Introduction
Renal cancer is among the top ten malignant tumors
in the world, with 400,000 newly diagnosed cases annually (1). Renal cell carcinoma (RCC) is a
malignant tumor that originates in the renal tubular epithelium,
accounting for >90% of renal cancer cases (2) and its incidence has gradually
increased in recent years (3).
Although great strides have been made on medical techniques where
local RCC can be surgically resected with a five-year overall
survival of 76% (4), health
management of metastatic RCC is quite challenging in clinical
practice (5). The occurrence and
development mechanism of RCC is closely linked to the epigenetics,
tumor heterogeneity and carcinogenesis, immune infiltration and
microenvironment (6), with ~3% of
cases having a family history (7).
A number of genes perform an abnormal expression in RCC, such as
the von Hippel-Lindau (VHL), which is often inactivated in sporadic
RCC by mutation or promoter hypermethylation (8), and BAP-1, which has important
implications for utilization of molecular testing, prognosis,
future therapeutics and distinguishing clear cell (cc)RCC from
other RCC (8,9). Therefore, early diagnosis and active
development of effective therapeutic targets have great
significance to the research and clinical diagnosis and treatment
of RCC.
lncRNAs are noncoding RNAs >200 nucleotides long,
serving as key regulators in the physiological and pathological
processes of human beings (10).
Increasing amounts of evidence has shown that abnormally expressed
lncRNAs in RCC are crucially involved in cancer development and
progression (11–14). lncRNA FTX is upregulated in
adenocarcinoma and gastric cancer, indicating that lncRNA FTX is an
oncogene (15,16). He et al (17) report that lncRNA FTX is upregulated
in RCC as an oncogene and its knockdown inhibits the migratory and
invasive capacities of RCC cells. However, the specific molecular
mechanism of lncRNA FTX in the malignant progression of RCC remains
to be elucidated.
Ubiquitin conjugating enzyme E2 C (UBE2C) is a
component of the ubiquitin proteasome system. As an essential
factor of the cell cycle regulatory E3, human anaphase promoting
complex/cyclosome (APC/C), UBE2C participates in the ubiquitination
modification of cyclins and mitosis-related factors, thereby
accelerating cell mitosis (18).
Previous studies have shown that the mRNA or protein level of UBE2C
is abnormally upregulated in a number of types of cancers with poor
clinical outcomes, including liver cancer, lung cancer, gastric
cancer, colon cancer and cervical cancer (19). UBE2C is a vital gene involved in
the development of RCC and is significant to mediate the
proliferation and migration of RCC cells (20). However, the molecular mechanism
between lncRNA FTX and UBE2C in regulating the development of RCC
remains to be elucidated.
In the present study, by detecting the expression
levels of lncRNA FTX in the clinical specimens of RCC, it was found
that FTX was significantly upregulated. Furthermore, lncRNA FTX was
predicted using the StarBase to exert the miRNA sponge effect on
miR-4429 to upregulate UBE2C. The present study further analyzed
the in vitro and in vivo functions of lncRNA FTX in
RCC by intervening the expression levels of lncRNA FTX, miR-4429
and UBE2C in RCC cells and establishing a xenograft model of RCC in
nude mice to explore the potential signaling and mechanisms of
lncRNA FTX/miR-4429/UBE2C axis.
Materials and methods
Sample collection
A total of six pairs of RCC and paracancerous
specimens that were ≤4 cm away from the margin of RCC lesions were
collected from patients with RCC (male to female ratio: 2:1) who
were surgically treated in the First Affiliated Hospital of Gannan
Medical University. All were patients with RCC hospitalized in the
First Affiliated Hospital of Gannan Medical University hospital
between March and June 2020 and aged 48–60 years-old, with the
median as 53. Renal cancer tissues and adjacent tissues were
removed after surgical treatment without radiotherapy or
chemotherapy. The fresh tissues were immediately stored in liquid
nitrogen and stored in a refrigerator at −80°C after removal and
then used for reverse transcription-quantitative (RT-q) PCR,
western blotting or immunohistochemistry (IHC). The specimens were
fixed in 4% paraformaldehyde for 24 h at a room temperature and
dehydrated and transparentized with gradient concentrations (30,
50, 75, 95 and 100%) of ethanol and xylene, and then embedded in
paraffin at 55°C to prepare 4-µm tissue sections. and some fresh
specimens were subjected to the isolation of total RNA and proteins
for RT-qPCR and western blotting, respectively. The present study
was approved by Ethics Review Committee of Scientific Research of
Gannan Medical College (number 2020075). All patients who
participated were informed of the content of the study and their
written informed consent was obtained.
Cell culture
RCC cell lines, including A498 (cat. no. BNCC338630;
BeNa Culture Collection), A704 (cat. no. BNCC342393; BeNa Culture
Collection), SN12C (cat. no. BNCC341858; BeNa Culture Collection),
769-P (cat. no. BNCC101643; BeNa Culture Collection), the human
renal fibroblast cell line KFB (cat. no. HTX2520; Otwo Biotech),
human proximal convoluted tubular cell line HK-2 (cat. no.
BNCC338012; BeNa Culture Collection) and the 293T cell line (cat.
no. MZ-0005; Ningbo Mingzhou Biotechnology Co., Ltd.) were used.
Briefly, the cells were seeded in a 10-mm culture dish and
cultivated in DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.)
and 1% penicillin and streptomycin in a humidified incubator
containing 5% CO2 and 95% air at 37°C. The culture
medium was replaced every 2 days. Cell passage was conducted using
trypsin at over 80% of confluence.
Cell transfection
miR-4429-mimics (5′-AAAAGCUGGGCUGAGAGGCG-3′) and
mimic-negative control (NC; 5′-AAGUGUCACCGAUUCAAGACG-3′; RiboBio
Co., Ltd., Guangzhou, China) and shUBE2C-1, shUBE2C-2, shUBE2C-3
and the empty vector was used as a shNC (Shanghai GeneChem Co.,
Ltd.) were used in cell transfection performed following the
recommendations of Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). Briefly, the cells
were seeded in a 12-well plate at a density of 5×104
cells/well and cultured to 80% of confluence. After 4-h starvation
in serum-free DMEM (Gibco; Thermo Fisher Scientific), 100 nM
miR-4429-mimics, mimic-NC or 2 µg shUBE2C was incubated with
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) for 20 min, followed by incubation with cells for
40 min at a room temperature. Fresh serum-containing DMEM (Gibco;
Thermo Fisher Scientific) was replaced at 24 h and the transfection
efficacy was evaluated by RT-qPCR.
RT-qPCR
The cells were cultured at 80% of confluence for RNA
extraction. RNA extraction, cDNA synthesis and qPCR were all
performed according to the manufacturer's protocols. Briefly, the
total RNAs in cells were isolated using TRIzol® (Thermo
Fisher Scientific, Inc.) and 1 µg of the total RNA was reversely
transcribed to cDNA using a Takara PrimeScript RT reagent kit.
