Introduction
Breast cancer (BC) is the most commonly diagnosed
cancer among female patients worldwide and accounted for 666,103
cancer-associated deaths in 2022 (1). BC encompasses multiple heterogeneous
subgroups at the molecular, histopathological and clinical levels
(2). BC comprises three major tumor
subtypes categorized according to presence or absence of estrogen
receptor (ER) and progesterone receptor (PR) expression and/or
amplification of human epidermal growth factor receptor 2 (HER2)
expression. Hormone receptor (HR)-positive/HER2-negative,
HER2-positive, and triple-negative (TN) BC (which lacks all three
standard molecular markers) represent 70, 15–20 and 15% of BC
cases, respectively (3). Surgery
and radiation therapy are considered local treatment, whereas
systemic treatment includes chemotherapy and hormone and targeted
therapy. Recent advancements, including use of adjunct
immunotherapy, support the use of a multimodal approach to improve
patient prognosis (4).
Current standards of treatment for BC depend on
tumor subtype, anatomical cancer staging and patient preference.
Hormone therapeutic agents are primary systemic therapy for ER- and
PR-positive BC (3,5). HER2-targeted therapy using monoclonal
antibodies, antibody-drug conjugates or tyrosine kinase inhibitors
is designed for HER2-positive BC, a subtype typically associated
with poor prognosis and chemotherapy resistance (3,6). The
introduction of anti-HER2 therapy has led to notable improvements
in survival of patients with HER2-positive BC in both early and
advanced stages (6).
However, acquisition of resistance to targeted drugs
is almost unavoidable, emphasizing the importance of biochemical
and pharmaceutical advances to improve treatment outcomes (7,8).
Moreover, crosstalk between HER2 and ER signaling pathways may
contribute to endocrine resistance (9,10). As
a result, exploring mechanisms underlying trastuzumab resistance
and developing innovative therapy are key for design and
implementation of precision medicine for BC.
Epithelial membrane proteins (EMPs) belong to the
growth arrest-specific 3 gene family and serve a key role in cell
migration, proliferation and differentiation (11). The 4-transmembrane glycoprotein
EMP3, an EMP family member, has received increased attention in
recent years because of its potential role in pathogenesis of human
cancer (11–14). The levels of EMP3 mRNA in BC are
significantly higher in cancer than in non-tumor tissue (15). EMP3 expression is associated with
the invasive phenotype of mammary carcinoma cell lines (16). Knocking down EMP3 in SK-BR-3 cells
inhibits tumor growth in vitro, suggesting that EMP3 may
function as an oncogene in human BC (17). In addition, EMP3 is upregulated in
association with epithelial-mesenchymal transition (EMT) (18) and is a signature EMT gene in
vitro (19). All six cancer
stem cell (CSC)-like BC cell lines are the ‘basal B’ subtype and
have high EMP3 expression (20).
However, contradictory results have reported that EMP3 serves as a
tumor suppressor via the negative regulation of DNA replication and
damage repair and stem-like properties in BC (21).
Further support for the role of EMP3 in the
pathogenesis of BC comes from the observation that EMP3 is
significantly upregulated in microarray profiling of HER2
overexpression in immortalized luminal human mammary epithelial
cells (22). Expression of EMP3 has
a dose-dependent association with HER2 status in vitro
(23). Overexpression of EMP3 in
primary BC is positively associated with HER2 protein expression,
high histological grade and nodal metastasis (15,16).
There is also functional crosstalk between HER2 and EMP3 in
vitro (24). Co-expression of
EMP3 and HER2 is significantly associated with poor disease- and
metastasis-free survival of patients with urothelial carcinoma of
the upper urinary tract. However, EMP3 positive cases presented
significantly better prognosis in HR+HER2+ BC (25), highlighting the need to clarify the
clinical role of EMP3 overexpression in BC.
The present study was designed to address the
potential role of EMP3 in the pathogenesis of BC and its
implications as a novel therapeutic target. In vitro
experiments using forward and reverse transfection approaches were
performed on cell lines. A SCID mouse model was created to examine
the impact of targeting EMP3 on BC in vivo. Finally,
expression of EMP3 protein was surveyed in a BC cohort to assess
its association with HER2 status and clinical outcome.
Materials and methods
Bioinformatics analysis
The potential use of EMP3 for the treatment of human
BC was analyzed in The Human Protein Atlas
(proteinatlas.org/ENSG00000142227-EMP3/cancer/breast+
cancer#BRCA_TCGA), cBioPortal (cbioportal.org/results/comparison?cancer_study_list=brca_tcga_gdc&Z_SCORE_THRESHOLD=2.0&RPPA_SCORE_THRESHOLD=2.0&profileFilter=mrna_seq_tpm_Zscores&case_set_id=brca_tcga_gdc_all&gene_list=EMP3&geneset_list=%20&tab_index=tab_visualize&Action=Submit&comparison_selectedGroups=%5B%22Unaltered%20group%22%2C%22Unprofiled%20group%22%2C%22EMP3%22%2C%22Altered%20group%22%5D&comparison_subtab=survival&comparison_overlapStrategy=Exclude&comparison_groupOrder=%5B%22Unprofiled%20group%22%2C%22EMP3%22%2C%22Altered%20group%22%2C%22Unaltered%20group%22%5D),
TNBC Database (https://rgcb.res.in/tnbcdb/tnbdbviewer.php?molecule=mRNA&ctype=EMP3),
and UALCAN(https://ualcan.path.uab.edu/cgi-bin/TCGA-survival1.pl?genenam=EMP3&ctype=BRCA)
and Genetic determinants of cancer patient outcome (https://tcga-survival.com/data-table?view=gene&gene=EMP3&filter=cancer_type&cancer_type=BRCA).
Cell lines and culture
TNBC cell lines (Hs578T and MDA-MB-231) were
chosen for EMP3 transfection experiments. MDA-MB-231 cells were
provided by Professor P.S. Chen (Department of Medical Laboratory
Science and Biotechnology, National Cheng Kung University, Taiwan)
and Hs578T cells were obtained from the Institute of Clinical
Medicine, National Cheng Kung University, Taiwan. MDA-MB-231 cells
were maintained in Leibovitz's L-15 medium (Invitrogen; Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS (HyClone;
Cytiva) and 1% penicillin/streptomycin (Cassion Laboratories) in 5%
CO2 at 37°C. Hs578T cells were cultured in high-glucose
DMEM (HyClone; Cytiva) supplemented with 0.01 mg/ml insulin, 10%
FBS and 1% penicillin/streptomycin in 5% CO2 at 37°C.
SK-BR-3 (cat. no. ATCC HTB-30) and MDA-MB-453 (cat. no. ATCC HTB
131) BC cell lines from Institute of Clinical Medicine, National
Cheng Kung University, Tainan, Taiwan were maintained in
high-glucose DMEM (Himsdia Labomlorier) and Leibovitz's L-15 medium
(Himsdia Labomlorier) with 10% FBS and 1% penicillin/streptomycin
in 5 or 0% CO2, respectively, at 37°C.
DNA constructs and retrovirus
preparation
Full-length EMP3 cDNA was amplified from HEK-293
cells using PCR with custom-designed primers (sense,
5′-gcttcgaattcatgtcactcctcttgc-3′ and antisense,
5′-ggtggatcccgctcccgcttccgtag-3′). The PCR was done using a thermal
cycler (G-Storm; GMI, Inc., Ramsey, MN). The cycling was as
follows: initial denaturation of 1 cycle at 98°C for 2 min,
followed by 30 cycles of denaturation at 98°C for 10 sec, annealing
at 65°C for 15 sec and extension at 72°C. The final one cycle of
extension was set at 72°C for 5 min. Then, full-length EMP3 cDNA
was cloned into the pMSCVpuro retroviral vector at 37°C for 14h
(Clontech), which was transfected into GP2-293 cells from the
Institute of Clinical Medicine, National Cheng Kung University,
Tainan, Taiwan via the calcium phosphate method for 24 h at 37°C as
previously described (24). Viral
supernatant was prepared by collecting GP2-293 culture medium 48 h
after transfection and centrifuging at 367 × g) at room temperature
for 5 min. Stably infected clones were selected by puromycin (5
mg/ml) and maintained at concentrations of 15 ng/µl. Two
overexpressed stable clones, EMP3-1 and EMP3-2, were selected from
Hs578T and MDA-MB-231 cell lines, respectively. Successful
overexpression of EMP3 was tested on MDA-MB231 cells
(HER2-negative) and confirmed by western blotting. The knockdown
experiment was performed as described previously (24). Briefly, small interfering (si)RNA
was obtained from Invitrogen (cat. no HSS103226; Thermo Fisher
Scientific, Inc.). Cells were transfected with 50 nM EMP3 siRNA
based on dose-dependent experiments (data not shown) or scramble
negative control using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.). The constructs of
pcDNA6.2-GW/EmGFP-miR-EMP3 were generated using BLOCK-iT Pol II miR
RNAi Expression Vector kits (Invitrogen; Thermo Fisher Scientific,
Inc.). Linearized vector pcDNA6.2-GW/EmGFP-miR was ligated to the
nucleotide sequence of EMP3 (5′-GCAGTAATGTCAGCGAGAATG-3′). The
oligonucleotide sequence of negative control was
5′-GAAATGTACTGCGCGTGGAGACGTTTTGGCCACTGACTGACGTCTCCACGCAGTACATTT-3′,
which does not target any known vertebrate gene.
