Introduction
Colorectal adenocarcinoma is the second leading
cause of malignancy-related mortality worldwide (1,2), with
a continuously increasing prevalence. Several factors and genes,
regulating a number of pathways, are involved in colorectal
tumorigenesis. Therefore, it is important to investigate the roles
of these factors and genes in colorectal adenocarcinoma to promote
early diagnosis, therapeutic development and cancer prevention.
A total of 86 differentially expressed sequence tags
(dbESTs) from human colorectal adenocarcinoma tissues were
previously identified by cDNA subtractive library construction and
cDNA microarray analysis (3,4). One
of these dbESTs, ES274071 (GenBank accession no. NM003011.3; gene
name, set) was selected for further investigation. The
set gene is located on human chromosome 9p34 and encodes a
277-amino acid protein with a molecular weight of 39 kDa. The
set gene was previously found to be involved in the
progression of leukemia and ovarian cancer. In a previous study by
our group, quantitative reverse transcription polymerase chain
reaction (qRT-PCR) analysis revealed that the mRNA level of
set in colorectal cancer tissues was higher compared to that
in the adjacent normal colorectal tissues, suggesting the
involvement of set in the development of human colorectal
adenocarcinoma (5).
The PI3K/Akt/mammalian target of rapamycin (mTOR)
pathway plays a pivotal role in the development and progression of
colorectal cancer by regulating cancer cell proliferation,
resistance to apoptosis, angiogenesis and metastasis (6). The mTOR protein is a key kinase
downstream of the growth factor receptor PI3K and the Akt signaling
pathway, which are both involved in the modulation of cell growth,
survival, metabolism and proliferation (7). Significant advancements have been made
in elucidating the role of mTOR in cancer development and
progression. Activation of the mTOR signaling pathway is often the
result of genetic alterations of negative regulators of mTOR, such
as phosphatase and tensin homolog (PTEN), tuberous sclerosis
complex (TSC) 1 and TSC2 (8). It
was demonstrated that the activation of the PI3K/Akt/mTOR pathway
correlates with tumor progression and poor survival in a variety of
tumors (9,10), suggesting that mTOR is a promising
molecular target for colorectal cancer therapy. The mTOR inhibitor
rapamycin is a natural macrolide antibiotic isolated from
Streptomyces hygroscopicus. This molecule binds to FKBP12
(FK506-binding protein) and the resulting complex inhibits the
protein kinase activity of mTOR. Rapamycin was originally used as
an antifungal and immunosuppressive agent. However, the subsequent
identification of the inherent antiproliferative properties of
rapamycin led to the investigation of this compound as a potential
anticancer agent (11). Therefore,
in order to elucidate the association of set with the
PI3K/Akt/mTOR pathway and their role in the development and
progression of colorectal cancer, the effect of rapamycin treatment
on set expression in colorectal cancer cells was
investigated.
Materials and methods
Cell lines and culture conditions
The SW480 and LoVo human colorectal carcinoma cell
lines (obtained from the American Type Culture Collection,
Manassas, VA, USA) were cultured in Dulbecco’s modified Eagle’s
medium (DMEM; HyClone Laboratories, Inc., Logan, UT, USA)
containing 10% fetal bovine serum, penicillin (100 IU/ml) and
streptomycin (100 μg/ml). Cells were grown at 37°C in a humidified
5% CO2 atmosphere. Experiments were performed using
cells harvested from exponentially growing cultures.
Drug
Rapamycin stock solutions (5 mg/ml; Fermentek Ltd.,
Jerusalem, Israel) were prepared in dimethyl sulfoxide (DMSO). The
solutions were stored at −20°C prior to use and were diluted in
DMEM to six different concentrations for subsequent
experiments.
In vitro cell assays
The rapamycin stock solutions were diluted in DMEM
to concentrations of 0.05, 0.1, 0.2, 0.5, 1 and 10 μM. DMSO was
used as the vehicle control, at a final concentration of 0.1%.
Cells were treated with rapamycin for 48 h.
RNA isolation
The TRIzol reagent (Invitrogen Life Technologies,
Carlsbad, CA, USA) was employed for total RNA isolation as
described in the manufacturer’s protocol. Total RNA yield was
determined by measuring the absorbance at 260 nm using a
spectrophotometer. The quality of RNA products was confirmed by the
presence of sharp bands representing the 28S and 18S rRNA molecules
in a 1% agarose gel, with an expected 28S:18S intensity ratio of
2:1.