Subsequently, qPCR was performed using a SYBR Premix Ex Taq II with
Tli RNaseH (Takara Bio, Inc.) on an ABI Prism 7500 system (Thermo
Fisher Scientific, Inc.). In brief, the total RNA induced with gDNA
eraser and reversely transcribed to cDNA in 42°C for 15 min and
then subjected to thermal cycles at 95°C for 15 min, followed by 40
cycles at 95°C for 5 sec, 60°C for 30 sec and 72°C for 40 sec and
finally annealing at 72°C for 10 min. The relative level was
calculated using the 2−ΔΔCq method with GAPDH or U6 as
the internal reference (21). All
the experiments were performed in triplicate and repeated three
times. The primer sequences are shown in Table I.
 | Table I.Sequences of primers used. |
Table I.
Sequences of primers used.
| Genes | Sequences |
|---|
| lncRNA FTX-F |
5′-GTGTCTCTCTCTCTCTCTCTCTT-3′ |
| lncRNA FTX-R |
5′-CCTCTTCAGCAGTAGCATAGTT-3′ |
| miR-4429-F |
5′-GGGAGAAAAGCTGGGCTGAG-3′ |
| miR-4429-R |
5′-GGCCAGGCAGTCTGAGTTG-3′ |
| UBE2C-F |
5′-CTGCCGAGCTCTGGAAAAAC-3′ |
| UBE2C-R |
5′-AGGAAAAATTAAAAAGACGACACAAG-3′ |
| Akt-F |
5′-CCTGAAGCTACTGGGCAAGGG-3′ |
| Akt-R |
5′-ACAAAGCAGAGGCGGTCGTG-3′ |
| CDK1-F |
5′-CCTAGCATCCCATGTCAAAAACTTGG-3′ |
| CDK1-R |
5′-TGATTCAGTGCCATTTTGCCAGA-3′ |
| CDK6-F |
5′-TGCACAGTGTCACGAACAGA-3′ |
| CDK6-R |
5′-ACCTCGGAGAAGCTGAAACA-3′ |
| GAPDH-F |
5′-AGAAGGCTGGGGCTCATTTG-3′ |
| GAPDH-R |
5′-AGGGGCCATCCACAGTCTTC-3′ |
| U6-F |
5′-CTCGCTTCGGCAGCACA-3′ |
| U6-R |
5′-AACGCTTCACGAATTTGCGT-3′ |
Western blotting
The total proteins in RCC cells or homogenate
tissues were isolated using RIPA buffer (Beyotime Institute of
Biotechnology) containing PMSF and a protease inhibitor. Protein
concentrations were measured using a BCA protein assay kit
(Beyotime Institute of Biotechnology). Later, 50 µg of protein
sample was loaded on 10% SDS-PAGE and transferred on PVDF membranes
at 300 mA. Nonspecific antigens on PVDF membranes were blocked by
immersing in TBST (1% Tween) containing 5% skimmed milk in room
temperature for 1 h. After immunoblotting with primary antibodies
(1:1,000) at 4°C overnight and secondary antibodies (1:1,000) at
room temperature for 1 h. Finally, the membrane was treated with
chemiluminescent horse radish peroxidase (HRP) substrate (cat. no.
WBKLS0500; MilliporeSigma) to visualize the protein bands and
exposure was performed using a Bio-Rad Universal Hood II Gel Doc
Imaging system (Bio-Rad Laboratories, Inc.) and the grey value was
analyzed using ImageJ 1.8.0.112 (National Institutes of Health).
The antibodies used in this study were purchased from ABclonal
Biotech Co., Ltd. and the catalog numbers were: UBE2C (cat. no.
A5499), AKT (cat. no. A17909), phosphorylated (p)-AKT (cat. no.
AP0140), CDK1 (cat. no. A0220), p-CDK1 (cat. no. AP0016), CDK6
(cat. no. A16357), p-CDK1 (cat. no. AP0326), GAPDH (cat. no.
A19056) and the secondary antibody HRP Rabbit Anti-Goat IgG (H+L)
(cat. no. AS029).
Hematoxylin & Eosin (H&E)
staining
RCC specimens were fixed in 4% paraformaldehyde for
24 h at room temperature and dehydrated and transparentized with
gradient concentrations (30, 50, 75, 95 and 100%) of ethanol and
xylene, and then embedded in paraffin at 55°C to prepare 4-µm
tissue sections for preparing tissue sections. Later, the sections
were incubated in xylene (20 min ×2), anhydrous ethanol (5 min ×2)
and 75% ethanol (5 min ×1). After washing in ddH2O, the
sections were stained using the H&E staining kit (Sangon
Biotech Co., Ltd.; cat. no. E607318). Briefly, the sections were
stained with hematoxylin for 5 min. After differentiation and
bluing, the sections were washed in ddH2O, dehydrated in
85 and 5% ethanol sequentially and stained with eosin for 5 min.
All the staining was carried out in a room temperature. Finally,
they were incubated in anhydrous ethanol for three times, xylene
twice and mounted using neutral gum for observation under a light
microscope (XSP-8CA; Shanghai Optical Instrument Co. Ltd.), five
fields of view were randomly selected for each tissue section and
observed at ×100 magnification.
Immunohistochemistry (IHC)
The RCC sections were incubated in xylene (15 min
×3), dehydrated in anhydrous ethanol (5 min ×2), 85% ethanol (5 min
×1) and 75% ethanol (5 min ×1) and washed in ddH2O.
Antigen retrieval was performed by pouring citrate buffer (pH 6.0;
Sangon Biotech Co., Ltd.; cat. no. E673000) on the sections at room
temperature for 15 min. After incubation in 3%
H2O2 in the dark at 37°C for 25 min and
blocking in 3% BSA (Sangon Biotech Co., Ltd.; cat. no. E661003) at
37°C for 30 min, the sections were incubated with primary
antibodies at 4°C overnight. On the next day, they were washed in
PBS (pH 7.4, 5 min ×3) and incubated with HRP-labeled secondary
antibodies at room temperature for 2 h. Later, the sections were
counterstained with DAB and hematoxylin at room temperature for 3
min. After dehydration in ddH2O, 75% ethanol (5 min ×1),
85% ethanol (5 min ×1), anhydrous ethanol (5 min ×2) and
n-butanol (5 min ×1) sequentially and permeabilization in
xylene (5 min ×1) at room temperature, neutral gum was used for
mounting. The positive staining of cells was observed under a light
microscope (XSP-8CA; Shanghai Optical Instrument Co. Ltd.), five
fields of view were randomly selected for each tissue section and
observed at ×100 magnification. The antibodies used in this
experiment were purchased from ABclonal Biotech Co., Ltd. and the
catalog numbers were: UBE2C (cat. no. A5499) and the secondary
antibody (cat. no. AS029).
