloid secretase pathway | MK04, KPCE |
| 4 | Angiogenesis | KPCE |
| 5 | Apoptosis signaling
pathway | KPCE |
| 6 | Cholecystokinin
receptor signaling map | KPCE |
| 7 | Cytoskeletal
regulation by Rho GTPase | MYH13 |
| 8 | DNA
replication | TOP2A |
| 9 | Epidermal growth
factor receptor signaling pathway | KPCE |
| 10 | Endothelin
signaling pathway | KPCE |
| 11 | Fibroblast growth
factor signaling pathway | KPCE |
| 12 | Gonadotropin
releasing hormone receptor pathway | KPCE |
| 13 | Heterotrimeric
G-protein signaling pathway: Gqα and
Goα-mediated pathway | KPCE |
| 14 | Histamine
H1 receptor-mediated signaling pathway | KPCE |
| 15 | Inflammation
mediated by chemokine and cytokine signaling pathway | KPCE, MYH13 |
| 16 | Muscarinic
acetylcholine receptor 1 and 3 signaling pathway | KPCE |
| 17 | Nicotinic
acetylcholine receptor 1 and 3 signaling pathway | MYH13 |
| 18 | Oxytocin
receptor-mediated signaling pathway | KPCE |
| 19 |
Thyrotropin-releasing hormone receptor
signaling pathway | KPCE |
| 20 | Vascular
endothelial growth factor signaling pathway | KPCE |
| 21 | Wnt signaling
pathway | KPCE, MYH13 |
In order to obtain maximum comparable classification
between the obtained proteins, DAVID enrichment analysis was
performed, which identified four clustering annotation groups with
medium classification stringency (Fig.
5). Markedly associated genes in functional annotations were
identified on the basis of threshold count ≥2 and EASE <0.1. It
has been observed that the majority of genes are associated with
cell-cell adhesion in the first cluster with enrichment score 1.96
and in the second cluster of nucleotide binding and kinase activity
with enrichment score 1.89. The majority of genes are associated
with cell adhesion and junction, nucleotide and ATP binding,
transferase and kinase activity, nucleus and transcription
regulation, cell and plasma membrane.
Interaction between protein
complexes
The selected genes were interrogated using the
STRING database for the protein-protein interaction network
analysis between the upregulated and downregulated proteins. STRING
generated an interconnected protein network with a medium
confidence level 0.04, developed three signaling modules including
two database-predicted nodes or genes following clustering using
the MCL. These three modules included PKCε protein regulating MAPK4
and PI4KA; apolipoprotein (APOB) interacting with transferrin
transfer protein; and chaperonin containing T-complex 1 (TCP1)
(CCT)8 gene interacting with STRING-predicted TCP1 subunits CCT2,
CCT3, CCT4 and CCT5, those again interact directly with DNA
topoisomerase IIα (TOP2A) and chromodomain helicase DNA-binding
protein 3 (CHD3) (Fig. 6).
Validation of selected proteins by
western blot analysis
Among the various protein spots identified using
MALDI-TOF-MS, four proteins, namely MAPK4, PI4K, PARP14 and PKCε,
were selected on the basis of MASCOT analysis, PANTHER and DAVID
database tool. These protein expressions were confirmed further by
western blot analysis. As presented in Fig. 7, the expression of MAPK4, PI4K,
PARP14 and PKCε in the presence of tangeretin was significantly
decreased compared with the untreated condition, thereby confirming
the involvement of these proteins from the differentially expressed
2DE analysis.
Discussion
Tangeretin is a polymethoxylated flavonoid with
anti-proliferative, anti-invasive, anti-metastasis and anticancer
properties (4,6,8). In
the present study, for the first time, proteomic analysis of AGS
gastric cancer cells treated with tangeretin was performed.
Proteomic analysis data indicated differential expression of 36
spots representing 16 different proteins which were identified
using MASCOT search engine analysis. PANTHER and DAVID
bioinformatics tools were used to identify the functional
properties of these differentially expressed proteins. Exploring
the protein-protein interaction networks may suggest novel
directions for future experimental research and provide
cross-species predictions for efficient interaction mapping. The
identified proteins represent several biological functions already
known in different cancer studies. However, attention towards
differential proteome expression analysis on gastric cancer using
flavonoids for the identification of functional biomarkers has not
been investigated to any great extent.
