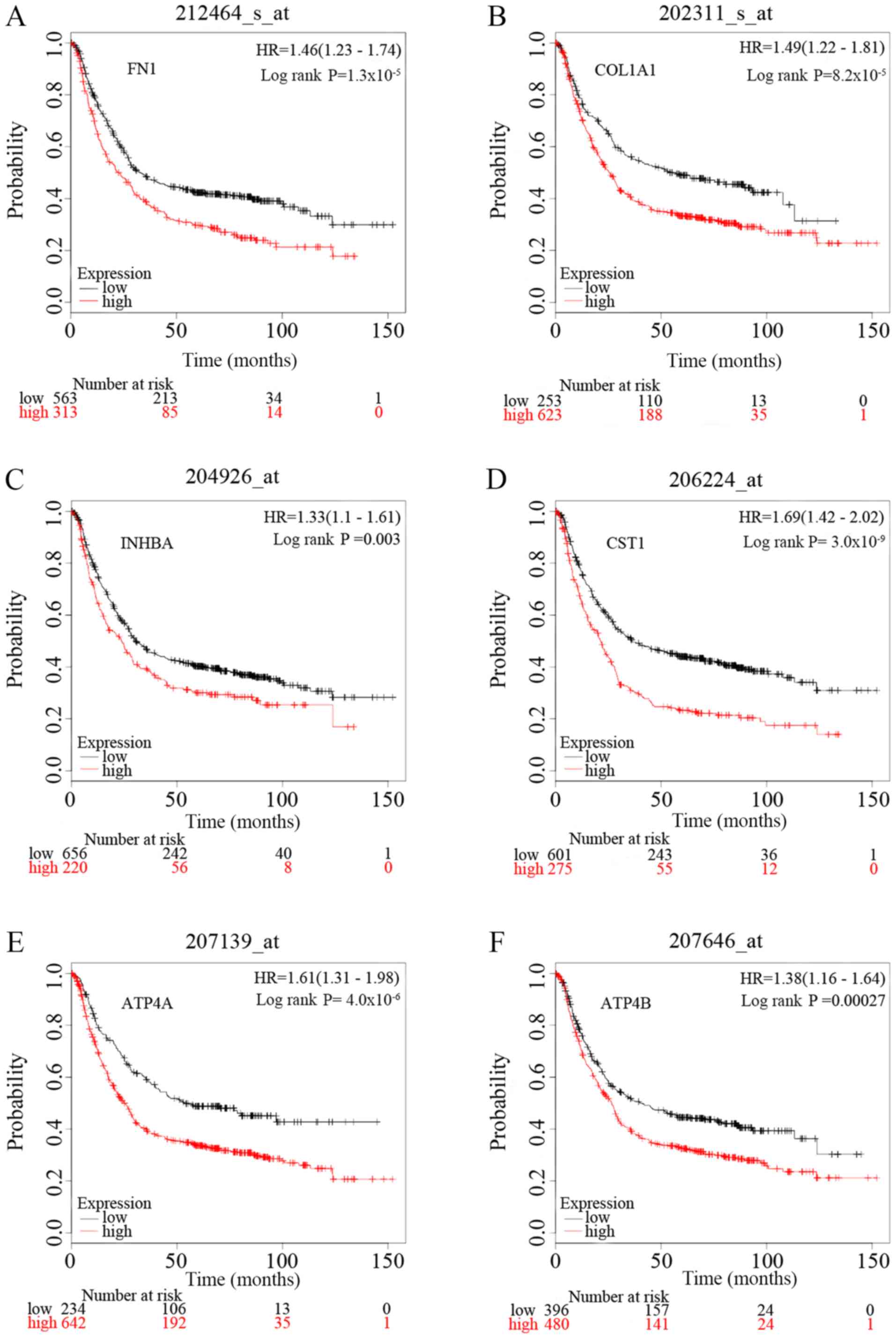
|
1
|
Shi J, Qu YP and Hou P: Pathogenetic
mechanisms in gastric cancer. World J Gastroenterol.