Lentivirus infection
The GV367 plasmid (Shanghai GeneChem Co., Ltd.) was
cut by AgeI/NheI and then the FTX sequence or short
hairpin (sh)FTX, shUBE2C1, shUBE2C2, shUBE2C3 sequences were
spliced to produce overexpressed lentivirus plasmids GV367-FTX and
knocked down plasmids GV367-shFTX (Shanghai GeneChem Co., Ltd.),
while the non-spliced GV367 was used as a blank control (sequences
were as listed: shFTX: 5′-CTGCTACGACACTGAATTC-3′; shUBE2C1:
5′-CCTGCAAGAAACCTACTCAAA-3′; shUBE2C2: 5′-CTTCTAGGAGAACCCAACA-3′;
shUBE2C3: 5′-TGATGTCAGGACCATTCT-3′; and the shNC:
5′-AATTCTCCGAACGTGTCACGT-3′.). Lentivirus packaging was performed
using the second-generation lentivirus packaging kit (GeneChem) at
37°C 15 min. The lentiviral plasmid, packaging vector and envelope
vector were mixed at a 4:3:2 ratio for a total DNA mass of 20 µg
and incubated with 1 ml of Lenti-Easy Packaging Mix (Shanghai
GeneChem Co., Ltd.) at 37°C for 15 min. The mixture was then
incubated for another 20 min in Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) and applied to a 293T
cell culture medium for 6 h at 37°C. 293T cells were seeded in a
12-well plate at a density of 2.5×105 cells/well and
cultured to 80% of confluence. Following the incubation in
serum-free DMEM for 4 h, the cells were transfected as mentioned
above with lentiviruses at 37°C for 3 days. The transfected cells
were filtered using a 0.45 µM mesh, which were concentrated by
ultracentrifugation at 70,000 × g at 4°C for 2 h. The supernatant
was collected for detecting viral titers. The A498 and A704 cell
lines were cultured to over 80% of confluence were cultured with
diluted lentiviruses at a multiplicity of infection of 5 and with
polybrene (MilliporeSigma) for 24 h and then fresh culture medium
was then used to replace the old medium and the GFP-labeled cells
with lentivirus transfection rate >80% at 72 h were screened
out, stable expressing clones were selected using 4 µg/ml
puromycin. The transfection efficacy of lncRNA FTX was finally
verified by RT-qPCR as described above.
Measurement of intracellular reactive
oxygen species (ROS)
To measure intracellular ROS, cells were stained
with DCF-DA and subjected to FACS analysis. Cells were plated in
6-well plates at 1.2-2.0×105 cells/well depending on the
proliferation rate of the cell line. After 24 h of incubation, the
cells were treated with vehicle (DMSO) or varying concentrations of
CDODA-Me (0.5, 1, 2.5, or 5 µM) at 37°C for 3 h and
CDODA-Me-induced intracellular ROS were estimated by adding 10 µM
DCF-DA.
Inhibition of ROS inductions by GSH was determined
by pretreatment with 5 mM GSH for 3 h alone and then in combination
with 2.5 µM CDODA-Me at 37°C for 3 h. Cells were then incubated
with 10 µM DCF-DA at 37°C for 30 min (at 2 h and 30 min to 3 h
treatment time point) at 37°C. Attached and floating cells were
then harvested by trypsinization and pelleted by centrifugation at
3,500 × g at 4°C for 5 min. The cells were then resuspended in
ice-cold PBS and DCF-DA fluorescence was detected using the FL1
channel of an FC500 flow cytometer (FACSCalibur; BD Biosciences).
All experiments were performed in triplicate; data were analyzed
using Cell Quest software version (Molecular Devices, LLC, version
5.1) and were shown as means ± SEMs.
Colony formation assay
Cells were seeded in a 6-well plate at a density of
5×102 cells/well and cultured for 15 days. Visible
colonies were fixed in 4% paraformaldehyde and stained using 0.2%
crystal violet at 37°C. The number of colonies (>50 cells) in
three replicates per sample was calculated. Images of colonies were
captured under an inverted microscope.
MTT assay
The cells were seeded in a 96-well plate at a
density of 3×103 cells/well in 200 µl of DMEM. Briefly,
15 µl of MTT solution (15 mg/ml, Sangon Biotech Co., Ltd.; cat. no.
A600799) was added at 6, 12, 18, 24, 30 and 36 h for 4-h cell
culture at 37°C. After lysing in 150 µl of DMSO for 10 min at room
temperature, the optical density at 490 nm was measured using an
HBS-1101 microplate reader (DeTie).
Flow cytometry for detecting cell
cycle progression
The cell cycle progression in RCC cells was measured
using a Cell Cycle Staining kit (Sangon Biotech Co., Ltd.; cat. no.
E607212). Briefly, the cells were washed in PBS, fixed in 70%
ice-cold ethanol and incubated using a commercial kit in the dark
at room temperature for 30 min. Cell cycle progression was measured
using FC500 flow cytometer (FACSCalibur; BD Biosciences) and
analyzed using FlowJo v10 (FlowJo LLC).
Wound healing assay
The A498 and A704 cells were seeded in a 12-well
plate at a density of 1×105 cells/well. A 1 ml sterile
pipette tip was used to make an artificial wound on a cell
monolayer and then FBS-free DMEM was replaced. At 0, 12 and 24 h,
cell migration was observed using an inverted microscope and
assessed by calculating the cell-free zone using ImageJ 1.8.0.112
software (National Institutes of Health, Bethesda).
Transwell assay
Matrigel (Corning Life Sciences; cat. no. 354234)
diluted in serum-free DMEM at a ratio of 1:5 was precoated on the
Transwell insert at 37°C and dried for 4 h. Cell suspension was
prepared in serum-free DMEM at 1×105 cells/ml and 200 µl
of suspension was added on the top chamber and 500 µl of DMEM
containing 10% FBS was added on the bottom. After cell culture for
24 h, the cells on the top chamber were wiped off using a cotton
swab. The penetrating cells were fixed in 4% paraformaldehyde at
room temperature for 20 min and stained using crystal violet at
room temperature for 10 min. After washing and air drying, the
cells were observed under a light microscope (XSP-8CA; Shanghai
Optical Instrument Co. Ltd.) and the migratory cells were
calculated in five randomly selected fields per well.
Dual-luciferase reporter assay
The targeting relationship was predicted by StarBase
3.0 (https://starbase.sysu.edu.cn/) and
TargetScan 7.1 (https://www.targetscan.org/vert_71/). Complementary
sequences in the UBE2C 3′UTR and promoter region of lncRNA FTX were
amplified and cloned in pGL3 3′UTR (Shanghai GeneChem Co., Ltd.) to
construct the wild-type (WT) plasmid pGL3-UBE2C-WT, while the
mutant-type (Mut) plasmid pGL3-UBE2C-Mut was constructed using a
site-directed mutagenesis kit (Sangon Biotech Co., Ltd.; cat. no.