PANTHER is part of the Gene Ontology Phylogenetic
Annotation Project. PANTHER classification categories involved
marked proteins enriched with protein binding; microtubule motor,
kinase and hydrolase activity. Tight control of cell proliferation
and morphogenesis in conjunction with programmed cell death
(apoptosis) is required to ensure normal tissue or cell patterning.
Imbalance in the cellular signal mechanism, promotes cell
proliferation, suppress apoptosis and enhance cell invasion.
Receptor protein binding such as with tyrosine kinases serves a
detrimental function in cancer cell development, with alterations
in the receptor tyrosine kinase potentially leading to the creation
of potent oncogenes (14).
Intermediate filaments and microtubule cytoskeleton protein binding
are the key functions that underpin cellular processes such as
disruption of cellular morphogenesis, inappropriate migration and
invasion, and genome instability by accompanying the progression of
disease (15). Studies have
revealed that G-proteins regulate a number of cellular functions
including cytoskeletal rearrangement, cell motility, intracellular
trafficking, transcriptional regulation, cell viability and
development (16). Microtubule
motor proteins regulate kinesin superfamily members and support a
number of cellular functions, including mitosis, meiosis and the
transport of macromolecules (17).
Kinases are important mediators of the signaling cascade and
oncogenic activation in cancer cells may be blocked by selective
kinase inhibitors (18).
Calcium-binding protein or calmodulin serves a major function in
eukaryote cell signaling, including cell proliferation, programmed
cell death and autophagy (19).
Similarly, in the present study, receptor binding protein MAPK4,
cytoskeleton microtubule actin cross-linking factor 1 (20) and microtubule-associated protein 6
are downregulated. In addition cell cycle regulator TOP2A;
intracellular cytoplasmic protein PI4KA, chaperone protein TCP1
subunit θ (TCPQ), kinase activity protein PKCε, PI4KA, cell
adhesion junction protein LM07, nuclear protein PARP14,
transcriptional repressor chromobox protein homolog 8 (21), DNA and RNA binding CHD3, G protein
and metalloprotease cytosolic carboxypeptidase 3 and transferrin
protein serrotransferrin (22) were
downregulated, whereas microtubule-associated
serine/threonine-protein kinase 4 and APOB were upregulated.
PKCε was identified to be major linking protein in
tangeretin-induced AGS cell death (23). Overexpression of PKCε has been
reported in a number of types of cancer. In glioma, skin carcinoma
and breast cancer, an increased level of PKCε induces cell invasion
and/or metastasis (24–26). Targeting PKCε is considered to be a
promising therapeutic method for cancer treatment. PKCε belongs to
the PKC family of proteins, considered to be key signaling
molecules in cellular functions (25,26).
Apoptosis, cell migration, proliferation, motility, chemoresistance
and differentiation are examples of the cellular processes
regulated by PKCs (27). PKCε
activates the Ras signaling cascade, which in turn leads to
activation of cyclin D1 promoter, thus promoting cell survival and
viability (28). PKCε has an
anti-apoptotic function as activation of PKCε activates
anti-apoptotic proteins B-cell lymphoma (Bcl) 2 proteins and
simultaneously suppressed the pro-apoptotic protein Bcl homology
3-interacting death agonist, thus inhibiting cell apoptosis
(29,30). It was also identified that
expression of PKCε is associated with resistance to chemotherapy in
prostate and breast carcinoma (25,31).
In the present study, it was observed that tangeretin
dose-dependently inhibited AGS cell viability. Furthermore, in the
proteomic analysis of AGS cells treated with tangeretin, it was
identified that PKCε expression was significantly downregulated.
This result was further confirmed by western blot analysis.
Furthermore, the PANTHER database also revealed that PKCε was the
major protein involved in apoptosis and angiogenesis pathway
(Table II). As PKCε has been
identified to be an anti-apoptotic protein, the result that
tangeretin treatment downregulated PKCε validates that tangeretin
induces apoptosis of AGS cells through the PKCε signaling
cascade.
PARP14 belongs to the PARP family of proteins.
Previous studies have identified a protective function for PARP14
in lymphocytes against apoptosis and in hepatoma cells in
vitro and in vivo (32,33).