First-strand cDNA synthesis
Total RNA isolated from each sample was treated with
DNase I to eliminate genomic DNA contamination prior to reverse
transcription (RT). The RT reaction was performed in a 20-μl volume
using the M-MuLV Reverse Transcriptase kit (Fermentas, Waltham, MA,
USA) for first-strand cDNA synthesis under the recommended
conditions. The synthesized cDNA product was either immediately
used for qRT-PCR or stored at −20°C for subsequent use.
qRT-PCR
The qRT-PCR reaction was performed following cDNA
amplification using SYBR Premix Ex Taq (Takara Bio Inc., Shiga,
Japan) and the primers were listed in Table I on a Bio-Rad C1000 real-time system
(Bio-Rad, Hercules, CA, USA) according to the manufacturer’s
instructions and international standards (12). For each reaction, 2 μl of cDNA
obtained from 1 μg of RNA template were used. The thermal cycling
conditions were as follows: an initial denaturation step at 95°C
for 30 sec, followed by 40 cycles of denaturation at 95°C for 5
sec, annealing at 62°C for 30 sec and elongation at 72°C for 30
sec. GAPDH was used as an internal control. The amplified
cDNA product was quantified using the ΔΔCt method. The
primers for set amplification were designed with Primer
Premier 5.0 software (Premier Biosoft International, Palo Alto, CA,
USA) to target its open reading frame.
 | Table IPrimers. |
Table I
Primers.
| Gene | Primer sequence
(5′-3′) |
|---|
| set | F:
GCTCAACTCCAACCACGAC |
| R:
TCCTCACTGGCTTGTTCATTA |
| GAPDH | F:
GGAAGGTGAAGGTCGGAGT |
| R:
TGAGGTCAATGAAGGGGTC |
Western blot assay
Cells were treated with 10 μM rapamycin for 48 h.
DMSO was used as the vehicle control, at a final concentration of
0.1%. Cell lysates were denatured in sample buffer containing
sodium dodecyl sulphate (SDS). The same amount of denatured protein
(30 μg) was loaded into each lane and proteins were separated by
12% SDS-PAGE. Following this step, the proteins were transferred to