B639281). WT plasmid pGL3-FTX-WT and Mut plasmid pGL3-FTX-Mut were
similarly constructed. The aforementioned vectors were mixed with
the mimics or mimic-NC and Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) for 20 min at room
temperature. Then, the cells were cotransfected with
wild-type/mutant-type plasmid and miR-4429-mimics/mimics-NC,
respectively, at 37°C in a humidified atmosphere containing 5% CO2
for 48 h, the plasmids was transfected at 500 ng per well and the
final concentration of mimic or mimic-NC was 20 nM. Relative
Firefly and Renilla luciferase activities were measured
using a Dual-Luciferase Reporter Gene Assay kit (Promega
Corporation). Relative luciferase activity was expressed as the
ratio of firefly luciferase activity to Renilla luciferase
activity.
Xenograft model of RCC in nude
mice
A total of 18 BALB/c male nude mice (5–6 weeks old;
~15 g; Guangdong Medical Laboratory Animal Center) were habituated
in a specific pathogen-free (SPF) environment with 12-h light/dark
cycle, at 22±2°C and in a humidity 30–70%. They had free access to
food and water. A suspension of RCC cells in PBS (1×108
cells/ml) was prepared. Prior to inoculation, the cell suspension
was mixed with Matrigel (Corning Life Sciences; cat. no. 354234) at
a ratio of 1:1 and then 100 µl of mixture was subcutaneously
injected into the mouse lateral abdomen. At four weeks later, the
mice were sacrificed to collect the subcutaneous tumors. When the
mice quickly lost >20% of their original body weight, could not
eat and drink, exhibited mental depression with hypothermia,
experienced body organ infection or severe liver failure, the
experiment was halted and the animals sacrificed. On day 30, a
total of 18 mice were sacrificed and no mice succumbed during the
experiment. The mice were sacrificed by intraperitoneal injection
of sodium pentobarbital with an injection dose of 200 mg/kg.
According to AVMA Guidelines for the Euthanasia of Animals: 2020
Edition, the death was confirmed by checking that their hearts had
stopped beating for 10 min and the respiratory arrest was also
observed. Their size was measured and they were weighed and
prepared for the following experiments. The experiments were
approved by Ethics Review Committee of Scientific Research of
Gannan Medical College (approval number 2020075).
Statistical analyses
Statistical analysis was performed using GraphPad
Prism 8 (GraphPad Software, Inc.). Data were normally distributed
and are presented as the mean ± SEM. Comparisons between multiple
groups were analyzed using one-way ANOVA followed by Tukey's post
hoc test or Bonferroni correction. All experiments were performed
in triplicate and repeated three times. P<0.05 was considered to
indicate a statistically significant difference.
Results
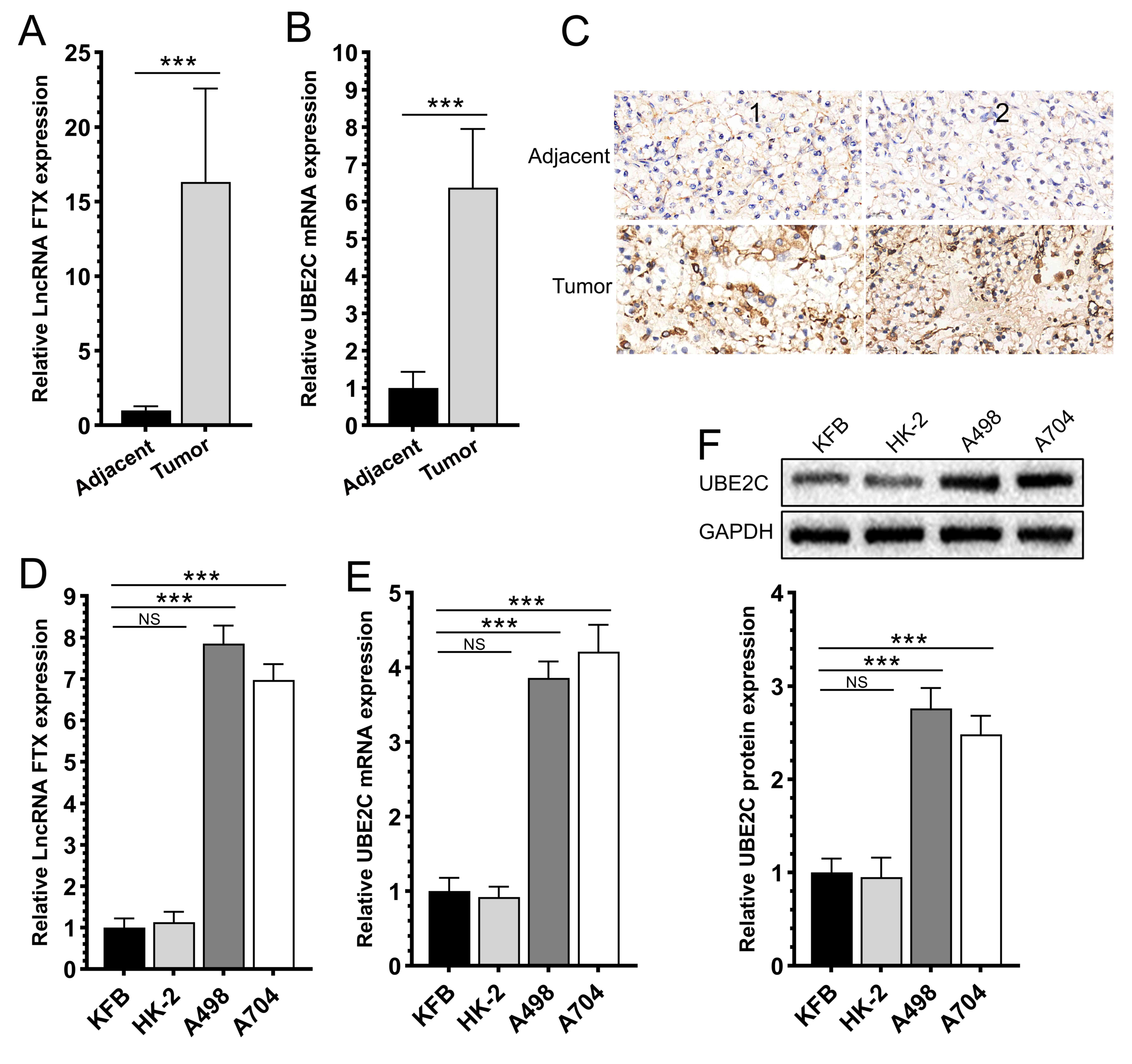
lncRNA FTX and UBE2C are upregulated
in RCC specimens and cell lines
A total of six pairs of RCC and paracancerous
specimens were examined for the expression levels of lncRNA FTX and
UBE2C. RT-qPCR data showed that lncRNA FTX and UBE2C mRNA
expression were significantly upregulated in RCC specimens than
that of paracancerous specimens (Fig.
1A and B). In addition, IHC data showed a significantly higher
positive expression of UBE2C in RCC specimens than paracancerous
specimens (Fig. 1C). Their
expression levels were further detected in RCC cell lines.