Western blotting
Hs578T and MDA-MB-231 cells were lysed in RIPA lysis
buffer supplemented with protease inhibitors (Thermo Fisher
Scientific, Inc.) to extract protein. Protein concentration was
determined using Bradford assay Briefly, a total of 30 µg/lane
protein was separated by SDS-PAGE through 6–12% gradient gels and
transferred to 0.2–0.45 µm PVDF membrane (Stratagene), which was
blocked with 5% non-fat milk in TBST (0.1% Tween-20). The membrane
was incubated with primary antibodies (GeneTex, Inc.) overnight at
4°C targeting human EMP3 (1:1,000, clone no. 43972, GeneTex, Inc.),
epidermal growth factor receptor (EGFR or HER1, 1:1,000, cat. no.
GTX100448), human epidermal growth factor 2 (HER2, 1:1,000,
Cat#GTX100509), human epidermal growth factor 3 (HER3, 1:1,000,
Cat#GTX50651), human epidermal growth factor 4 (HER4, 1:1,000,
Cat#GTX111276), progesterone receptor (PR) (1:1,000,
Cat#GTX22765;), estrogen receptor (ER, 1:1,000, Cat. no.
GTX127978), focal adhesion kinase (FAK; 1:1,000, Cat#GTX100764),
Src proto-oncogene, non-receptor tyrosine kinase (SRC, 1:1,000,
Cat#GTX132369), phosphorylated (p-)SRC (1:1,000, Cat#GTX13347), rho
associated coiled-coil containing protein kinase 1 (ROCK1, 1:1,000,
Cat#GTX629972), ROCK2 (1:1,000, Cat#GTX102619), α-actinin (1:1,000,
Cat#GTX103219), JAK2, Cell Signaling Technology, Inc., 1:1,000,
Cat#3230), p-JAK2 (1:1,000, Cat#GTX132784), signal transducer and
activator of transcription 3 (STAT3, 1:1,000, Cat#GTX104616),
p-STAT3 (1:1,000, Cat#GTX118000), RAS Proto-Oncogene, GTPase (RAS,
1:1,000, cat. no. GTX132480), raf-1 proto-oncogene,
serine/threonine kinase (Raf-1, 1:1,000, Cat#GTX111588),
extracellular signal-regulated kinase 1 and 2 (ERK1/2, 1:1,000,
Cat#GTX134462), p-ERK1/2 (1:1,000, Cat#GTX132783), SRY-box
transcription factor 2 (SOX2, 1:1,000m Cat#GTX101507),
octamer-binding transcription factor 4 (OCT4, 1:1,000,
Cat#GTX101497), twist family bhlh transcription factor 1 (Twist1,
1:1,000, Cat#GTX60776), snail family transcriptional repressor 1
(Snail, 1:1,000, Cat#GTX638370), snail family transcriptional
repressor 2 (Slug, 1:1,000, Cat#GTX128796), zinc finger e-box
binding homeobox 1 (ZEB1, 1:1,000, cat. no. GTX638294;), E-cadherin
(1:1,000, Cat#GTX100443), vimentin (1:1,000, Cat#GTX100619) and
human β-actin (1:5,000, Cat#GTX109639). Following washing with TBST
three times, the membranes were incubated with horseradish
peroxidase-conjugated secondary antibodies (anti-rabbit or
anti-mouse IgG; 1:5,000, Cat# GTX213110, GeneTex, Inc.) for 1 h at
room temperature. Detection of protein bands was performed using an
enhanced chemiluminescence system (cat#GTX400006, GeneTex). The
densitometry was performed using ImageJ (1.41o National Institutes
of Health).
MTT assay
MTT assay was performed to assess the impact of EMP3
on BC cell proliferation in vitro. A total of 1,000
transfected cells was seeded in 100 µl medium in each well of a
96-well plate and cultured at 37°C for 24, 48, 72 and 96 h. Then,
the cells were incubated with 50 µl MTT (Cyrusbioscience, Inc.)
reagent/well in the dark at 37°C for 4 h and the formazan crystals
were dissolved in DMSO and quantified by measuring absorbance at
590 nm using microplate reader (SpectraMax i3X, Molecular Devices).
A total of four independent experiments were performed in all
assays. For chemosensitivity, cells were treated with appropriate
concentrations of trastuzumab (HERCEPTIN®, 440 mg/20 ml,
Genentech, Inc.) and incubated for 3 days at 37°C. MTT assay was
performed in quintuplicate to obtain the average.
Cell migration and invasion assay
Migration and invasion experiments were performed
using Transwell membrane filter inserts (cat. no. #3422, Corning
Costar, Inc.; 8 µm pore size) at 37°C. In brief, 2.5×104
cells (Hs578T or MDA-MB-231) were seeded into DMEM plated in upper
chamber of a 24-well Transwell plate with or without Matrigel for
invasion and migration assays, respectively. The lower chamber was
filled with complete medium (L-15 or DMEM) with 10% FBS. The
migration and invasion assays were performed at 37°C for 72 and 48
h, respectively. Migrated cells on the lower surface were stained
with 2% crystal violet at room temperature for 10 min, and those
that did not penetrate the filter were removed. The number of
migrating or invading cells was counted under a light microscope
from five randomly selected fields of view/well) in a single
chamber (magnification, ×200).
Gap closure assay
To assess cell migration in vitro, culture
inserts (Ibidi GmbH) consisting of two wells separated by a
500-µm-thick wall were used. The insert was placed into one well of
six-well plates and pressed to ensure tight adhesion. Stable
EMP3-overexpressing Hs578T and MDA-MB-231 cells
(2×105/100 µl) were seeded into each well and incubated
overnight until 90% confluence in DMEM-High glucose (HyClone),
supplemented with 0.01 mg/ml insulin, 5% FBS (HyClone) at 37°C. The
inserts were removed, and the width between the gaps visible under
a light microscope (cat. no. TE300, Nikon) was recorded at 0, 5, 10
and 15 for HS578T and 0, 5, 10, and 20 h for MDA-MB-231 cells
(magnification, ×100). The area covered by cells was measured to
quantify cell migration in vitro. The closure of the
cell-free gap for each treatment was determined by comparing the
results to those at 0 h (magnification, ×200).
In vivo xenograft model
A total of 240 Six-week-old male NOD/SCID mice
weight, 20 to 25 grams was purchased from the National Cheng Kung
University Laboratory Animal Centre and maintained in a
pathogen-free facility under isothermal conditions with regular
photoperiods. The animals had free access to sterile water and
food. The experimental protocol adhered to regulations of the
Animal Protection Act of Taiwan (26) and was approved by the NCKU
Laboratory Animal Care and User Committee (approval no. 108075).
The temperature for mice ranged from 20–24°C, and the humidity
maintained between 40–60%. The common light cycle is 12/12-h
light/dark cycle. The mice were subcutaneously (s.c.) injected with
1×107 BC cells (SKBR3) in 50 µl normal saline on the
left and right flanks. A total of 6 mice (three mice in each group,
SKBR3/Vec, and SKBR3/EMP3) (12 tumors) were obtained in each
experimental group. Body weight and tumor size were measured every
week. If the weight of a single tumor exceeds 10% of body weight,
or the average diameter of the tumor in adult mice exceeds 20 mm,
the animal needs to be euthanized. The maximum tumor volume and
tumor diameter measured were 520 mm3 and 15 mm,
respectively. All mice were euthanized by stepwise escalation
(30–50% of chamber volume, 0.02 MPa) of carbon dioxide inhalation
and cervical dislocation. Death was verified by pupil dilation as
well as cessation of breathing and heartbeat. The tumors were
resected, photographed and weighed on day 90 post-injection.
Immunohistochemistry (IHC)
IHC was performed to examine expression of HER1,
HER2 and EMP3 in xenografts using primary antibodies targeting
human EMP3 (GeneTex, Inc., 1:1,000), HER1 (Zeta Corporation, 1:200)
and HER2 (Zeta Corporation, 1:200). Briefly, tissue was fixed with
10% neutral formalin at room temperature overnight and
paraffin-embedded, Sections of 4 mm thickness were deparaffinized
with xylene and pre-treated with Epitope Retrieval Solution 2 (EDTA
buffer, pH 9.0, Dako Denmark A/S, Inc.) at 100°C for 30 min. The
blocking was carried out by protein blocking buffer (RE7102-CE,
Leica Biosystems) at room temperature for 30 min and super blocking
buffer (AAA-IFU, ScyTek Laboratories, Inc.) at room temperature for
9 min. Then, sections were reacted with primary antibody at room
temperature for 30 min, followed by incubation at room temperature
for 8 min using a ready-to-use Bond Polymer Refine Detection kit
containing peroxide block for quenching (DS9800, Cat#68086, Leica
Biosystems), secondary antibody, polymer reagent and DAB chromogen.
The DAB was developed for 10 min. Counterstaining was performed
with hematoxylin for 5 min at room temperature. The histology was
examined light microscope (Nikon; magnification, ×40 and ×100).
Immunocytochemistry
Cells were plated onto glass coverslips and fixed
with 4% paraformaldehyde for 20 min at room temperature, blocked
with 5% rabbit serum (Neuromics; Cat# SER003) for 60 min at room
temperature, and stained with primary antibody for Ki-67 (1:200
dilution, Cat#sc-23900, Santa Cruz Biotechnology, Inc.) at room
temperature for 1 h. A high-sensitivity DAB system (Liquid DAB+
Substrate kit K3468, Dako) was used for immunocytochemical
staining. The slides were counterstained with Mayer's hematoxylin
at room temperature for 1 min, dehydrated, and then mounted.
Specific primary antibody replaced with PBS in tissue sections was
used as a negative control. The cytology was examined light
microscope (Nikon) (magnification, ×40, ×100, ×200 and ×400).