20:13804–13819. 2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Van Cutsem E, Sagaert X, Topal B,
Haustermans K and Prenen H: Gastric cancer. Lancet. 388:2654–2664.
2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
In H, Solsky I, Palis B, Langdon-Embry M,
Ajani J and Sano T: Validation of the 8th Edition of the AJCC TNM
staging system for gastric cancer using the national cancer
database. Ann Surg Oncol. 24:3683–3691. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Peng H, Deng Y, Wang L, Cheng Y, Xu Y,
Liao J and Wu H: Identification of potential biomarkers with
diagnostic value in pituitary adenomas using prediction analysis
for microarrays method. J Mol Neurosci. 69:399–410. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Wu Y, Jamal M, Xie T, Sun J, Song T, Yin
Q, Li J, Pan S, Zeng X, Xie S and Zhang Q: Uridine-cytidine kinase
2 (UCK2): A potential diagnostic and prognostic biomarker for lung
cancer. Cancer Sci. 110:2734–2747. 2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Chen Z, Zhou Y, Luo R, Liu K and Chen Z:
Trophinin-associated protein expression is an independent
prognostic biomarker in lung adenocarcinoma. J Thorac Dis.
11:2043–2050. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Li D, Lin C, Li N, Du Y, Yang C, Bai Y,
Feng Z, Su C, Wu R, Song S, et al: PLAGL2 and POFUT1 are regulated
by an evolutionarily conserved bidirectional promoter and are
collaboratively involved in colorectal cancer by maintaining
stemness. EBioMedicine. 45:124–138. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Yong L, YuFeng Z and Guang B: Association
between PPP2CA expression and colorectal cancer prognosis tumor
marker prognostic study. Int J Surg. 59:80–89. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Troiano G, Guida A, Aquino G, Botti G,
Losito NS, Papagerakis S, Pedicillo MC, Ionna F, Longo F, Cantile
M, et al: Integrative histologic and bioinformatics analysis of
BIRC5/Survivin expression in oral squamous cell carcinoma. Int J
Mol Sci. 19(E2664)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
D'Errico M, de Rinaldis E, Blasi MF, Viti
V, Falchetti M, Calcagnile A, Sera F, Saieva C, Ottini L, Palli D,
et al: Genome-wide expression profile of sporadic gastric cancers
with microsatellite instability. Eur J Cancer. 45:461–469.
2009.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wang Q, Wen YG, Li DP, Xia J, Zhou CZ, Yan
DW, Tang HM and Peng ZH: Upregulated INHBA expression is associated
with poor survival in gastric cancer. Med Oncol. 29:77–83.
2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Li L, Zhu Z, Zhao Y, Zhang Q, Wu X, Miao
B, Cao J and Fei S: FN1, SPARC, and SERPINE1 are highly expressed
and significantly related to a poor prognosis of gastric
adenocarcinoma revealed by microarray and bioinformatics. Sci Rep.
9(7827)2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000.PubMed/NCBI View Article : Google Scholar
|
|
15
|
The Gene Ontology Consortium: The Gene
Ontology Resource: 20 years and still GOing strong. Nucleic Acids
Res 47: D330.D338, 2019.
|
|
16
|
Kanehisa M: ‘Post-genome Informatics’,
Oxford University Press (2000). https://www.kanehisa.jp/docs/archive/PGI-contents.html.
|
|
17
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Xu W, Wang Y, Wang Y, Lv S, Xu X and Dong
X: Screening of differentially expressed genes and identification
of NUF2 as a prognostic marker in breast cancer. Int J Mol Med.
44:390–404. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613.
2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol 8 (Suppl 4): S11, 2014.
|
|
23
|
Liu Y, Cui S, Li W, Zhao Y, Yan X and Xu
J: PAX3 is a biomarker and prognostic factor in melanoma: Database
mining. Oncol Lett. 17:4985–4993. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Nagy Á, Lánczky A, Menyhárt O and Győrffy
B: Validation of miRNA prognostic power in hepatocellular carcinoma
using expression data of independent datasets. Sci Rep.