PVDF membranes (Millipore, Billerica, MA, USA). After blocking for
1 h in Tris-buffered saline containing 0.1% Tween-20 and 3% bovine
serum albumin, the membranes were incubated overnight at 4°C with
an anti-SET primary antibody (1:200; Santa Cruz Biotechnology,
Inc., Santa Cruz, CA, USA). The membranes were then incubated with
an appropriate horseradish peroxidase-conjugated secondary antibody
and the corresponding protein product was visualized using ECL
reagent (Thermo Fisher Scientific, Waltham, MA, USA).
Statistical analysis
Statistical analysis was performed using the t-test
with SPSS software, version 19.0 (SPSS Inc., Chicago, IL, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
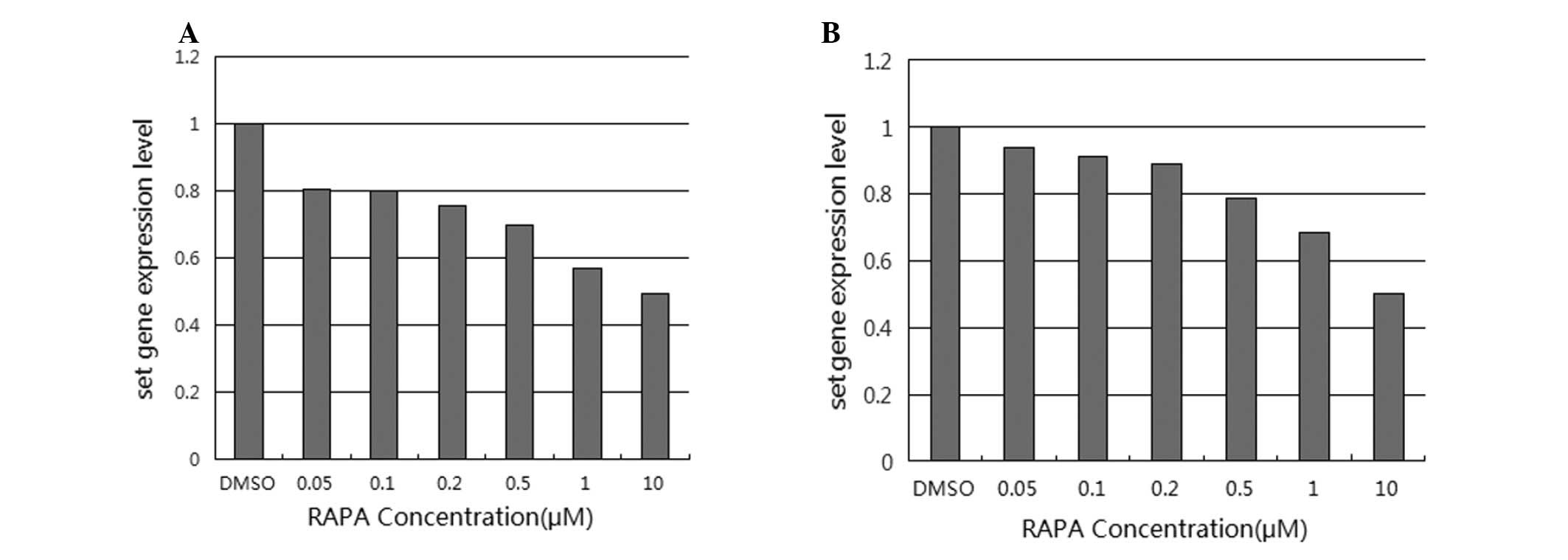
As shown in Fig. 1,
the set mRNA expression decreased with the increasing
concentration of rapamycin, as compared to cells treated with DMSO
alone. In SW480 cells, the set mRNA expression levels
relative to the control DMSO were 80.46, 80.18, 75.85, 69.95, 57.2
and 49.65%, for rapamycin concentrations ranging from 0.05 to 10
μM. In this case, only the downregulation in expression observed
with the two highest concentrations (1 and 10 μM) were considered
statistically significant according to the t-test analysis
(P=0.033). The corresponding values in LoVo cells were 93.8, 91.41,
89.18, 78.73, 68.43 and 50.35%, respectively; for this cell type,
only the downregulation in response to the highest concentration
(10 μM) was considered statistically significant according to the
t-test analysis (P=0.027).
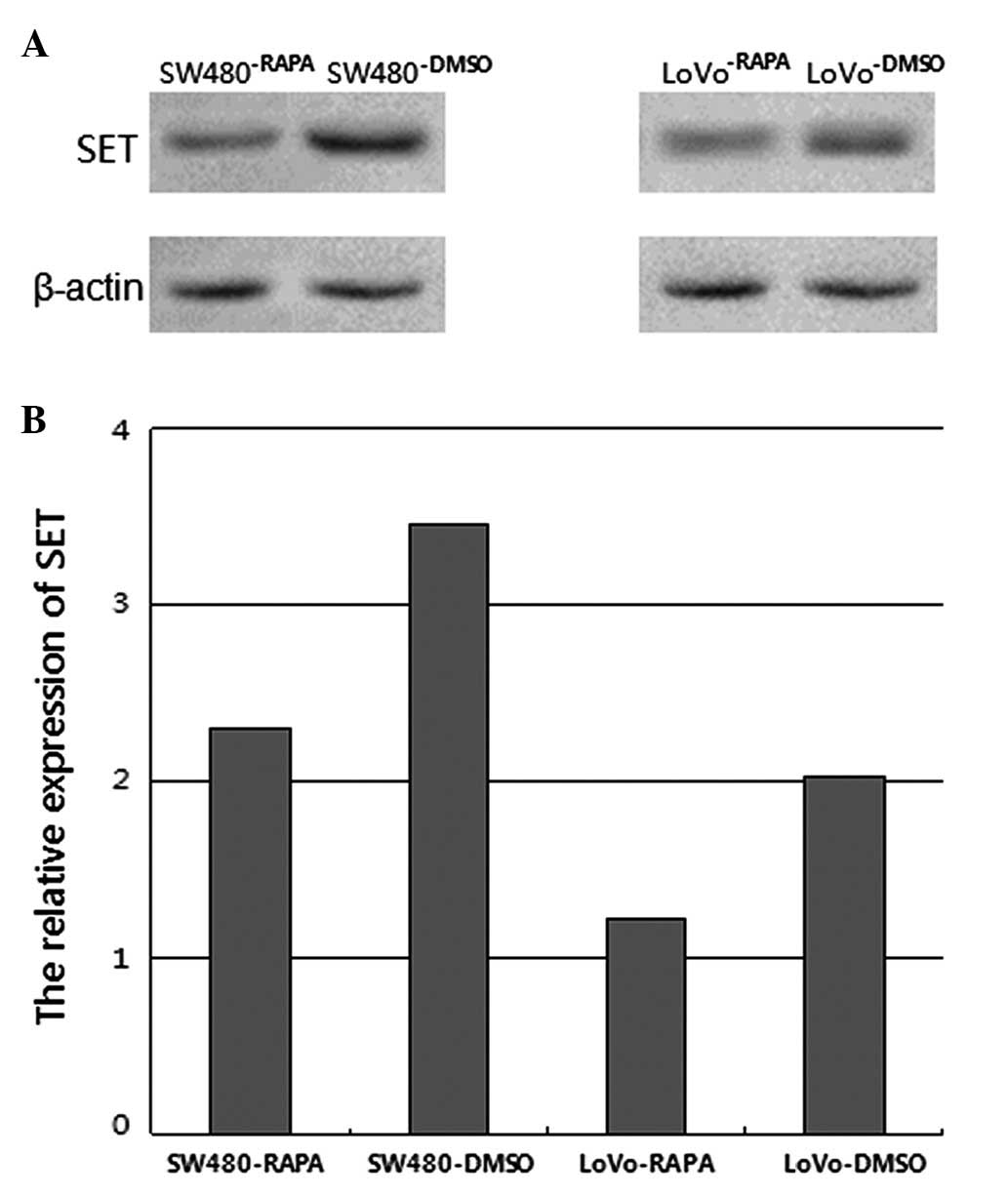
As shown in Fig. 2,
following treatment with 10 μM rapamycin for 48 h, the SET protein
expression was downregulated in SW480 and LoVo cells treated with
rapamycin, as compared to the cells treated with the vehicle
control. The relative expression levels of SET were 46.45%
(P=0.023) and 39.93%, respectively (P=0.022).
Discussion
In a previous study, qPCR analysis demonstrated that