PARP14 acts as a downstream protein of the c-Jun N-terminal kinase
(JNK) signaling pathway, thus further leading to cell survival in
cancer cells. Barbarulo et al (34) concluded that JNK2 promotes myeloma
cell survival via PARP14 which in turn inhibits JNK1, resulting in
suppression of apoptosis in myeloma cells. In the present study, it
was observed that tangeretin significantly inhibits PARP14 in
tangeretin-treated AGS cells. The downregulation of PARP14
expression was further confirmed by western blot analysis. Thus, a
significant decrease in PARP14 in tangeretin-treated AGS cells
validated the inhibition of AGS cell viability and further
induction of apoptosis.
MAPK4 belongs to the atypical MAPK family of
proteins. Unlike the classical MAPK family proteins, the conserved
T-X-Y motif is replaced by an S-E-G motif in MAPK4 (35). Mitogen-activated protein
kinase-activated protein kinase 5 (MK5) is one of the best
characterized downstream substrates of MAPK4. Overexpression of
MAPK4 typically leads to activation of MK5 (36). MK5 is known to serve a function in
tumor initiation and development. A function of the MAPK4/MK5
signaling pathway in insulin-like growth factor 2-binding protein
1-induced tumor cell migration has previously been identified
(37). Suppression of MAPK4, being
an upstream target of MK5, may inhibit tumor initiation and
development. In the present study, MAPK4 expression was
significantly suppressed in tangeretin-treated AGS cells,
indicating its function as an antitumor marker.
Typically, cancer cells exhibit increased expression
of PI4K. PI4K has been identified as a substrate for
phosphoinositide 3-kinase and for producing secondary messengers
(38). Knockdown of PI4K inhibits
cell proliferation and induces apoptosis in breast cancer cells
(39). It has been observed that
PI4K acts as a mediator of resistance to cisplatin, thus inhibiting
apoptosis in cancer cells (40).
Thus, suppressing PI4K inhibits cell proliferation and induces
apoptosis. Similarly, in the present study, it was observed that
tangeretin inhibited PI4KA in AGS cancer cells, indicating the
involvement of oncogenic protein degradation.
The results of the present study demonstrated that
tangeretin-treated human AGS cancer cells exhibit decreased
viability and induction of cell death. Furthermore, proteomic
changes in the cellular response towards tangeretin treatment in
AGS cells have been identified. Differently expressed proteins
identified functional genes that have been altered and
significantly decreased. PKCε, MAPK4, PI4K and PARP14 proteins
promote cell survival, tumor growth or development and suppression
of apoptosis. The results of the present study demonstrated that
tangeretin-induced cell death is regulated by the KPCE gene.
Targeting KPCE may be a promising therapeutic marker in treating
gastric cancer, thus tangeretin may be a useful therapeutic drug in
gastric cancer treatment.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Research Foundation of Korea funded by Ministry of Science Industry
and Technology (grant nos. 2012M3A9B8019303 and
2017R1A2B4003974).
Availability of data and materials
All the data generated or analyzed during this study
are included in this published article.
Authors' contributions
SY, EHK, WSL and GSK conceived and designed the
experiments. SY, SR and SMK performed the experiments and analyzed
the data. VVGS, HJL, SEH, JDH, SJL and JAK contributed with
reagents and data interpretation. SY, SR, JDH, SJL and JAK
contributed in drafting and writing the manuscript. GSK, EHK and
WSL drafted and critically reviewed the manuscript for important
intellectual content. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nagini S: Carcinoma of the stomach: A
review of epidemiology, pathogenesis, molecular genetics and
chemoprevention. World J Gastrointest Oncol. 4:156–169. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Luo C, Du Z, Wei X, Chen G and Fu Z:
Bisdemethoxycurcumin attenuates gastric adenocarcinoma growth by
inducing mitochondrial dysfunction. Oncol Lett. 9:270–274. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ishii K, Tanaka S, Kagami K, Henmi K,
Toyoda H, Kaise T and Hirano T: Effects of naturally occurring
polymethyoxyflavonoids on cell growth, p-glycoprotein function,
cell cycle, and apoptosis of daunorubicin-resistant T
lymphoblastoid leukemia cells. Cancer Invest. 28:220–229. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Manthey JA and Guthrie N:
Antiproliferative activities of citrus flavonoids against six human
cancer cell lines. J Agric Food Chem. 50:5837–5843. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Conesa Martinez C, Ortega Vicente V,
Gascón Yáñez MJ, Baños Alcaraz M, Canteras Jordana M,
Benavente-García O and Castillo J: Treatment of metastatic melanoma
B16F10 by the flavonoids tangeretin, rutin, and diosmin. J Agric
Food Chem. 53:6791–6797. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Morley KL, Ferguson PJ and Koropatnick J:
Tangeretin and nobiletin induce G1 cell cycle arrest but not
apoptosis in human breast and colon cancer cells. Cancer Lett.