8(9227)2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Rausei S, Ruspi L, Galli F, Pappalardo V,
Di Rocco G, Martignoni F, Frattini F, Rovera F, Boni L and Dionigi
G: Seventh tumor-node-metastasis staging of gastric cancer:
Five-year follow-up. World J Gastroenterol. 22:7748–7753.
2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Chen X, Leung SY, Yuen ST, Chu KM, Ji J,
Li R, Chan AS, Law S, Troyanskaya OG, Wong J, et al: Variation in
gene expression patterns in human gastric cancers. Mol Biol Cell.
14:3208–3215. 2003.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Cho JY, Lim JY, Cheong JH, Park YY, Yoon
SL, Kim SM, Kim SB, Kim H, Hong SW, Park YN, et al: Gene expression
signature-based prognostic risk score in gastric cancer. Clin
Cancer Res. 17:1850–1857. 2011.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Cui J, Chen Y, Chou WC, Sun L, Chen L, Suo
J, Ni Z, Zhang M, Kong X, Hoffman LL, et al: An integrated
transcriptomic and computational analysis for biomarker
identification in gastric cancer. Nucleic Acids Res. 39:1197–207.
2011.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Serra O, Galán M, Ginesta MM, Calvo M,
Sala N and Salazar R: Comparison and applicability of molecular
classifications for gastric cancer. Cancer Treat Rev. 77:29–34.
2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Gilkes DM, Semenza GL and Wirtz D: Hypoxia
and the extracellular matrix: Drivers of tumour metastasis. Nat Rev
Cancer. 14:430–439. 2014.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Climent M, Pera M, Aymar I, Ramón JM,
Grande L and Nogués X: Bone health in long-term gastric cancer
survivors: A prospective study of high-dose vitamin D
supplementation using an easy administration scheme. J Bone Miner
Metab. 36:462–469. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhou ZH, Ji CD, Xiao HL, Zhao HB, Cui YH
and Bian XW: Reorganized collagen in the tumor microenvironment of
gastric cancer and its association with prognosis. J Cancer.
8:1466–1476. 2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Jang M, Koh I, Lee SJ, Cheong JH and Kim
P: Droplet-based microtumor model to assess cell-ECM interactions
and drug resistance of gastric cancer cells. Sci Rep.
7(41541)2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Liu X and Chu KM: E-cadherin and gastric
cancer: Cause, consequence, and applications. Biomed Res Int.
2014(637308)2014.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Hayashi Y, Bardsley MR, Toyomasu Y,
Milosavljevic S, Gajdos GB, Choi KM, Reid-Lombardo KM, Kendrick ML,
Bingener-Casey J, Tang CM, et al: Platelet-derived growth factor
receptor-α regulates proliferation of gastrointestinal stromal
tumor cells with mutations in KIT by stabilizing ETV1.
Gastroenterology. 149:420–432.e16. 2015.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Rahbari NN, Kedrin D, Incio J, Liu H, Ho
WW, Nia HT, Edrich CM, Jung K, Daubriac J, Chen I, et al: Anti-VEGF
therapy induces ECM remodeling and mechanical barriers to therapy
in colorectal cancer liver metastases. Sci Transl Med.
8(360ra135)2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Andersen MK, Rise K, Giskeødegård GF,
Richardsen E, Bertilsson H, Størkersen Ø, Bathen TF, Rye M and
Tessem MB: Integrative metabolic and transcriptomic profiling of
prostate cancer tissue containing reactive stroma. Sci Rep.
8(14269)2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Bao Y, Wang L, Shi L, Yun F, Liu X, Chen
Y, Chen C, Ren Y and Jia Y: Transcriptome profiling revealed
multiple genes and ECM-receptor interaction pathways that may be
associated with breast cancer. Cell Mol Biol Lett.