Expression of EMP3 in primary BC
cohort
A retrospective study of a BC cohort, who underwent
surgical treatment with modified radical mastectomy or
breast-conserving surgery and sentinel lymph node biopsy between
April 2005 and October 2014, was recruited under the approval of
the Institutional Review Board of the National Cheng Kung
University Hospital (approval no. A-ER-107-441). All samples were
from female patients recruited from National Cheng Kung University
Hospital, Tainan, TAIWAN. The study population comprised 166
patients with median age of 53 years (range, 29–83 years). Clinical
demographic data, including TNM status and clinical information,
were obtained by chart review. Each case was independently reviewed
by two pathologists for diagnosis. The mean follow-up time was
108.28±37.24 months.
IHC staining of EMP3 protein was performed to
examine its association with clinicopathological indicators and
patient outcomes. The breast tissue was fixed with 10% neutral
formalin overnight at room temperature. Then, it was cut for
dehydration and paraffin embedding. Immunostaining was performed on
formalin-fixed paraffin-embedded tissue using a standard
avidin-biotin complex-peroxidase procedure with an automated
stainer (BenchMark Ultra Auto-Stainer; Ventana) (24). Briefly, 4-µm tissue sections from
each tumor were subjected to heat-induced epitope retrieval using
CC1 cell conditioning solution (Ventana). The slides were treated
with 3 % hydrogen peroxide in PBS at 4°C for 30 min, and then 10%
normal goat serum (Neuromics, Edina, MN; Cat# SER003) at room
temperature for 60 min. The slides were incubated with rabbit
polyclonal EMP3 antibody (LTK BioLaboratories) or PATHWAY
anti-HER2/Neu (4B5) rabbit monoclonal antibody (Ventana) at a
dilution of 1:10. The immunostained proteins were visualized using
a Roche OptiView DAB IHC Detection kit (Ventana). Anti-rabbit
poly-HRP secondary antibody (Leica Biosystems) was added at room
temperature for 30 min. Sections were counterstained with
hematoxylin at room temperature for 5 min. Samples treated with PBS
instead of specific primary antibody were used as a negative
control. Both positive reference (BC known to express EMP3) and
negative controls (normal liver) were included.
The histology was examined (by light microscope
(Nikon) (magnification, ×40 and ×100). HER2 protein expression was
scored according to 2018 American Society of Clinical Oncology and
the College of American Pathologists for the testing and reporting
of biomarkers in cancer HER2 testing in BC guidelines (27). EMP3 staining intensity was graded as
negative, +, ++ and +++ by two pathologists using smooth muscle
cells of small arteries as the internal reference (28). ++ or +++ was considered to indicate
EMP3 upregulation.
Statistical analysis
The data were derived from triplicate experiments.
All data are presented as the mean ± SD by GraphPad prism 6
software (GraphPad Software, Inc.; Dotmatics). The differences
between categorical variables were analyzed with one-way ANOVA
followed by post hoc Tukey's test. Disease-free survival was
calculated from the date of surgery to the date of disease
recurrence. The significance of various covariates in survival was
assessed using univariate analysis with log-rank test. The survival
curve was calculated using the Kaplan-Meier method (SPSS 17.0 for
Windows (SPSS Inc., All tests were two-sided and P<0.05 was
considered to indicate a statistically significant difference.
Results
Bioinformatics analysis
To investigate the potential role of EMP3 in human
BC, bioinformatics analysis was performed using The Human Protein
Atlas, Gene Expression Profiling Interactive Analysis, cBioPortal,
UALCAN and OncoLnc. EMP3 expression tended to be positively
associated with poor overall survival in cBioPortal (data not
shown) but this was not significant. High EMP3 expression was
associated with better survival in The Human Protein Atlas and
UALCAN (data not shown) datasets. High EMP3 expression tended to be
associated with better overall survival in Gene Expression
Profiling Interactive Analysis dataset, but no significant
difference was observed for disease-free survival (data not shown).
However, analysis of the OncoLnc data did not reveal a prognostic
value for EMP3 expression (data not shown).
Biological effects induced by EMP3
overexpression in BC cells
To clarify the biological role of EMP3, stable
EMP3-1 and EMP3-2 clones were established in HS578T and MDA-MB-231
BC cell lines, respectively. These stable clones exhibited higher
levels of EMP3 protein expression compared with parental cells, as
shown by western blotting. Expression of EMP3 protein in stable
EMP3-1 and EMP3-2 clones was moderate and high, respectively
(Fig. 1A). The protein expression
profiles, including those of ER, PR and HER family members and
EMT-related markers, were assessed in vector (Vec) control cells
and stable EMP3-overexpressing clones (EMP3-1 and EMP3-2; Fig. 1A). The morphology of stable
EMP3-overexpressing cells was examined via light microscopy
(Fig. 1B). Both Hs578T and
MDA-MB-231 cells became slender in shape following transfection
with EMP3. The viability was significantly increased in stable
EMP3-1- and EMP3-2-overexpressing clones, both in the Hs578T and
MDA-MB-231 cell lines (Fig.
1B).
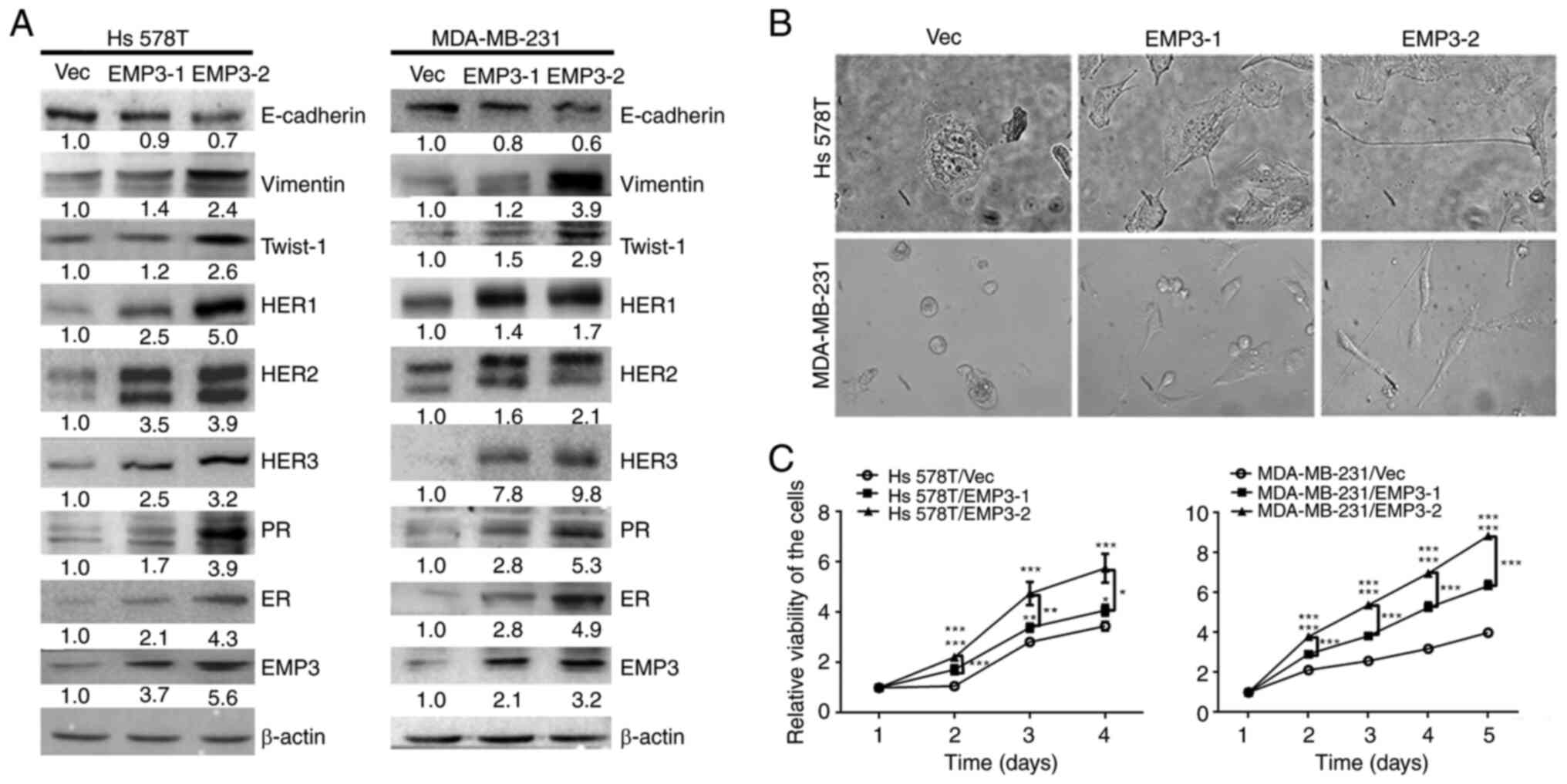 | Figure 1.Protein expression profiles,
morphology and proliferation of EMP3-overexpressing BC cells.
Stable clones of EMP3-1 and EMP3-2 were established from HS578T and
MDA-MB-231 BC cell lines, respectively. (A) Protein expression
profiles were examined for EMP3, ER, PR, HER family members and
EMT-related markers in stable EMP3-overexpressing cells compared
with Vec control cells via western blotting. (B) Both stable EMP3-1
and stable EMP3-2 cells became slender in shape (fibroblastoid)
following EMP3 overexpression, particularly stable EMP3-2 clones.
(C) EMP3 overexpression significantly increased the viability of
tumor cells, with a greater rate observed in stable EMP3-2 clones.