set gene expression was significantly upregulated in human
colorectal adenocarcinoma tissues, as compared to adjacent normal
colorectal tissues. Although there was no significant correlation
between set gene expression and patient characteristics such
as gender, age, Dukes’ stage, or tumor differentiation, a higher
rate was observed in Dukes’ C (70.59%) and D stages (71.43%)
compared to Dukes’ B stage (50%). This observation was in
accordance with the findings of Ouellet on human ovarian cancer
(13). The inhibition of set
gene expression effectively suppressed cell proliferation and
promoted cell apoptosis, suggesting that the set gene is
involved in the development of human colorectal adenocarcinoma
(5). In the present study, we
demonstrated that rapamycin treatment was able to induce set
downregulation in human colorectal adenocarcinoma cell lines at the
mRNA and protein levels.
The set gene was initially identified by von
Lindern et al(14) in a
patient with acute undifferentiated leukemia. A translocation of
chromosome 9 resulted in the generation of the SET-CAN fusion
protein. The set gene is crucial in the progression of acute
undifferentiated leukemia, possibly by activating CAN in the
nucleus and stimulating the transformation potential of SET-CAN.
Cristóbal et al(15)
suggested that set overexpression may be a key mechanism
responsible for the inhibition of protein phosphatase 2A in acute
myeloid leukemia. Almeida et al(16) assessed the effect of the
overexpression of the SET protein, a histone acetylation modulator
accumulated in head and neck squamous cell carcinoma, on the gene
regulation and protein activity of aldehyde dehydrogenase 2 and
glutathione S-transferase P1. Leopoldino et al(17) reported that accumulated set
may act via the Akt/PTEN pathway as a cell survival signal or as an
oxidative stress sensor promoting cell death. The studies mentioned
above indicated that set may be involved in the development
of several malignant tumors.