251:168–178. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pan MH, Chen WJ, Lin-Shiau SY, Ho CT and
Lin JK: Tangeretin induces cell-cycle G1 arrest through inhibiting
cyclin-dependent kinases 2 and 4 activities as well as elevating
Cdk inhibitors p21 and p27 in human colorectal carcinoma cells.
Carcinogenesis. 23:1677–1684. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dong Y, Cao A, Shi J, Yin P, Wang L, Ji G,
Xie J and Wu D: Tangeretin, a citrus polymethoxyflavonoid, induces
apoptosis of human gastric cancer AGS cells through extrinsic and
intrinsic signaling pathways. Oncol Rep. 31:1788–1794. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hong ML, Jiang N, Gopinath S and Chew FT:
Proteomics technology and therapeutics. Clin Exp Pharmacol Physiol.
33:563–568. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Grover A, Shandilya A, Bisaria VS and
Sundar D: Probing the anticancer mechanism of prospective herbal
drug Withaferin A on mammals: A case study on human and bovine
proteasomes. BMC Genomics. 11 Suppl 4:S152010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lu Z, Song Q, Yang J, Zhao X, Zhang X,
Yang P and Kang J: Comparative proteomic analysis of anti-cancer
mechanism by periplocin treatment in lung cancer cells. Cell
Physiol Biochem. 33:859–868. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shevchenko A, Wilm M, Vorm O and Mann M:
Mass spectrometric sequencing of proteins silver-stained
polyacrylamide gels. Anal Chem. 68:850–858. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39:D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sangwan V and Park M: Receptor tyrosine
kinases: Role in cancer progression. Curr Oncol. 13:191–193.
2006.PubMed/NCBI
|
|
15
|
Hall A: The cytoskeleton and cancer.
Cancer Metastasis Rev. 28:5–14. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Takai Y, Sasaki T and Matozaki T: Small
GTP-binding proteins. Physiol Rev. 81:153–208. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu Y and Feng YM: The role of kinesin
family proteins in tumorigenesis and progression: Potential
biomarkers and molecular targets for cancer therapy. Cancer.
116:5150–5160. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Paul MK and Mukhopadhyay AK: Tyrosine
kinase-Role and significance in cancer. Int J Med Sci. 1:101–115.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Berchtold MW and Villalobo A: The many
faces of calmodulin in cell proliferation, programmed cell death,
autophagy, and cancer. Biochim Biophys Acta. 1843:398–435. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Miao Z, Ali A, Hu L, Zhao F, Yin C, Chen
C, Yang T and Qian A: Microtubule actin cross-linking factor 1, a
novel potential target in cancer. Cancer Sci. 108:1953–1958. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang G, Tang J, Zhan W, Zhang R, Zhang M,
Liao D, Wang X, Wu Y and Kang T: CBX8 suppresses tumor metastasis
via repressing snail in esophageal squamous cell carcinoma.
Theranostics. 7:3478–3488. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
de Resende MF, Vieira S, Chinen LT,
Chiappelli F, da Fonseca FP, Guimarães GC, Soares FA, Neves I,
Pagotty S, Pellionisz PA, et al: Prognostication of prostate cancer
based on TOP2A protein and gene assessment: TOP2A in prostate
cancer. J Transl Med. 11:362013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Byun S, Lee KW, Jung SK, Lee EJ, Hwang MK,
Lim SH, Bode AM, Lee HJ and Dong Z: Luteolin inhibits protein
kinase C(epsilon) and c-Src activities and UVB-induced skin cancer.
Cancer Res. 70:2415–2423. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pal D, Outram SP and Basu A: Upregulation
of PKCη by PKCε and PDK1 involves two distinct mechanisms and
promotes breast cancer cell survival. Biochim Biophys Acta.