*P<0.05, **P<0.01, ***P<0.001 vs. EMP3-1 or −2. EMP,
epithelial membrane protein; PR, progesterone receptor; ER,
estrogen receptor; HER, human epidermal growth factor receptor; BC,
breast cancer; Vec, vector. |
Crosstalk of EMP3 with HER family in
human BC cells in vitro
Expression of all HER family members (HER1-HER3), PR
and ER increased in both Hs578T and MDA-MB-231 cell lines stably
overexpressing EMP3 (Fig. 1A).
Therefore, overexpression of EMP3 may upregulate expression of
HER1-HER3 and HR in human BC in vitro (Fig. 1A). These results indicated that EMP3
can functionally crosstalk with HER superfamily and HR during BC
tumorigenesis.
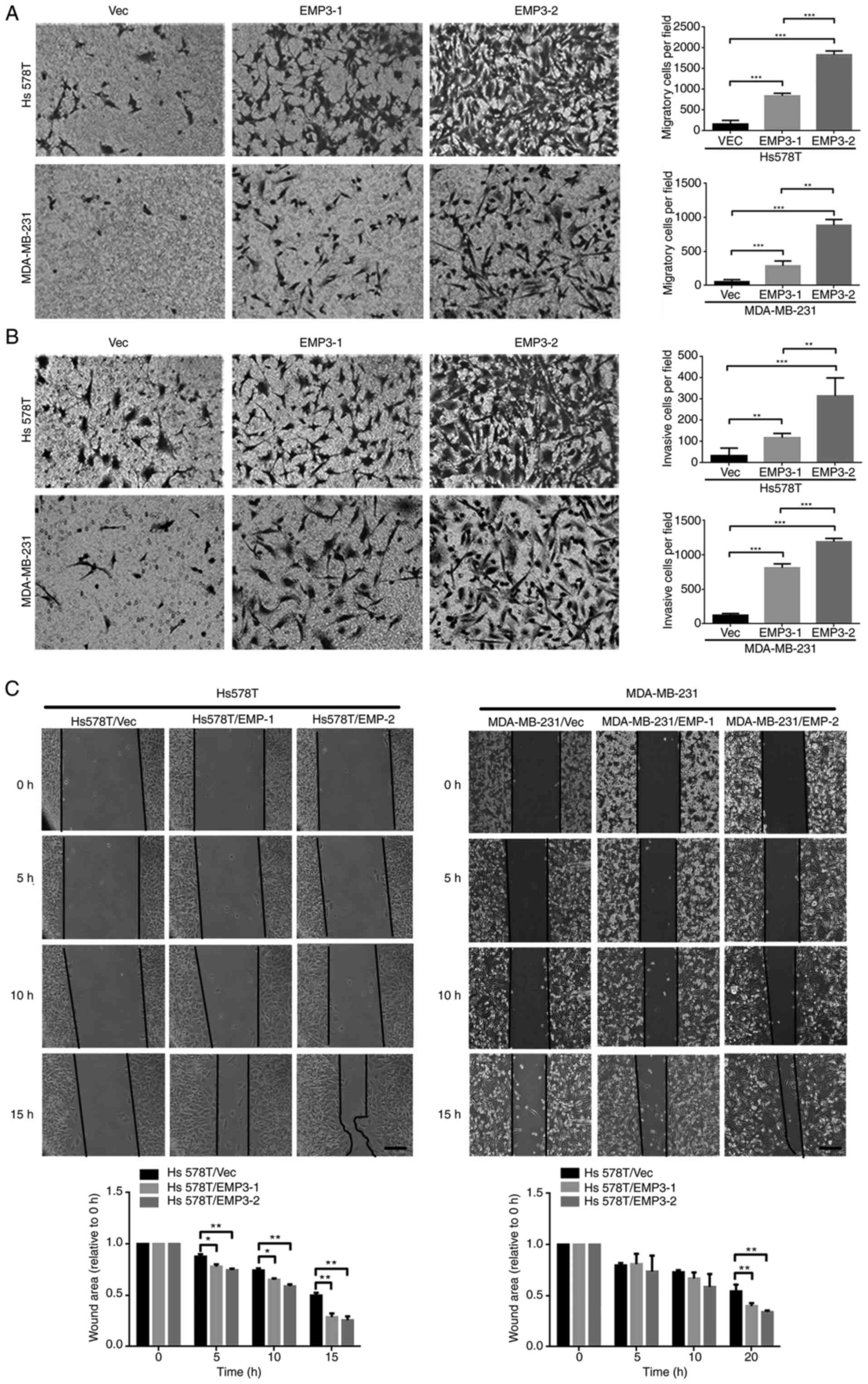
EMP3 overexpression promotes cell
migration and invasion
Overexpression of EMP3 upregulated protein
expression of fibronectin, twist1/2 and vimentin but suppressed
E-cadherin expression in both Hs578T and MDA-MB-231 cell lines
(Fig. 1A). However, expression of
Snail, Slug and ZEB1 was not upregulated (data not shown). EMP3
overexpression in EMP3 stable cells, defined as two-fold higher
expression, compared with vector control may contribute to
migration and invasion of TNBC cells in vitro via EMT.
Stable clones of EMP3-1 and EMP3-2 were subjected to cell
migration, invasion and gap closure assays. Compared with Vec
control cells, stable EMP3-overexpressing Hs578T and MDA-MB-231
cells exhibited increased migration (Fig. 2A) and invasion (Fig. 2B). In addition, EMP3-2 stable clones
had increased ability to migrate and invade in vitro
compared with EMP3-1 stable clones of Hs578T and MDA-MB-231 cell
lines, respectively (Fig. 2A).
The impact of EMP3 on cell migration was confirmed
via gap closure assay. Stable EMP3-1 and EMP3-2 cells showed
greater gap closure than Vec control cells (Fig. 2C). This was observed in both Hs578T
and MDA-MB-231 cell lines. Together, these findings suggested that
EMP3 overexpression exerted pro-oncogenic effects on BC in
vitro.
Intracellular signaling pathways
upregulated by EMP3 overexpression in human BC
There is crosstalk between EMP2 and integrins αV and
β3 in regulation of urothelial cell adhesion and migration during
tumorigenesis in vitro (11). Therefore, involvement of this
signaling pathway was assessed in BC development. Western blotting
showed that components of FAK-c/SRC/RhoA, SRC/JAK2/STAT3,
SRC/Ras/Raf-1 and ERK1/2 pathways were upregulated in Hs578T and
MDA-MB-231 EMP3 stable cells compared with Vec control cells
(Fig. 3A). Protein expression
(JAK2, STAT3, RAS, RAF, p-ERK, ERK, SRC, p-SRC, α-actinin ROCK1 and
ROCK2) was quantified. All of the signal proteins in Hs578T cells
were significantly higher in EMP3-2 stable cells compared with
vector control (Fig. 3B and C).
Overall, EMP3 upregulated proteins associated with JAK2/STAT3,
RAS/RAF/ERK and FAK/SRC/α-actinin/ROCK1/2 signaling pathways in BC
in vitro.
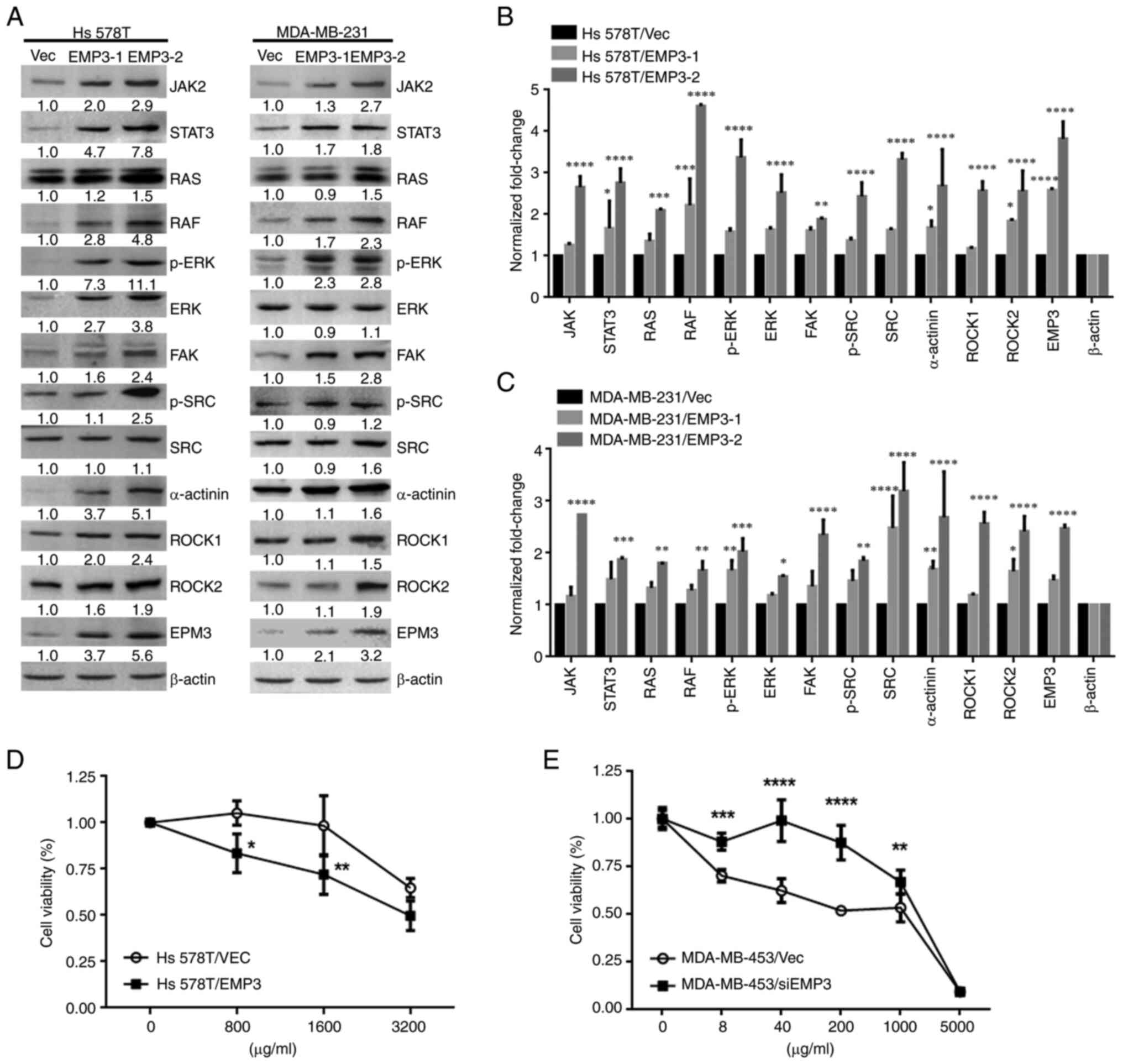 | Figure 3.Effects of EMP3 on signaling pathways
and chemoresistance in breast cancer cells in vitro. (A)
Protein expression of JAK2, STAT3, RAS, RAF, p-ERK, ERK, FAK,
p-SRC, SRC, α-actinin, ROCK1, ROCK2, and EMP3. β-actin was used as
a loading control in (B) Hs578T and (C) MDA-MB-231 cell lines.