The mTOR protein is a serine/threonine protein
kinase involved in a nutrient-sensitive signaling pathway that is
crucial in regulating cell growth and proliferation. The mTOR
pathway is activated in various cell processes, including
tumorigenesis, insulin resistance, adipogenesis, angiogenesis and
T-lymphocyte activation. In addition, the mTOR pathway is
associated with various human diseases, such as cancer, obesity and
type 2 diabetes (7). The activation
of the PI3K/Akt/mTOR pathway inhibits apoptosis induced by several
types of stimuli, thereby promoting cell cycle progression, cell
survival and proliferation, which are important for tumor invasion
and metastasis. Moreover, the role of the PI3K/Akt/mTOR pathway in
neovascularization may be another mechanism through which it
promotes tumorigenesis. The PI3K/Akt/mTOR pathway was also found to
be involved in reactive oxygen species-induced and integrin
β3-mediated migration and invasion of colorectal cancer cells
(18). Kim et al(19) reviewed the key components of the
PI3K/Akt/mTOR pathway, their molecular alterations and the
inhibitors targeting the mTOR pathway in colorectal cancer.
It was reported that rapamycin is able to induce
apoptosis, suggesting a potential role of the mTOR pathway in
regulating cell survival (20).
Moreover, rapamycin has been shown to be effective in the clinical
treatment of several types of cancer. For example, Boffa et
al(21) demonstrated that
rapamycin treatment may inhibit cancer cell growth and metastatic
progression of non-small-cell lung carcinoma. Samkari et
al(22) demonstrated that
rapamycin treatment may induce the expression of the anti-apoptotic
protein survivin in neuroblastoma cells. Kim et al(19) suggested that mTOR inhibitors may be
attractive antitumor drugs, which may be used in combination with
other treatment strategies.
In the present study, we demonstrated that rapamycin
treatment was able to induce set downregulation in the SW480
and LoVo human colorectal adenocarcinoma cell lines at the mRNA and
protein levels. The downregulation of set mRNA expression
was observed with rapamycin treatment at all six tested
concentrations, ranging from 0.05 to 10 μM. However, in SW480
cells, only the downregulation in response to the highest
concentrations (1 and 10 μM) was considered statistically
significant. The same effect was observed in LoVo cells, with only
the downregulation in response to the highest concentration (10 μM)
being statistically significant. Thus, the effective concentration
of rapamycin for significant set downregulation was 10 μM in
the two cell lines. In the present study, we have demonstrated that
rapamycin was able to inhibit the expression of the set gene
in human colorectal adenocarcinoma cell lines. The dose-dependent
association between set gene expression and rapamycin
treatment suggests the potential involvement of set in the
PI3K/Akt/mTOR pathway in colorectal adenocarcinoma cells, possibly
through the regulation of mTOR activity. The set gene may
act as an oncogene in colorectal adenocarcinoma by inhibiting cell
apoptosis, accelerating cell cycle progression and promoting
metastasis through the activation of the PI3K/Akt/mTOR pathway,
which is opposite to the effects of rapamycin. Furthermore, the
downregulation of set by rapamycin treatment suggests that
rapamycin may be used as an antineoplastic drug in colorectal
adenocarcinoma therapy. The PI3K/Akt/mTOR pathway may be a
diagnostic marker and molecular target in colorectal
adenocarcinoma. However, further investigation is required to
comprehensively analyze the functions of set and the
PI3K/Akt/mTOR pathway in carcinogenesis.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant no. 81072023).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Herrinton LJ, Liu L, Levin TR, Allison JE,
Lewis JD and Velayos F: Incidence and mortality of colorectal
adenocarcinoma in persons with inflammatory bowel disease from 1998
to 2010. Gastroenterology. 143:382–389. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen Y, Zhang Y, Zhou Z, Wang G and Yi Z:
Identification of differentially expressed genes in human
colorectal adenocarcinoma. World J Gastroenterol. 12:1025–1032.
2006.
|
|
4
|
Zhang C and Chen Y: Electronic cloning and
validating of the suppression subtractive hybridization EST
ES274070 of human colorectal adenocarcinoma. US Chin J Lymphol
Oncol. 6:83–88. 2007.
|
|
5
|
Jiang Q, Zhang C, Zhu J, Chen Q and Chen
Y: The set gene is a potential oncogene in human colorectal
adenocarcinoma and oral squamous cell carcinoma. Mol Med Rep.