Consistently, the mRNA level of lncRNA FTX and UBE2C was
significantly higher in RCC cell lines compared with those of human
renal fibroblast cell line KFB and human proximal convoluted
tubular cell line HK-2 (Fig. 1D and
E). The protein level of UBE2C was also upregulated in RCC cell
lines (Fig. 1F). Collectively,
lncRNA FTX and UBE2C were significantly upregulated in RCC
specimens and cell lines, indicating their potential roles in the
development of RCC.
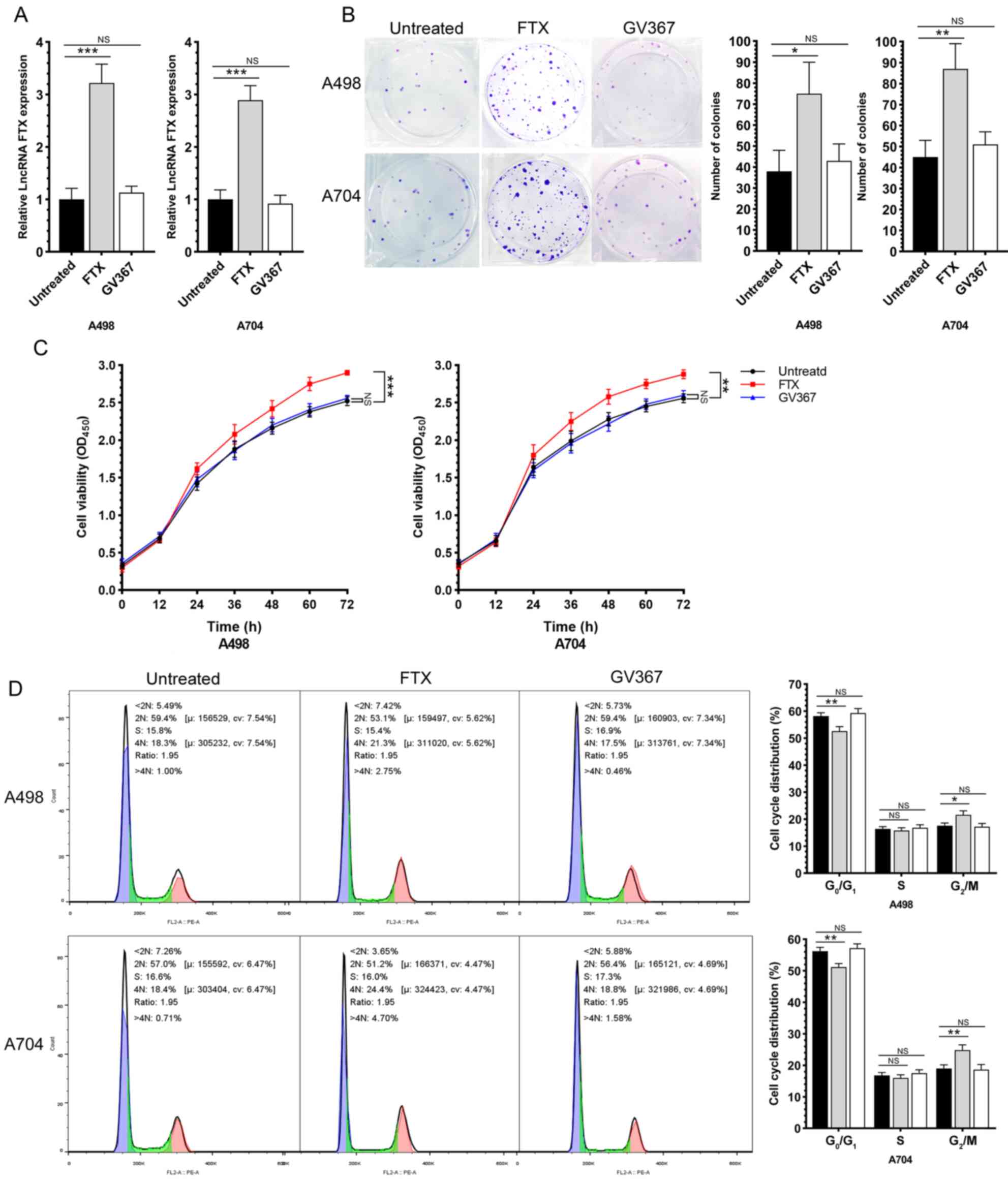
lncRNA FTX promotes the proliferation,
viability, cell cycle progression, migration and invasion of RCC
cells
After intervening lncRNA FTX by lentivirus
transfection, the malignant phenotypes of RCC cells were evaluated.
First, the transfection efficacy of lentivirus was validated by
RT-qPCR (Fig. 2A). A colony
formation assay proved that the overexpression of lncRNA FTX
significantly enhanced the proliferative capacity of RCC cells
(Fig. 2B). In addition, the
overexpression of lncRNA FTX increased the RCC cell viability as
revealed by the MTT assay (Fig.
2C). Compared with those of controls, the cell ratio in
G0/G1 phase was significantly reduced, while
that in G2/M phases increased, in RCC cells
overexpressing lncRNA FTX, indicating that the overexpression of
lncRNA FTX significantly accelerated the cell cycle progression of
RCC (Fig. 2D). Wound healing assay
and Transwell assay results showed the role of lncRNA FTX in
promoting the migratory and invasive capacities (Fig. 3A and B). While the intracellular
ROS test showed that there are no significantly differences among
the groups that treated by lentivirus (Fig. 3C). These data indicated that lncRNA
FTX served as an oncogene that promoted the proliferation,
viability, cell cycle progression, migration and invasion of RCC
cells.
lncRNA FTX exerts the miRNA sponge
effect on miR-4429 to upregulate UBE2C
As predicted using the online tools StarBase 3.0 and
TargetScan 7.1, a binding site exists between lncRNA FTX and
miR-4429 and another binding site exists between miR-4429 and UBE2C
3′UTR (Fig. 4A). A dual-luciferase
reporter assay validated that lncRNA FTX could specifically bind to
miR-4429 and the latter could specifically bind to its downstream
target UBE2C (Fig. 4B and C). The
overexpression of lncRNA FTX significantly downregulated miR-4429
and upregulated UBE2C in RCC cells (Fig. 4D-G). Notably, the transfection of
miR-4429-mimics could significantly downregulate the expression of
UBE2C and effectively reverse the effects on UBE2C of overexpressed
lncRNA FTX (Fig. 4H-J). The
results showed that lncRNA FTX could bind with miR-4429 and
miR-4429 could directly bind with the 3′-UTR sequence of UBE2C and
the above results suggested that there is a signaling lncRNA
FTX/mIR-4429/UBE2C in RCC.
Knockdown of UBE2C effectively
reverses the role of lncRNA FTX in promoting the malignant
phenotype of RCC cells
shUBE2C-1, shUBE2C-2, shUBE2C-3 and shNC were
synthesized and their transfection efficacies were detected.