Growth curves of (D) trastuzumab-treated Hs578T and (E)
EMP3-knockdown MDA-MB-453 cell lines were generated via MTT assay.
*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 vs. EMP3-1
or −2 stable cells or between these two stable cells. EMP,
epithelial membrane antigen; RAF, raf-1 proto-oncogene,
serine/threonine kinase; p-, phosphorylated; FAK, focal adhesion
kinase; ROCK, rho-associated protein kinase; SRC, Src
proto-oncogene, non-receptor tyrosine kinase; Vec, vector; si,
small interfering. |
EMP3 alters chemosensitivity of BC
cells to trastuzumab in vitro
As EMP3 was demonstrated to upregulate expression of
HER1-HER3, as well as proteins associated with JAK2/STAT3,
RAS/RAF/ERK and FAK/SRC/α-actinin/ROCK1/2 signaling pathways in the
Hs578T and MDA-MB-231 BC cell lines, the potential of EMP3 in
modulating drug resistance of BC was investigated. Trastuzumab is a
monoclonal anti-HER2 antibody for patients with early or metastatic
HER2-positive BC (29). Therefore,
cell viability was examined in response to trastuzumab in the
presence of EMP3 overexpression or knockdown. MTT assay revealed
that Hs578T EMP3 stable cells with higher HER2 expression were more
sensitive to trastuzumab than Hs578T/Vec control cells (Fig. 3D). By contrast, knocking down EMP3
in MDA-MB-453 cells conferred resistance to trastuzumab compared
with MDA-MB-453/Vec control cells (Fig.
3E). Together, these findings indicated that altered EMP3
expression modified the sensitivity of BC cells to trastuzumab
treatment in vitro.
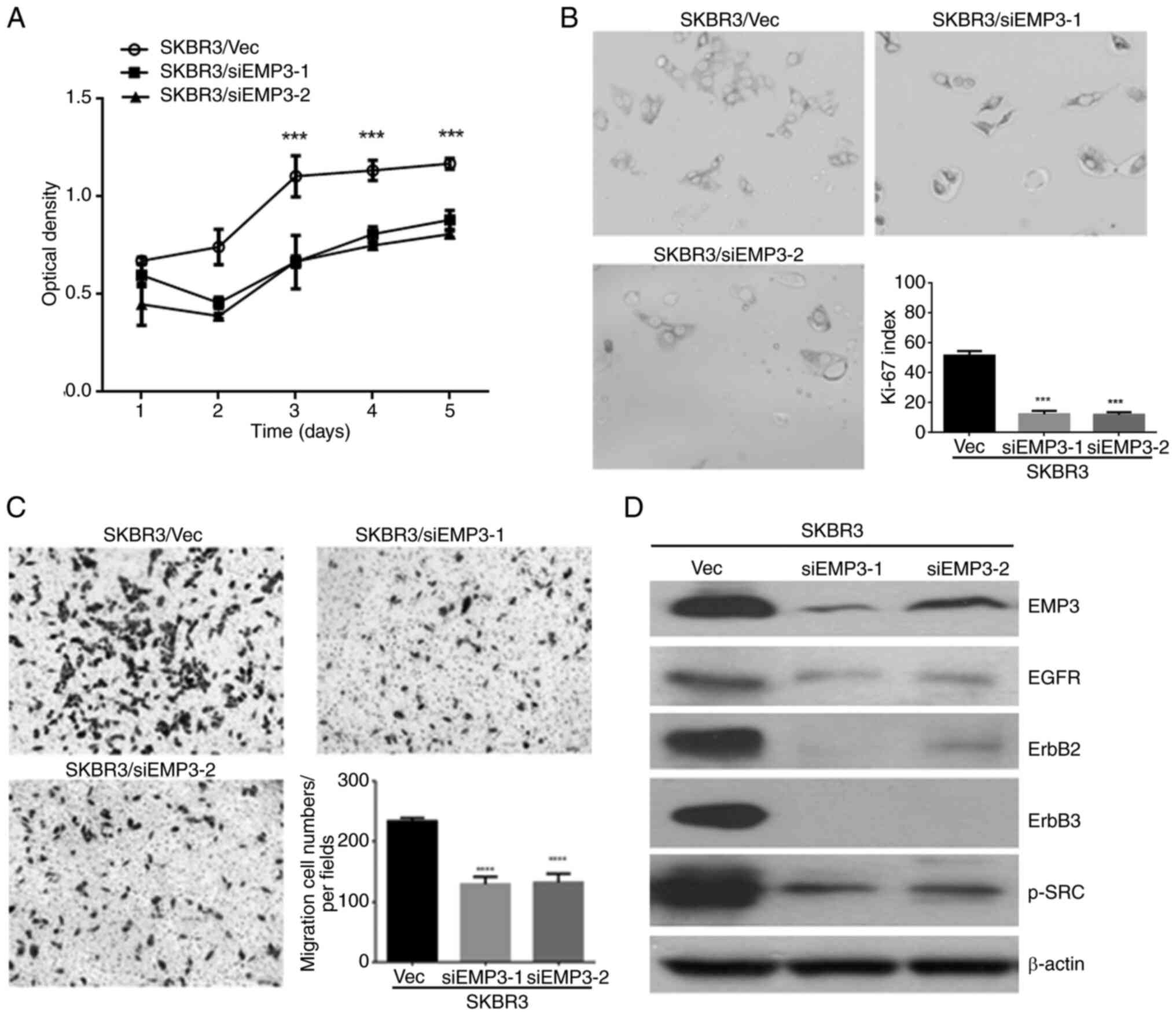
EMP3 as a potential novel therapeutic
target for human BC in vitro
As aforementioned EMP3 was demonstrated to confer
pro-tumorigenic effects on human BC in vitro and upregulate
HER receptor expression and multiple signaling pathways. To clarify
the potential of EMP3 as a therapeutic target for human BC, the
SK-BR-3 cell line was assessed because of its endogenous expression
of HER1-HER3 and EMP3. Therefore, EMP3-knockdown cell lines,
SKBR3/siEMP3-1 and SKBR3/siEMP3-2, and a vector control cell line
(SKBR3/Vec) were established from SK-BR-3 BC cells. MTT assay
(Fig. 4A) and Ki-67 index
measurement (Fig. 4B) revealed that
the proliferation of SK-BR-3 cells with EMP3 knockdown was
significantly suppressed compared with that of control cells.
Compared with SKBR3/Vec control cells, cell migration in
vitro was inhibited after EMP3 was knocked down, as
demonstrated by Transwell migration assay (Fig. 4C). Furthermore, expression of HER1,
HER2, HER3 and p-SRC was downregulated in SKBR3/siEMP3-1 and
SKBR3/siEMP3-2 stable cells compared with that in SKBR3/Vec control
cells (Fig. 4D). These results
suggested EMP3 as a novel therapeutic target for human BC with
co-expression of HER2 and EMP3.