1830:4040–4045. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pan Q, Bao LW, Kleer CG, Sabel MS,
Griffith KA, Teknos TN and Merajver SD: Protein kinase C epsilon is
a predictive biomarker of aggressive breast cancer and a validated
target for RNA interference anticancer therapy. Cancer Res.
65:8366–8371. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Selzer E, Okamoto I, Lucas T, Kodym R,
Pehamberger H and Jansen B: Protein kinase C isoforms in normal and
transformed cells of the melanocytic lineage. Melanoma Res.
12:201–209. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gutcher I, Webb PR and Anderson NG: The
isoform-specific regulation of apoptosis by protein kinase C. Cell
Mol Life Sci. 60:1061–1070. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kampfer S, Windegger M, Hochholdinger F,
Schwaiger W, Pestell RG, Baier G, Grunicke HH and Uberall F:
Protein kinase C isoforms involved in the transcriptional
activation of cyclin D1 by transforming Ha-Ras. J Biol Chem.
276:42834–42842. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ding L, Wang H, Lang W and Xiao L: Protein
kinase C-epsilon promotes survival of lung cancer cells by
suppressing apoptosis through dysregulation of the mitochondrial
caspase pathway. J Biol Chem. 277:35305–35313. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
McJilton MA, Van Sikes C, Wescott GG, Wu
D, Foreman TL, Gregory CW, Weidner DA, Ford Harris O, Lasater
Morgan A, Mohler JL, et al: Protein kinase Cepsilon interacts with
Bax and promotes survival of human prostate cancer cells. Oncogene.
22:7958–7968. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu D, Foreman TL, Gregory CW, McJilton MA,
Wescott GG, Ford OH, Alvey RF, Mohler JL and Terrian DM: Protein
kinase cepsilon has the potential to advance the recurrence of
human prostate cancer. Cancer Res. 62:2423–2429. 2002.PubMed/NCBI
|
|
32
|
Cho SH, Ahn AK, Bhargava P, Lee CH,
Eischen CM, McGuinness O and Boothby M: Glycolytic rate and
lymphomagenesis depend on PARP14, an ADP ribosyltransferase of the
B aggressive lymphoma (BAL) family. Proc Natl Acad Sci USA.
108:15972–15977. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Iansante V, Choy PM, Fung SW, Liu Y, Chai
JG, Dyson J, Del Rio A, D'Santos C, Williams R, Chokshi S, et al:
PARP14 promotes the Warburg effect in hepatocellular carcinoma by
inhibiting JNK1-dependent PKM2 phosphorylation and activation. Nat
Commun. 6:78822015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Barbarulo A, Iansante V, Chaidos A, Naresh
K, Rahemtulla A, Franzoso G, Karadimitris A, Haskard DO, Papa S and
Bubici C: Poly(ADP-ribose) polymerase family member 14 (PARP14) is
a novel effector of the JNK2-dependent pro-survival signal in
multiple myeloma. Oncogene. 32:4231–4242. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Aberg E, Perander M, Johansen B, Julien C,
Meloche S, Keyse SM and Seternes OM: Regulation of MAPK-activated
protein kinase 5 activity and subcellular localization by the
atypical MAPK ERK4/MAPK4. J Biol Chem. 281:35499–35510. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Perander M, Al-Mahdi R, Jensen TC, Nunn
JA, Kildalsen H, Johansen B, Gabrielsen M, Keyse SM and Seternes
OM: Regulation of atypical MAP kinases ERK3 and ERK4 by the
phosphatase DUSP2. Sci Rep. 7:434712017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Stöhr N, Köhn M, Lederer M, Glass M,
Reinke C, Singer RH and Hüttelmaier S: IGF2BP1 promotes cell
migration by regulating MK5 and PTEN signaling. Genes Dev.
26:176–189. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Balla A and Balla T: Phosphatidylinositol
4-kinases: Old enzymes with emerging functions. Trends Cell Biol.
16:351–361. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chu KM, Minogue S, Hsuan JJ and Waugh MG:
Differential effects of the phosphatidylinositol 4-kinases, PI4KIIα
and PI4KIIIβ, on Akt activation and apoptosis. Cell Death Dis.
1:e1062010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Waugh MG: Phosphatidylinositol 4-kinases,
phosphatidylinositol 4-phosphate and cancer. Cancer Lett.
325:125–131. 2012. View Article : Google Scholar : PubMed/NCBI
|