RT-qPCR and western blotting confirmed the best transfection
efficacy of shUBE2C-2 (Fig. 5A and
B). Subsequently, the RCC cells were co-transfected with
si-UBE2C-2 and overexpression plasmid of lncRNA FTX. A series of
functional experiments further indicated that the knockdown of
UBE2C effectively reversed the enhanced proliferative, migratory
and invasive capacities, viability and accelerated the cell cycle
progression in RCC cells overexpressing lncRNA FTX (Figs. 5C-E and 6A and B). In addition, the expression of
lncRNA FTX, miR-4429, UBE2C and the upregulated phosphorylation
levels of AKT, CDK1 and CDK6 by the overexpression of lncRNA FTX
were downregulated by the knockdown of UBE2C (Fig. 7A and B). It was suggested that
lncRNA FTX promoted the progression of RCC by exerting the miRNA
sponge effect on miR-4429, thus upregulating UBE2C.
lncRNA FTX significantly affects the
size and differentiation of subcutaneous RCC in nude mice
A-498 cells with lncRNA FTX overexpression or
knockdown by lentivirus transfection were first subjected to
RT-qPCR for validating the transfection efficacy (Fig. 8A). The transfected A-498 cells
cultured to 80% confluence were subcutaneously injected into nude
mice. The mice were sacrificed at day 30 for collecting tumor
tissues. Compared with those of controls, the subcutaneous tumor
tissues were larger and heavier in mice implanted with A-498 cells
overexpressing lncRNA FTX, while those with lncRNA FTX knockdown
had smaller and lighter subcutaneous tumor tissues (Fig. 8B and C). Tumor tissues were then
prepared for sections and isolation of the total RNA and protein.
It was found that the mRNA levels of lncRNA FTX and UBE2C were
upregulated and miR-4429 was downregulated in subcutaneous tumor
tissues collected from mice implanted with A-498 cells
overexpressing lncRNA FTX. The mRNA and protein levels of UBE2C
were upregulated. In mice implanted with A-498 cells with lncRNA
FTX knockdown, downregulated UBE2C was detected in subcutaneous
tissues (Fig. 8D-H). It was
concluded that the in vivo overexpression of lncRNA FTX
significantly influenced the expression level of UBE2C and the
phosphorylation of AKT and CDK6 in tumor tissues, accelerating the
carcinogenesis by implanting A-498 cells in nude mice.
Discussion
RCC is a globally prevalent type of cancer. More
than 140,000 deaths of RCC are reported every year, ranking 13th
among the cancer mortality in the world (22). Although the early nonmetastatic RCC
can be surgically treated, the recurrence rate of RCC is ≤40%.
Metastatic RCC has a poor prognosis and low survival rate (23). Novel biomarkers for RCC
microenvironment and immunotherapy have been widely analyzed.
Targeted drugs including sunitinib (24) and bevacizumab (25) that inhibit VEGF and its receptor
VEGFR, sirolimus (26) that
inhibits mTOR and nivolumab (27)
used in immune therapies have been used for the treatment of
metastatic RCC. It is important to actively search for new
biomarkers and therapeutic targets of RCC.
lncRNA FTX has been extensively evaluated as a vital
regulator of cancer cells. Chen et al (28) report that FTX is abnormally
overexpressed in osteosarcoma (OS) tissues and cells that promote
the proliferation and invasion and inhibits the apoptosis of OS
cells by regulating the miR-214-5p/SOX4 axis, indicating that FTX
might be a potential therapeutic target for OS. Zhao et al
(29) show that lncRNA FTX
promotes the progression of colorectal cancer (CRC) by regulating
the miR-192-5p/EIF5A2 axis as an oncogene, providing new insights
into the pathogenesis of CRC. Jin et al (30) reveal a novel ceRNA axis
FTX/miR-200a-3p/FOXA2 in lung cancer cells, validating the
therapeutic potential of FTX in lung cancer. In addition, FTX is
upregulated in liver cancer, RCC, glioma and hematological cancer
(31) and the regulatory effects
of FTX on the invasion and migration of RCC cells has been
confirmed (17). The present study
consistently showed the presence of upregulated lncRNA FTX in RCC
specimens. In addition, it showed that the overexpression of lncRNA
FTX promoted the proliferation, cell cycle progression, migration
and invasion of RCC cells and upregulated UBE2C, p-AKT, p-CDK1 and
p-CDK6. In addition, the in vivo overexpression of lncRNA
FTX in RCC cells significantly enlarged the size and increased the
weight of subcutaneous tissues in nude mice. Taken together, the
findings demonstrated the vital function of lncRNA FTX in
aggravating the progression of RCC.
miRs have been previously confirmed to regulate the
expression levels of RCC-related proteins, which serve as effective
biomarkers for diagnosing, accurately determining the molecular
classification and predicting the prognosis of RCC (32). miR-4429 is widely involved in the
progression of various types of tumors. Wang et al (33) found that miR-4429 inhibits the
growth of prostate cancer cells by downregulating DLX1 and
inactivating the Wnt/β-catenin pathway. Li et al (34) demonstrated that miR-4429 blocks the
proliferation, migration, invasion and epithelial-mesenchymal
transition (EMT) process of CRC cells by targeting FOXM1 to
downregulate SMAD3. Cai et al (35) confirm that the silencing of lncRNA
SNHG12 and overexpression of miR-4429 significantly inhibit the
proliferation, migration and invasion of RL95-2 cells by
downregulating MMP2 and MMP9 and targeting of the SNHG12/miR-4429
axis and can be used as a potential therapeutic target for
endometrial cancer in the future. Pan et al (36) also report that miR-4429 suppresses
ccRCC tumor progression and EMT by targeting CDK6. The present
study predicted and later confirmed that lncRNA FTX specifically
targeted miR-4429 and the latter could specifically bind to UBE2C
3′UTR using an online tool and dual-luciferase reporter assay,
respectively. lncRNA FTX overexpression was able to downregulate
miR-4429 and enhance the transcription level of UBE2C. In addition,
the overexpression of miR-4429 significantly reversed the promotive
effect of lncRNA FTX on the progression of RCC and expression level
of UBE2C. It is suggested that the regulatory function of lncRNA
FTX in the malignant progression of RCC depends on the miRNA sponge
effect on miR-4429 to upregulate UBE2C.
UBE2C has been widely studied in cancers due to its
carcinogenic effect. Wang et al (37) found that UBE2C is highly expressed
in gastric cancer tissues. Silencing of UBE2C not only inhibits the
colony formation of gastric cancer cells, but also DNA
biosynthesis. In addition, miR-300 inhibits the progression of
gastric cancer by reducing the abundance of UBE2C mRNA. Hu et
al (38) report that the
knockdown of UBE2C inhibits the proliferation and invasion of
prostate cancer cells, which is a direct target of miR-381-3p.