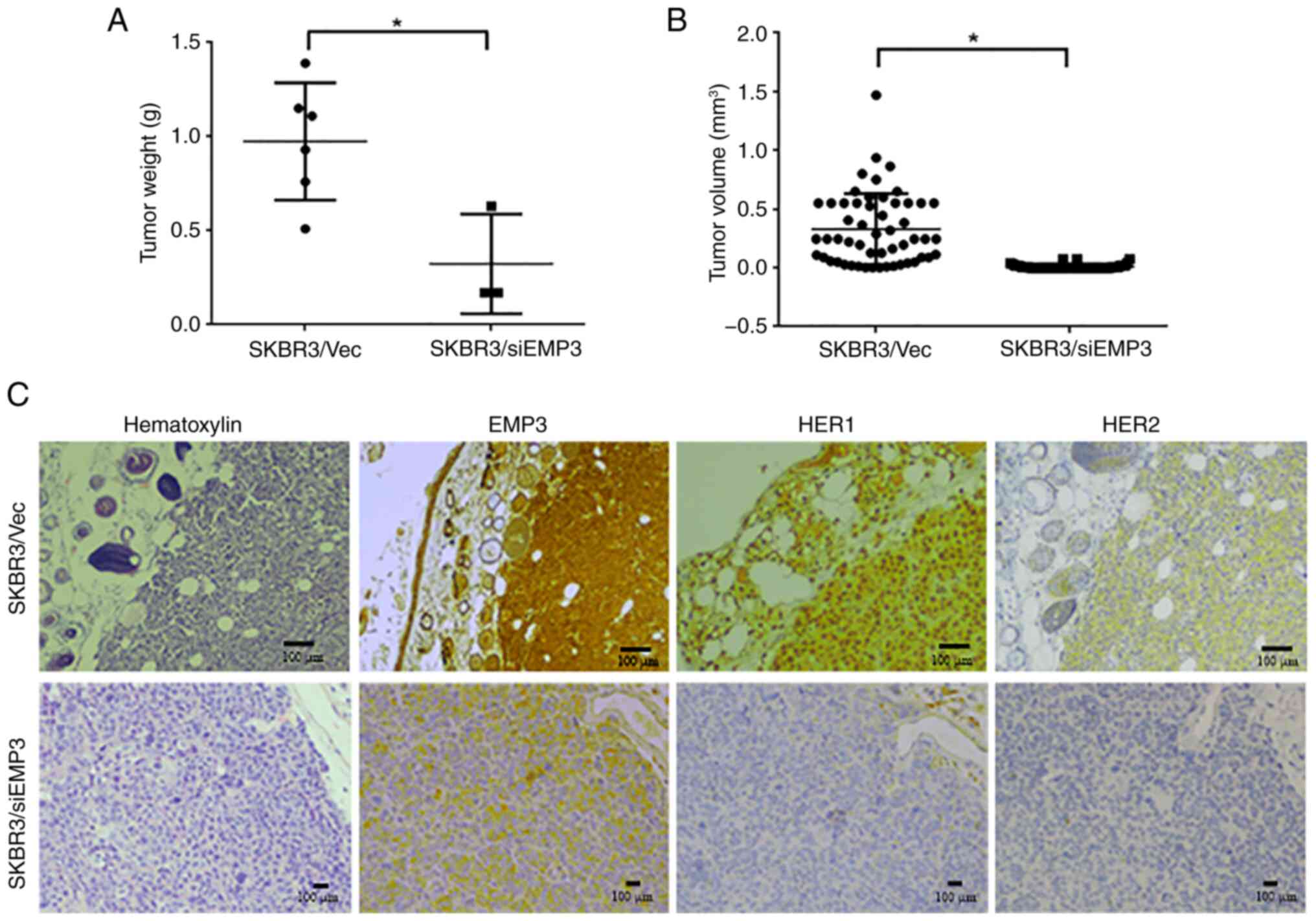
Tumorigenicity of EMP3 in a xenograft
NOD/SCID mouse model
To verify the clinical implications of targeting
EMP3 in BC, a xenograft NOD/SCID mouse model was constructed using
EMP3-knockdown and vector control SK-BR-3 cell lines. The weights
of xenografts derived from SKBR3/siEMP3 stable cells were
significantly lower than those derived from SKBR3/Vec control cells
(Fig. 5A). The tumor volume derived
from SKBR3/siEMP3 cells was also lower than that derived from
SKBR3/Vec control cells (Fig. 5B).
In terms of biomarker expression in vivo, strong membrane
expression of EMP3, HER1 and HER2 was demonstrated in tumors
derived from SKBR3/Vec control cells, whereas expression of these
proteins was decreased in tumors derived from SKBR3/siEMP3 stable
cells (Fig. 5C).
Association of EMP3 with
clinicopathological indicators of BC
Expression of EMP3 was investigated in BC cases from
the NCKUH biobank (Table SI). EMP3
protein was detected in the cytoplasm and/or membrane of cancer
cells. In addition, its expression was heterogeneous in tumors,
with higher expression usually occurring at the invasive front
(Fig. 5C). EMP3 expression was
detected in 102 of 166 cases (61.4%) of BC; however, its expression
intensity was not associated with tumor stage, nodal metastasis,
ER, PR or HER2 status or clinical outcome. EMP3 was upregulated in
72 cases (43.4%) in the BC cohort. Because of crosstalk between
EMP3 and HER2 in vitro, co-expression of these biomarkers
was analyzed in relation to clinicopathological indicators and
patient outcomes (Table I). The
incidence of EMP3(−)/HER2(−), EMP3(−)/HER2(+), EMP3(+)/HER2(−), and
EMP3(+)/HER2(+) in the BC cohort was 69 (41.6), 25 (15.1), 54
(32.5) and 18 (10.8%) cases, respectively. Co-expression of EMP3
and HER2 was positively associated with ER expression and tended to
be associated with nodal metastasis, however this was not
significant. In the absence of HER2 amplification, EMP3(+)/ER(+)
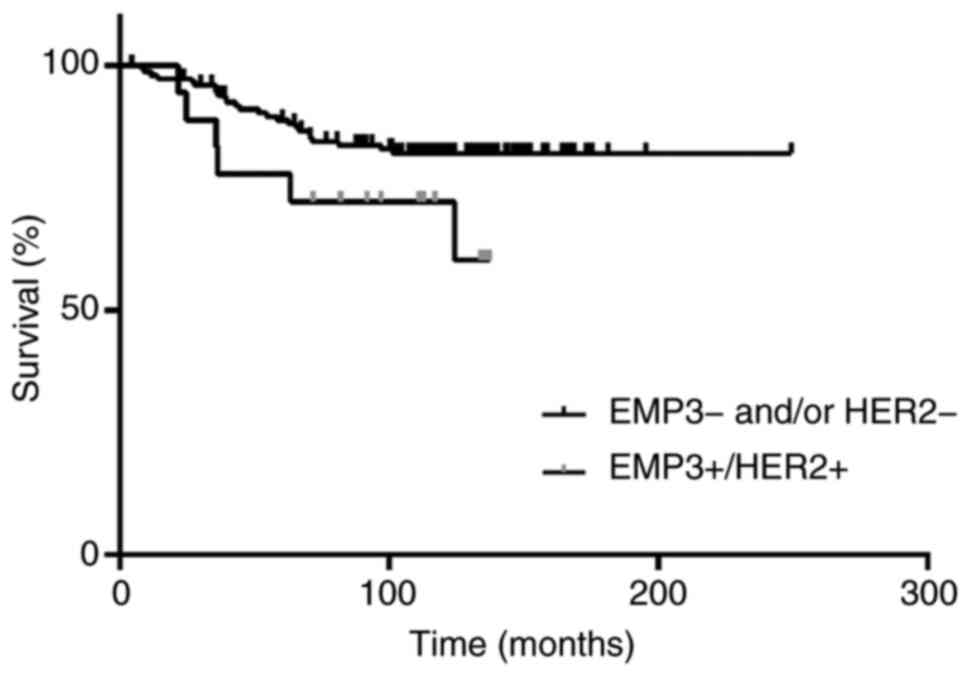
and EMP3(+)/PR(+) patterns were detected in 46 (27.7) and 30
(18.1%) cases of BC, respectively. Patients with EMP3(+)/HER2(+) BC
tended to have a higher risk of recurrence (data not shown) and
lower disease-free survival than patients with other expression
patterns (Fig. 6), however this was
not significant.
 | Table I.Association of EMP3 and HER2
expression with clinicopathological indicators and patient outcome
(n = 166) |
Table I.
Association of EMP3 and HER2
expression with clinicopathological indicators and patient outcome
(n = 166)
|
|
EMP3/HER2a |
|
|---|
|
|
|
|
|---|
| Indicator | -/- (n=69) | -/+ (n=25) | +/- (n=54) | +/+ (n=18) | P value |
|---|
| Anatomic stage |
|
|
|
| 0.896 |
| I | 14 (20) | 3 (12) | 12 (22) | 3 (17) |
|
| II | 40 (58) | 17 (68) | 33 (61) | 10 (55) |
|
|
III | 15 (22) | 5 (20) | 9 (17) | 5 (28) |
|
| pT status |
|
|
|
| 0.742 |
|
pT1 | 22 (32) | 5 (20) | 14 (26) | 7 (39) |
|
|
pT2 | 43 (62) | 18 (72) | 35 (65) | 11 (61) |
|
| pT3 or
pT4 | 4 (6) | 2 (8) | 5 (9) | 0 (0) |
|
| Nodal status |
|
|
|
| 0.085 |
|
Negative | 32 (46) | 16 (64) | 37 (69) | 10 (56) |
|
|
Positive | 37 (54) | 9 (36) | 17 (31) | 8 (44) |
|
| ER status |
|
|
|
| 0.028 |
|
Negative | 18 (26) | 10 (40) | 8 (15) | 8 (44) |
|
|
Positive | 51 (74) | 15 (60) | 46 (85) | 10 (56) |
|
| PR status |
|
|
|
| 0.568 |
|
Negative | 33 (48) | 14 (56) | 24 (44) | 11 (61) |
|
|
Positive | 36 (52) | 11 (44) | 30 (56) | 7 (39) |
|
| Recurrence |
|
|
|
| 0.432 |
|
Negative | 57 (83) | 21 (84) | 45 (83) | 12 (67) |
|
|
Positive | 12 (17) | 4 (16) | 9 (17) | 6 (33) |
|
| Survival |
|
|
|
|
|
|
Alive | 57 (83) | 21 (84) | 45 (83) | 13 (72) | 0.716 |
|
Dead | 12 (17) | 4 (16) | 9 (17) | 5 (28) |
|
Discussion
EMP3 is a transmembrane signaling protein that
serves a key role in the regulation of apoptosis, differentiation
and invasion of cancer cells (11).