24(38)2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zhu H, Chen H, Wang J, Zhou L and Liu S:
Collagen stiffness promoted non-muscle-invasive bladder cancer
progression to muscle-invasive bladder cancer. Onco Targets Ther.
12:3441–3457. 2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Jin GH, Xu W, Shi Y and Wang LB: Celecoxib
exhibits an anti-gastric cancer effect by targeting focal adhesion
and leukocyte transendothelial migration-associated genes. Oncol
Lett. 12:2345–2350. 2016.PubMed/NCBI View Article : Google Scholar
|
|
41
|
He WQ, Gu JW, Li CY, Kuang YQ, Kong B,
Cheng L, Zhang JH, Cheng JM and Ma Y: The PPI network and clusters
analysis in glioblastoma. Eur Rev Med Pharmacol Sci. 19:4784–4790.
2015.PubMed/NCBI
|
|
42
|
Miryala SK, Anbarasu A and Ramaiah S:
Discerning molecular interactions: A comprehensive review on
biomolecular interaction databases and network analysis tools.
Gene. 642:84–94. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Yan H, Zheng G, Qu J, Liu Y, Huang X,
Zhang E and Cai Z: Identification of key candidate genes and
pathways in multiple myeloma by integrated bioinformatics analysis.
J Cell Physiol. 234:23785–23797. 2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Zhang H, Sun Z, Li Y, Fan D and Jiang H:
MicroRNA-200c binding to FN1 suppresses the proliferation,
migration and invasion of gastric cancer cells. Biomed
Pharmacother. 88:285–292. 2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Kun-Peng Z, Chun-Lin Z, Xiao-Long M and
Lei Z: Fibronectin-1 modulated by the long noncoding RNA
OIP5-AS1/miR-200b-3p axis contributes to doxorubicin resistance of
osteosarcoma cells. J Cell Physiol. 234:6927–6939. 2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Liao YX, Zhang ZP, Zhao J and Liu JP:
Effects of fibronectin 1 on cell proliferation, senescence and
apoptosis of human glioma cells through the PI3K/AKT signaling
pathway. Cell Physiol Biochem. 48:1382–1396. 2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Cai X, Liu C, Zhang TN, Zhu YW, Dong X and
Xue P: Down-regulation of FN1 inhibits colorectal carcinogenesis by
suppressing proliferation, migration, and invasion. J Cell Biochem.
119:4717–4728. 2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Yan P, He Y, Xie K, Kong S and Zhao W: In
silico analyses for potential key genes associated with gastric
cancer. PeerJ. 6(e6092)2018.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Jiang K, Liu H, Xie D and Xiao Q:
Differentially expressed genes ASPN, COL1A1, FN1, VCAN and MUC5AC
are potential prognostic biomarkers for gastric cancer. Oncol Lett.
17:3191–3202. 2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Sun S, Wang Y, Wu Y, Gao Y, Li Q,
Abdulrahman AA, Liu XF, Ji GQ, Gao J, Li L, et al: Identification
of COL1A1 as an invasion-related gene in malignant astrocytoma. Int
J Oncol. 53:2542–2554. 2018.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Liu J, Shen JX, Wu HT, Li XL, Wen XF, Du
CW and Zhang GJ: Collagen 1A1 (COL1A1) promotes metastasis of
breast cancer and is a potential therapeutic target. Discov Med.
25:211–223. 2018.PubMed/NCBI
|
|
52
|
Huang C, Yang X, Han L, Fan Z, Liu B,
Zhang C and Lu T: The prognostic potential of alpha-1 type I
collagen expression in papillary thyroid cancer. Biochem Biophys
Res Commun. 515:125–132. 2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Zhang QN, Zhu HL, Xia MT, Liao J, Huang
XT, Xiao JW and Yuan C: A panel of collagen genes are associated
with prognosis of patients with gastric cancer and regulated by
microRNA-29c-3p: An integrated bioinformatics analysis and
experimental validation. Cancer Manag Res. 11:4757–4772.