Icaritin downregulates UBE2C and upregulates miR-381-3p in human
prostate cancer cells. Zhang et al (39) confirm that UBE2C is upregulated in
rectal cancer and that the knockdown of UBE2C significantly
inhibits the growth of xenograft tumor in mice. Previous studies
support the oncogenic role of UBE2C in RCC, although the specific
mechanism remains to be elucidated (20,39).
The present study also identified upregulated UBE2C in RCC
specimens. Notably, the knockdown of UBE2C effectively reversed the
promoted proliferation, viability, cell cycle progression,
migration and invasion induced by the overexpression of lncRNA FTX,
as well as upregulating p-AKT, p-CDK1 and p-CDK6. It is suggested
that UBE2C was responsible for aggravating the malignant
progression of RCC through the FTX/miR-4429/UBE2C axis. In
addition, implantation of A-498 cells overexpressing lncRNA FTX in
nude mice resulted in enlarged subcutaneous tissues with heavier
tumor weight compared with those of controls. lncRNA FTX also
regulated UBE2C, p-AKT, p-CDK1 and p-CDK6 in subcutaneous tissues.
Taken together, the in vivo findings indicate that lncRNA
FTX significantly influenced the UBE2C level and phosphorylation of
AKT, CDK1 and CDK6 and promoted the carcinogenesis of implanting
A-498 cells in nude mice. The effects of lncRNA FTX in promoting
the malignant phenotype of RCC could be significantly reversed by
knockdown of UBE2C, which indicated that the axis lncRNA
FTX/miR-4429/UBE2C could significantly influence the proliferation,
viability, cell cycle progression, migration and invasion of RCC
cells and hence are promising biomarkers and therapeutic targets
for RCC.
In general, the present study examined the
abnormally high expression of lncRNA FTX in RCC, its promoting
effects and mechanism on RCC malignant phenotype. The results
showed that lncRNA FTX could significantly promote the cell
viability, proliferation, migration and invasion of RCC. lncRNA FTX
acted as a molecular sponge of miR-4429 and promoted the process of
RCC through the FTX/mir-4429/UBE2C axis. However, due to the
limited number of the clinical samples, the correlation of clinical
data could not be well demonstrated, which might be the main
limitation of the present study. In addition, the present study did
not test the expression level of apoptosis-related genes, which
would be another limitation and a future research perspective.
In brief, lncRNA FTX and UBE2C are significantly
upregulated in RCC specimens. Through the miRNA sponge effect on
miR-4429 to upregulate UBE2C, lncRNA FTX markedly promoted the
proliferation, viability, cell cycle progression, migration and
invasion of RCC cells, thus aggravating the malignant progression
of RCC. The findings indicated that lncRNA FTX and UBE2C are
promising biomarkers and therapeutic targets for RCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by Natural Science Foundation of
Jiangxi Province (grant nos. 20192BAB205075 and 20202BABL206119),
Scientific research project of Gannan Medical University (grant no.
ZD201830) and Youth project of Jiangxi Provincial Department of
Education (grant no. GJJ201544).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZC, and LW made substantial contributions to the
conception or design of the work and agreed to be accountable for
all aspects of the work in ensuring that questions related to the
accuracy and integrated of any part of the work are appropriately
investigated and resolved. ZC, MZ and YuL performed the
experiments. TD, ZL and LW performed the acquisition, analysis, and
interpretation of data. TD, YaL and ZZ drafted the manuscript and
revised it critically for important intellectual content; ZC and LW
confirm the authenticity of all the raw data; all the authors
approved the final version to be published.
Ethics approval and consent to
participate
This research was approved by the Ethics Review
Committee of Scientific Research of Gannan Medical College
(approval number 2020075).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Padala SA and Barsouk A, Thandra KC,
Saginala K, Mohammed A, Vakiti A, Rawla P and Barsouk A:
Epidemiology of renal cell carcinoma. World J Oncol. 11:79–87.
2020. View Article : Google Scholar
|
|
2
|
Shen H, Luo G and Chen Q: Long noncoding
RNAs as tumorigenic factors and therapeutic targets for renal cell
carcinoma. Cancer Cell Int. 21:1102021. View Article : Google Scholar
|
|
3
|
Sun M, Thuret R, Abdollah F, Lughezzani G,
Schmitges J, Tian Z, Shariat SF, Montorsi F, Patard JJ, Perrotte P
and Karakiewicz PI: Age-adjusted incidence, mortality, and survival
rates of stage-specific renal cell carcinoma in North America: A
trend analysis. Eur Urol. 59:135–141. 2011. View Article : Google Scholar
|
|
4
|
Weyerer V, Strissel PL, Stohr C, Eckstein
M, Wach S, Taubert H, Brandl L, Geppert CI, Wullich B, Cynis H, et
al: Endogenous Retroviral-K envelope is a novel tumor antigen and
prognostic indicator of renal cell carcinoma. Front Oncol.
11:6571872021. View Article : Google Scholar
|
|
5
|
Tacconi EMC, Tuthill M and Protheroe A:
Review of adjuvant therapies in renal cell carcinoma: Evidence to
date. Onco Targets Ther. 13:12301–12316. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hsieh JJ, Purdue MP, Signoretti S, Swanton
C, Albiges L, Schmidinger M, Heng DY, Larkin J and Ficarra V: Renal
cell carcinoma. Nat Rev Dis Primers. 3:170092017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Maher ER: Hereditary renal cell carcinoma
syndromes: Diagnosis, surveillance and management. World J Urol.
36:1891–1898. 2018. View Article : Google Scholar
|
|
8
|
Gallan AJ, Parilla M, Segal J, Ritterhouse
L and Antic T: BAP1-Mutated clear cell renal cell carcinoma. Am J
Clin Pathol. 155:718–728. 2021. View Article : Google Scholar
|
|
9
|
Kim BJ, Kim JH, Kim HS and Zang DY:
Prognostic and predictive value of VHL gene alteration in renal
cell carcinoma: A meta-analysis and review. Oncotarget.
8:13979–13985. 2017. View Article : Google Scholar
|
|
10
|
Ciomborowska-Basheer J, Staszak K, Kubiak
MR and Makalowska I: Not so dead genes-retrocopies as regulators of
their disease-related progenitors and hosts. Cells. 10:9122021.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Q, Tian Y, Hu G, Liang Y, Bai W and Li
H: Highly expressed antisense noncoding RNA in the INK4 locus
promotes growth and invasion of renal clear carcinoma cells via the
β-catenin pathway. Oncol Res. 25:1373–1382. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang G, Li H and Hou Y: LncRNA MAGI2-AS3
inhibits tumor progression and angiogenesis by regulating ACY1 via
interacting with transcription factor HEY1 in clear cell renal cell
carcinoma. Cancer Gene Ther. 29:585–596. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cheng T, Shuang W, Ye D, Zhang W, Yang Z,
Fang W, Xu H, Gu M, Xu W and Guan C: SNHG16 promotes cell
proliferation and inhibits cell apoptosis via regulation of the
miR-1303-p/STARD9 axis in clear cell renal cell carcinoma. Cell
Signal. 84:1100132021. View Article : Google Scholar
|
|
14
|
Zhang J, Jin S, Xiao W, Zhu X, Jia C and
Lin Z: Long noncoding RNA LINC00641 promotes renal cell carcinoma
progression via sponging microRNA-340-5p. Cancer Cell Int.