The present showed that EMP3 exerted regulatory effects on the
oncogenic HER family and HR in BC in vitro. Moreover, EMP3
activated the expression of proteins associated with JAK2/STAT3,
RAS/RAF/ERK and FAK/SRC/α-actinin/ROCK1/2 pathways in BC in
vitro, consistent with our previous report and other studies
(24,30,31).
In addition to roles in proliferation, migration and invasion in
vitro, these pathways have also been reported to be involved in
EMT and/or trastuzumab resistance in BC (32–34).
The present results support the potential role of EMP3-dependent
HER1/HER3 regulation in pathogenesis of BC (11,15,17).
Compared with xenografts derived from SK-BR-3/Vec
control cells, levels of HER1, HER2, HER3, and p-SRC expression in
xenografts derived from SK-BR-3/siEMP3 stable cells were notably
lower, in conjunction with lower tumor weight and smaller size. To
the best of our knowledge, the present study is the first to
demonstrate the potential of EMP3 as a novel therapeutic target for
human BC.
As a membrane scaffolding protein, EMP3 forms a
network with other transmembrane proteins on the cell surface to
promote signaling cascades (35).
In human sarcoma, melanoma and embryonic kidney cell line models
(11–17), EMP3 serves a key role in organizing
signaling complexes at the membrane and is required for maintaining
elevated levels of mitogenic signaling as well as cell survival.
Using isocitrate dehydrogenase-wild glioblastoma as a model,
Martija et al (36) proposed
that EMP3 facilitates TBC1D5 recruitment into maturing endosomes,
where the complex inactivates member RAS oncogene family (RAB7) and
restricts progression of internalized EGFR cargoes toward lysosomal
degradation. EMP3-dependent stabilization of EGFR sustains its
downstream signaling via CDK2. These mechanisms ensure sustained
proliferation, apoptosis resistance and decreased susceptibility to
targeted kinase inhibitors. Here, immunoprecipitation (data not
shown) revealed that EMP3 interacted strongly with HER2 and weakly
with EGFR at the cell membrane of Hs578T/EMP3 stable cells. The
present results concur with the mechanism proposed by Martija et
al (36).
Although EMP3 serves as an oncogene in BC, previous
studies report a tumor-suppressive function (21) and lack of prognostic significance
for BC (25). This discrepancy
might be explained by unique experimental conditions and dissimilar
scoring of immunohistochemistry using different antibodies on
limited tissue microarrays.
The present study demonstrated that targeting EMP3
could simultaneously downregulate multiple oncogenes and
pleiotropic signaling pathways. This strategy is simple compared
with current combination therapies or dual-target inhibitors in the
context of immunotherapy and molecular-targeted therapy focused on
HER family members (37). Whether
dual targeting of EMP3 and HER2 exerts synergistic effects requires
clarification. EMP3 is enriched in macrophages and specialized
macrophages, such as Kupffer cells in the liver and Hofbauer cells
in the placenta (28). Using
mouse-derived macrophages, Kusumoto et al (38) demonstrated that EMP3-overexpressing
macrophages inhibit induction of alloreactive cytotoxic T
lymphocytes, suggesting a role for EMP3 in inhibition of T
cell-mediated immunity. Whether a systemic EMP3-targeted approach
for BC also affects the tumor microenvironment, particularly in
terms of immune cell infiltration or stromal interactions (39–44),
requires clarification.
A clinical cohort study revealed a significant
association of EMP3 and HER2 co-expression with ER-positive status
in BC (17,20), supporting the present in
vitro observations. Conversely, ~40% of HER2-amplified BC cases
also demonstrated EMP3 upregulation and should be considered for
EMP3-targeted therapy. The present data suggested patients with BC
with co-expressed EMP3 and HER2, ER or PR may be good candidates
for EMP3-targeted therapy. In the present BC cohort, co-expression
of EMP3 and HER2 tended to be associated with nodal metastasis
(15,16), suggesting the potential role of EMP3
in metastatic progression of BC.
However, EMP3 expression alone was not significantly
associated with patient outcome. Further investigation in a larger
cohort using monoclonal antibodies and an optimized scoring system
is key to clarify the potential of EMP3 as an independent
prognostic biomarker, whether for all types or specific subsets of
BC. The reason for the difference in the prognostic value of EMP3
in BC dataset analysis is unclear. Potential explanations may be
variation in co-expressed biomarkers in the BC cohort and antibody
selection with interpretation criteria.
The present study confirmed the role of EMP3 in
regulation of HER family signaling in BC. Overexpression of HER1
(45) or HER3 (46,47) is
associated with primary resistance to trastuzumab (44–46).
The successful in vitro modulation of chemosensitivity to
trastuzumab by targeting EMP3 supports its role as a potential
co-targeting candidate in the design of HER2-based cancer therapy.
A multi-institutional cohort study reported the significance of
combined HER3 and HER1 expression score in prediction of resistance
to adjuvant chemotherapy in TNBC (48). In addition, 12 of the present
patients with TNBC had negative (1+) to equivocal (2+) HER2
expression in the absence of amplification (data not shown).
Accordingly, EMP3 warrants exploration as a potential drug target
for TNBC. Moreover, expression of HER3 confers HER2-mediated
tamoxifen resistance (49).
EMP3-targeted agents may have potential as HER2- or HR-targeting
follow-up therapy for patients who fail to respond to contemporary
regimens.
In conclusion, EMP3 served multiple roles in
regulation of the HER family and HR expression via activation of
mitogenic signaling pathways in BC in vitro. The potential
of EMP3-targeted therapy for BC was supported by chemosensitivity
assays and in vitro and in vivo experiments. Further
investigations are key for determining the benefit of EMP3-targeted
therapy for patients with BC with co-expressed EMP3 and HER2.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by Taiwan National Science and
Technology Council (grant nos. NSC 105-2320-B-006-029-MY3 and
NSC-108-2320-B-006-050-MY3) and China Medical University Hospital
(grant nos. C1130502012 and C1130528047).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YWW, CTL and NHC conceived and designed the project.
YLT, YLC and CAC performed experiments. YWW acquired the data. HYC,
YLC, HWC and NHC provided technical support. HYC, JYW, CLH and NHC
analyzed and interpreted the data. HWC interpreted data. HYC, YWW,
NHC wrote the manuscript. All authors read and approved the final
manuscript. NHC and CTL confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
The present study was approved by the National Cheng
Kung University Hospital Laboratory Animal Care and User Committee
(IACUC Approval No. 108075).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
BC
|
breast cancer
|
|
HER2
|
human epidermal growth factor 2
|
|
HR
|
hormone receptor
|
|
PR
|
progesterone receptor
|
|
EMP
|
epithelial membrane protein
|
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Testa U, Castelli G and Pelosi E: Breast
cancer: A molecularly heterogenous disease needing subtype-specific
treatments. Med Sci (Basel). 8:182020.PubMed/NCBI
|
|
3
|
Waks AG and Winer EP: Breast cancer
treatment: A review. JAMA. 321:288–300. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Harbeck N, Penault-Llorca F, Cortes J,
Gnant M, Houssami N, Poortmans P, Ruddy K, Tsang J and Cardoso F:
Breast cancer. Nat Rev Dis Primers. 5:662019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Spring LM, Gupta A, Reynolds KL, Gadd MA,
Ellisen LW, Isakoff SJ, Moy B and Bardia A: Neoadjuvant endocrine
therapy for estrogen receptor-positive breast cancer: A systematic
review and meta-analysis. JAMA Oncol. 2:1477–1486. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Swain SM, Shastry M and Hamilton E:
Targeting HER2-positive breast cancer: Advances and future
directions. Nat Rev Drug Discov. 22:101–126. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tesch ME and Gelmon KA: Targeting HER2 in
breast cancer: Latest developments on treatment sequencing and the
introduction of biosimilars. Drugs. 80:1811–1830. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hanker AB, Sudhan DR and Arteaga CL:
Overcoming endocrine resistance in breast cancer. Cancer Cell.
37:496–513. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nayar U, Cohen O, Kapstad C, Cuoco MS,
Waks AG, Wander SA, Painter C, Freeman S, Persky NS, Marini L, et
al: Acquired HER2 mutations in ER+ metastatic breast
cancer confer resistance to estrogen receptor-directed therapies.
Nat Genet. 51:207–216. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pegram M, Jackisch C and Johnston SRD:
Estrogen/HER2 receptor crosstalk in breast cancer: Combination
therapies to improve outcomes for patients with hormone
receptor-positive/HER2-positive breast cancer. NPJ Breast Cancer.
9:452023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang YW, Cheng HL, Ding YR, Chou LH and
Chow NH: EMP1, EMP 2, and EMP3 as novel therapeutic targets in
human cancer. Biochim Biophys Acta Rev Cancer. 1868:199–211. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li L, Xia S, Zhao Z, Deng L, Wang H, Yang
D, Hu Y, Ji J, Huang D and Xin T: EMP3 as a prognostic biomarker
correlates with EMT in GBM. BMC Cancer. 24:892024. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ma Q, Zhang Y, Liang H, Zhang F, Liu F,
Chen S, Hu Y, Jiang L, Hao Y, Li M and Liu Y: EMP3 as a key
downstream target of miR-663a regulation interferes with MAPK/ERK
signaling pathway to inhibit gallbladder cancer progression. Cancer
Lett. 575:2163982023. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim EK and Koo JS: Expression of
epithelial membrane protein (EMP) 1, EMP 2, and EMP 3 in thyroid
cancer. Histol Histopathol. 37:51–61. 2022.PubMed/NCBI
|
|
15
|
Zhou W, Jiang Z, Li X, Xu F, Liu Y, Wen P,
Kong L, Hou M and Yu J: EMP3 overexpression in primary breast
carcinomas is not associated with epigenetic aberrations. J Korean
Med Sci. 24:97–103. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Evtimova V, Zeillinger R and Weidle UH:
Identification of genes associated with the invasive status of
human mammary carcinoma cell lines by transcriptional profiling.