4:993–999. 2011.PubMed/NCBI
|
|
6
|
Zoncu R, Efeyan A and Sabatini DM: mTOR:
from growth signal integration to cancer, diabetes and ageing. Nat
Rev Mol Cell Biol. 12:21–35. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Laplante M and Sabatini DM: mTOR signaling
at a glance. J Cell Sci. 122:3589–3594. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Feng Z, Zhang H, Levine AJ and Jin S: The
coordinate regulation of the p53 and mTOR pathways in cells. Proc
Natl Acad Sci USA. 102:8204–8209. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gulhati P, Cai Q, Li J, et al: Targeted
inhibition of mammalian target of rapamycin signaling inhibits
tumorigenesis of colorectal cancer. Clin Cancer Res. 15:7207–7216.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chiang GG and Abraham RT: Targeting the
mTOR signaling network in cancer. Trends Mol Med. 13:433–442. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Miyake N, Chikumi H, Takata M, Nakamoto M,
Igishi T and Shimizu E: Rapamycin induces p53-independent apoptosis
through the mitochondrial pathway in non-small cell lung cancer
cells. Oncol Rep. 28:848–854. 2012.PubMed/NCBI
|
|
12
|
Bustin SA, Benes V, Garson JA, Hellemans
J, et al: The MIQE guidelines: minimum information for publication
of quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ouellet V, Le Page C, Guyot MC, Lussier C,
Tonin PN, Provencher DM and Mes-Masson AM: SET complex in serous
epithelial ovarian cancer. Int J Cancer. 119:2119–2126. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
von Lindern M, Van Baal S, Wiegant J, Raap
A, Hagemeijer A and Grosveld G: Can, a putative oncogene associated
with myeloid leukemogenesis, may be activated by fusion of its 3′
half to different genes: characterization of the set gene. Mol Cell
Biol. 12:3346–3355. 1992.PubMed/NCBI
|
|
15
|
Cristóbal I, Garcia-Orti L, Cirauqui C,
Cortes-Lavaud X, García-Sánchez MA, Calasanz MJ and Odero MD:
Overexpression of SET is a recurrent event associated with poor
outcome and contributes to protein phosphatase 2A inhibition in
acute myeloid leukemia. Haematologica. 97:543–550. 2012.PubMed/NCBI
|
|
16
|
Almeida LO, Goto RN, Pestana CR, Uyemura
SA, Gutkind S, Curti C and Leopoldino AM: SET overexpression
decreases cell detoxification efficiency: ALDH2 and GSTP1 are
downregulated, DDR is impaired and DNA damage accumulates. FEBS J.
279:4615–4628. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Leopoldino AM, Squarize CH, Garcia CB, et
al: Accumulation of the SET protein in HEK293T cells and mild
oxidative stress: cell survival or death signaling. Mol Cell
Biochem. 363:65–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lei Y, Huang K, Gao C, et al: Proteomics
identification of ITGB3 as a key regulator in reactive oxygen
species-induced migration and invasion of colorectal cancer cells.
Mol Cell Proteomics. 10:M110.0053972011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim DD and Eng C: The promise of mTOR
inhibitors in the treatment of colorectal cancer. Expert Opin
Investig Drugs. 21:1775–1788. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Thimmaiah KN, Easton J, Huang S, Veverka
KA, Germain GS, Harwood FC and Houghton PJ: Insulin-like growth
factor I-mediated protection from rapamycin-induced apoptosis is
independent of Ras-Erk1-Erk2 and phosphatidylinositol 3′-kinase-Akt
signaling pathways. Cancer Res. 63:364–374. 2003.PubMed/NCBI
|
|
21
|
Boffa DJ, Luan F, Thomas D, Yang h, Sharma
VK, Lagman M and Suthanthiran M: Rapamycin inhibits the growth and
metastatic progression of non-small cell lung cancer. Clin Cancer
Res. 10:293–300. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Samkari A, Cooper ZA, Holloway MP, Liu J
and Altura RA: Rapamycin induces the anti-apoptotic protein
survivin in neuroblastoma. Int J Biochem Mol Biol. 3:28–35.
2012.PubMed/NCBI
|