21:2102021. View Article : Google Scholar
|
|
15
|
Zhang F, Wang XS, Tang B, Li PA, Wen Y and
Yu PW: Long non-coding RNA FTX promotes gastric cancer progression
by targeting miR-215. Eur Rev Med Pharmacol Sci. 24:3037–3048.
2020.
|
|
16
|
Huo X and Wang H, Huo B, Wang L, Yang K,
Wang J, Wang L and Wang H: FTX contributes to cell proliferation
and migration in lung adenocarcinoma via targeting miR-335-5p/NUCB2
axis. Cancer Cell Int. 20:892020. View Article : Google Scholar
|
|
17
|
He X, Sun F, Guo F, Wang K, Gao Y, Feng Y,
Song B, Li W and Li Y: Knockdown of long noncoding RNA FTX inhibits
proliferation, migration, and invasion in renal cell carcinoma
cells. Oncol Res. 25:157–166. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hao Z, Zhang H and Cowell J:
Ubiquitin-conjugating enzyme UBE2C: Molecular biology, role in
tumorigenesis, and potential as a biomarker. Tumour Biol.
33:723–730. 2012. View Article : Google Scholar
|
|
19
|
Xie C, Powell C, Yao M, Wu J and Dong Q:
Ubiquitin-conjugating enzyme E2C: A potential cancer biomarker. Int
J Biochem Cell Biol. 47:113–117. 2014. View Article : Google Scholar
|
|
20
|
Chen Z and Wang L: The clinical
significance of <em>UBE2C</em> gene in progression of
renal cell carcinoma. Eur J Histochem. 65:31962021. View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Capitanio U, Bensalah K, Bex A, Boorjian
SA, Bray F, Coleman J, Gore JL, Sun M, Wood C and Russo P:
Epidemiology of renal cell carcinoma. Eur Urol. 75:74–84. 2019.
View Article : Google Scholar
|
|
23
|
Larroquette M, Peyraud F, Domblides C,
Lefort F, Bernhard JC, Ravaud A and Gross-Goupil M: Adjuvant
therapy in renal cell carcinoma: Current knowledges and future
perspectives. Cancer Treat Rev. 97:1022072021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Motzer RJ, Hutson TE, Tomczak P,
Michaelson MD, Bukowski RM, Rixe O, Oudard S, Negrier S, Szczylik
C, Kim ST, et al: Sunitinib versus interferon alfa in metastatic
renal-cell carcinoma. N Engl J Med. 356:115–124. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Escudier B, Pluzanska A, Koralewski P,
Ravaud A, Bracarda S, Szczylik C, Chevreau C, Filipek M, Melichar
B, Bajetta E, et al: Bevacizumab plus interferon alfa-2a for
treatment of metastatic renal cell carcinoma: A randomised,
double-blind phase III trial. Lancet. 370:2103–2111. 2007.
View Article : Google Scholar
|
|
26
|
Yang S, de Souza P, Alemao E and Purvis J:
Quality of life in patients with advanced renal cell carcinoma
treated with temsirolimus or interferon-alpha. Br J Cancer.
102:1456–1460. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Motzer RJ, Escudier B, McDermott DF, Arén
Frontera O, Melichar B, Powles T, Donskov F, Plimack ER, Barthélémy
P, Hammers HJ, et al: Survival outcomes and independent response
assessment with nivolumab plus ipilimumab versus sunitinib in
patients with advanced renal cell carcinoma: 42-month follow-up of
a randomized phase 3 clinical trial. J Immunother Cancer.
8:e0008912020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen H, Liu T, Ouyang H, Lin S, Zhong H,
Zhang H and Yang Y: Upregulation of FTX promotes osteosarcoma
tumorigenesis by increasing SOX4 expression via miR-214-5p. Onco
Targets Ther. 13:7125–7136. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhao K, Ye Z, Li Y, Li C, Yang X, Chen Q
and Xing C: LncRNA FTX contributes to the progression of colorectal
cancer through regulating miR-192-5p/EIF5A2 axis. Onco Targets
Ther. 13:2677–2688. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jin S, He J, Zhou Y, Wu D, Li J and Gao W:
LncRNA FTX activates FOXA2 expression to inhibit non-small-cell
lung cancer proliferation and metastasis. J Cell Mol Med.
24:4839–4849. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lin Y, Sheng Y, Chen J, Hu C, Zhou Z and
Yuan C: The function of LncRNA FTX in several common cancers. Curr
Pharm Des. 27:2381–2386. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tsiakanikas P, Giaginis C, Kontos CK and
Scorilas A: Clinical utility of microRNAs in renal cell carcinoma:
Current evidence and future perspectives. Expert Rev Mol Diagn.
18:981–991. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang J, Xie S, Liu J, Li T, Wang W and Xie
Z: MicroRNA-4429 suppresses proliferation of prostate cancer cells
by targeting distal-less homeobox 1 and inactivating the
Wnt/β-catenin pathway. BMC Urol. 21:402021. View Article : Google Scholar
|
|
34
|
Li H, Liang W, Zhang H, Shui Y and Zhang
Z: MicroRNA-4429 restrains colorectal cancer cell invasion and
migration via regulating SMAD3-induced epithelial-mesenchymal
transition. J Cell Physiol. 236:5875–5884. 2021. View Article : Google Scholar
|
|
35
|
Cai P, Wu M, Zhang B, Wu S, Wei H and Wei
L: Long noncoding RNA SNHG12 regulates cell proliferation, invasion
and migration in endometrial cancer by targeting miR4429. Mol Med
Rep. 22:2842–2850. 2020.PubMed/NCBI
|
|
36
|
Pan H, Hong Y, Yu B, Li L and Zhang X:
miR-4429 inhibits tumor progression and epithelial-mesenchymal
transition via targeting CDK6 in clear cell renal cell carcinoma.
Cancer Biother Radiopharm. 34:334–341. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang Y, Huang F, Liu M and Zhao Q: UBE2C
mRNA expression controlled by miR-300 and HuR determines its
oncogenic role in gastric cancer. Biochem Biophys Res Commun.
534:597–603. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hu J, Wu X, Yang C, Rashid K, Ma C, Hu M,
Ding Q and Jiang H: Anticancer effect of icaritin on prostate
cancer via regulating miR-381-3p and its target gene UBE2C. Cancer
Med. 8:7833–7845. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang Y, Tian S, Li X, Ji Y, Wang Z and
Liu C: UBE2C promotes rectal carcinoma via miR-381. Cancer Biol
Ther. 19:230–238. 2018. View Article : Google Scholar : PubMed/NCBI
|