Tumour Biol. 24:189–198. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hong XC, Fen YJ, Yan GC, Hong H, Yan CH,
Bing LW and Zhong YH: Epithelial membrane protein 3 functions as an
oncogene and is regulated by microRNA-765 in primary breast
carcinoma. Mol Med Rep. 12:6445–6450. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Blick T, Hugo H, Widodo E, Waltham M,
Pinto C, Mani SA, Weinberg RA, Neve RM, Lenburg ME and Thompson EW:
Epithelial mesenchymal transition traits in human breast cancer
cell lines parallel the CD44(hi/)CD24 (lo/-) stem cell phenotype in
human breast cancer. J Mammary Gland Biol Neoplasia. 15:235–252.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kohn KW, Zeeberg BM, Reinhold WC and
Pommier Y: Gene expression correlations in human cancer cell lines
define molecular interaction networks for epithelial phenotype.
PLoS One. 9:e992692014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Isbilen M, Mert Senses K and Osmay Gure A:
Predicting chemotherapy sensitivity profiles for breast cancer cell
lines with and without stem cell-like features. Curr Signal
Transduct Ther. 8:268–273. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou K, Sun Y, Dong D, Zhao C and Wang W:
EMP3 negatively modulates breast cancer cell DNA replication, DNA
damage repair, and stem-like properties. Cell Death Dis.
12:8442021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mackay A, Jones C, Dexter T, Silva RL,
Bulmer K, Jones A, Simpson P, Harris RA, Jat PS, Neville AM, et al:
cDNA microarray analysis of genes associated with ERBB2 (HER2/neu)
overexpression in human mammary luminal epithelial cells. Oncogene.
22:2680–2688. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chakrabarty A, Bhola NE, Sutton C, Ghosh
R, Kuba MG, Dave B, Chang JC and Arteaga CL: Trastuzumab-resistant
cells rely on a HER2-PI3K-FoxO-survivin axis and are sensitive to
PI3K inhibitors. Cancer Res. 73:1190–1200. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang YW, Li WM, Wu WJ, Chai CY, Liu HS,
Lai MD and Chow NH: Potential significance of EMP3 in patients with
upper urinary tract urothelial carcinoma: Crosstalk with
ErbB2-PI3K-Akt pathway. J Urol. 192:242–251. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cha YJ and Koo JS: Expression and role of
epithelial membrane proteins in tumorigenesis of hormone
receptor-positive breast cancer. J Breast Cancer. 23:385–397. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Animal Protection Act. Ministry of
Agriculture, ROC (Taiwan), . 2021.https://law.moj.gov.tw/ENG/LawClass/LawAll.aspx?pcode=M0060027
|
|
27
|
Wolff AC, Hammond MEH, Allison KH, Harvey
BE, Mangu PB, Bartlett JMS, Bilous M, Ellis IO, Fitzgibbons P,
Hanna W, et al: Human epidermal growth factor receptor 2 testing in
breast cancer: American society of clinical oncology/college of
American pathologists clinical practice guideline focused update.
Arch Pathol Lab Med. 142:1364–1382. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gemmete JJ and Mukherji SK: Trastuzumab
(herceptin). AJNR Am J Neuroradiol. 32:1373–1374. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hsieh YH, Hsieh SC, Lee CH, Yang SF, Cheng
CW, Tang MJ, Lin CL, Lin CL and Chou RH: Targeting EMP3 suppresses
proliferation and invasion of hepatocellular carcinoma cells
through inactivation of PI3K/Akt pathway. Oncotarget.
6:34859–34874. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Christians A, Poisel E, Hartmann C, von
Deimling A and Pusch S: Characterization of the epithelial membrane
protein 3 interaction network reveals a potential functional link
to mitogenic signal transduction regulation. Int J Cancer.
145:461–473. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kar R, Jha NK, Jha SK, Sharma A, Dholpuria
S, Asthana N, Chaurasiya K, Singh VK, Burgee S and Nand P: A
‘NOTCH’ deeper into the epithelial-to-mesenchymal transition (EMT)
program in breast cancer. Genes (Basel). 10:9612019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee KL, Kuo YC, Ho YS and Huang YH:
Triple-negative breast cancer: Current understanding and future
therapeutic breakthrough targeting cancer stemness. Cancers
(Basel). 11:13342019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Derakhshani A, Rezaei Z, Safarpour H,
Sabri M, Mir A, Sanati MA, Vahidian F, Gholamiyan Moghadam A,
Aghadoukht A, Hajiasgharzadeh K and Baradaran B: Overcoming
trastuzumab resistance in HER2-positive breast cancer using
combination therapy. J Cell Physiol. 235:3142–3156. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Martija AA and Pusch S: The
multifunctional role of EMP3 in the regulation of membrane
receptors associated with IDH-wild-type glioblastoma. Int J Mol
Sci. 22:52612021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Martija AA, Krauß A, Bächle N, Doth L,
Christians A, Krunic D, Schneider M, Helm D, Will R, Hartmann C, et
al: EMP3 sustains oncogenic EGFR/CDK2 signaling by restricting
receptor degradation in glioblastoma. Acta Neuropathol Commun.
11:1772023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kumagai S, Koyama S and Nishikawa H:
Antitumour immunity regulated by aberrant ERBB family signalling.
Nat Rev Cancer. 21:181–197. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kusumoto Y, Okuyama H, Shibata T, Konno K,
Takemoto Y, Maekawa D, Kononaga T, Ishii T, Akashi-Takamura S,
Saitoh SI, et al: Epithelial membrane protein 3 (Emp3)
downregulates induction and function of cytotoxic T lymphocytes by
macrophages via TNF-α production. Cell Immunol. 324:33–41. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sun M, Jiang H, Liu T, Tan X, Jiang Q, Sun
B, Zheng Y, Wang G, Wang Y, Cheng M, et al: Structurally defined
tandem-responsive nanoassemblies composed of dipeptide-based
photosensitive derivatives and hypoxia-activated camptothecin
prodrugs against primary and metastatic breast tumors. Acta Pharm
Sin B. 12:952–966. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zeng R, Li Y, Li Y, Wan Q, Huang Z, Qiu Z
and Tang D: Smartphone-based photoelectrochemical immunoassay with
simpleCo9S8@ZnIn2S4 for
point-of-care diagnosis of breast cancer biomarker. Research (Wash
D C). 2022:98315212022.PubMed/NCBI
|
|
41
|
Chen H, Li H, Shi W, Qin H and Zheng L:
The roles of m6A RNA methylation modification in cancer stem cells:
New opportunities for cancer suppression. Cancer Insight. 1:1–18.
2022.
|
|
42
|
Yin Y, Yan Y, Fan B, Huang W, Zhang J, Hu
HY, Li X, Xiong D, Chou SL, Xiao Y and Wang H: Novel combination
therapy for triple-negative breast cancer based on an intelligent
hollow carbon sphere. Research (Wash DC). 6:00982023.PubMed/NCBI
|
|
43
|
Ji X, Tian X, Feng S, Zhang L, Wang J, Guo
R, Zhu Y, Yu X, Zhang Y, Du H, et al: Intermittent F-actin
perturbations by magnetic fields inhibit breast cancer metastasis.
Research (Wash DC). 6:00802023.PubMed/NCBI
|
|
44
|
Zheng C, Zhang D, Kong Y, Niu M, Zhao H,
Song Q, Feng Q, Li X and Wang L: Dynamic regulation of drug
biodistribution by turning tumors into decoys for biomimetic
nanoplatform to enhance the chemotherapeutic efficacy of breast
cancer with bone metastasis. Exploration (Beijing). 3:202201242023.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cheng H, Ballman K, Vassilakopoulou M,
Dueck AC, Reinholz MM, Tenner K, Gralow J, Hudis C, Davidson NE,
Fountzilas G, et al: EGFR expression is associated with decreased
benefit from trastuzumab in the NCCTG N9831 (alliance) trial. Br J
Cancer. 111:1065–1071. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yang L, Li Y, Shen E, Cao F, Li L, Li X,
Wang X, Kariminia S, Chang B, Li H and Li Q: NRG1-dependent
activation of HER3 induces primary resistance to trastuzumab in
HER2-overexpressing breast cancer cells. Int J Oncol. 51:1553–1562.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Watanabe S, Yonesaka K, Tanizaki J,
Nonagase Y, Takegawa N, Haratani K, Kawakami H, Hayashi H, Takeda
M, Tsurutani J and Nakagawa K: Targeting of the HER2/HER3 signaling
axis overcomes ligand-mediated resistance to trastuzumab in
HER2-positive breast cancer. Cancer Med. 8:1258–1268. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ogden A, Bhattarai S, Sahoo B, Mongan NP,
Alsaleem M, Green AR, Aleskandarany M, Ellis IO, Pattni S, Li XB,
et al: Combined HER3-EGFR score in triple-negative breast cancer
provides prognostic and predictive significance superior to
individual biomarkers. Sci Rep. 10:30092020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu B, Ordonez-Ercan D, Fan Z, Edgerton
SM, Yang X and Thor AD: Downregulation of erbB3 abrogates
erbB2-mediated tamoxifen resistance in breast cancer cells. Int J
Cancer. 120:1874–1882. 2007. View Article : Google Scholar : PubMed/NCBI
|