2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Seder CW, Hartojo W, Lin L, Silvers AL,
Wang Z, Thomas DG, Giordano TJ, Chen G, Chang AC, Orringer MB, et
al: Upregulated INHBA expression may promote cell proliferation and
is associated with poor survival in lung adenocarcinoma. Neoplasia.
11:388–396. 2009.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Yang H, Wu J, Zhang J, Yang Z, Jin W, Li
Y, Jin L, Yin L, Liu H and Wang Z: Integrated bioinformatics
analysis of key genes involved in progress of colon cancer. Mol
Genet Genomic Med. 7(e00588)2019.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Chen ZL, Qin L, Peng XB, Hu Y and Liu B:
INHBA gene silencing inhibits gastric cancer cell migration and
invasion by impeding activation of the TGF-β signaling pathway. J
Cell Physiol. 234:18065–18074. 2019.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Katayama Y, Oshima T, Sakamaki K, Aoyama
T, Sato T, Masudo K, Shiozawa M, Yoshikawa T, Rino Y, Imada T and
Masuda M: Clinical significance of INHBA gene expression in
patients with gastric cancer who receive curative resection
followed by adjuvant S-1 chemotherapy. In Vivo. 31:565–571.
2017.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Dai DN, Li Y, Chen B, Du Y, Li SB, Lu SX,
Zhao ZP, Zhou AJ, Xue N, Xia TL, et al: Elevated expression of CST1
promotes breast cancer progression and predicts a poor prognosis. J
Mol Med (Berl). 95:873–886. 2017.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Tian A, Pu K, Li B, Li M, Liu X, Gao L and
Mao X: Weighted gene coexpression network analysis reveals hub
genes involved in cholangiocarcinoma progression and prognosis.
Hepatol Res. 49:1195–1206. 2019.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Kim J, Bae DH, Kim JH, Song KS, Kim YS and
Kim SY: HOXC10 overexpression promotes cell proliferation and
migration in gastric cancer. Oncol Rep. 42:202–212. 2019.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Jiang J, Liu HL, Tao L, Lin XY, Yang YD,
Tan SW and Wu B: Let-7d inhibits colorectal cancer cell
proliferation through the CST1/p65 pathway. Int J Oncol.
53:781–790. 2018.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Oh SS, Park S, Lee KW, Madhi H, Park SG,
Lee HG, Cho YY, Yoo J and Dong Kim K: Extracellular cystatin SN and
cathepsin B prevent cellular senescence by inhibiting abnormal
glycogen accumulation. Cell Death Dis. 8(e2729)2017.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Lin S, Lin B, Wang X, Pan Y, Xu Q, He JS,
Gong W, Xing R, He Y, Guo L, et al: Silencing of ATP4B of ATPase
H+/K+ transporting beta subunit by intragenic
epigenetic alteration in human gastric cancer cells. Oncol Res.
25:317–329. 2017.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Fei HJ, Chen SC, Zhang JY, Li SY, Zhang
LL, Chen YY, Chang CX and Xu CM: Identification of significant
biomarkers and pathways associated with gastric carcinogenesis by
whole genome-wide expression profiling analysis. Int J Oncol.
52:955–966. 2018.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Lozano-Pope I, Sharma A, Matthias M, Doran
KS and Obonyo M: Effect of myeloid differentiation primary response
gene 88 on expression profiles of genes during the development and
progression of Helicobacter-induced gastric cancer. BMC Cancer.
17(133)2017.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Di Mario F and Goni E: Gastric acid
secretion: Changes during a century. Best Pract Res Clin
Gastroenterol. 28:953–65. 2014.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Saha A, Hammond CE, Beeson C, Peek RM Jr
and Smolka AJ: Helicobacter pylori represses proton pump expression
and inhibits acid secretion in human gastric mucosa. Gut.
59:874–881. 2010.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Friis-Hansen L: Achlorhydria is associated
with gastric microbial overgrowth and development of cancer:
Lessons learned from the gastrin knockout mouse. Scand J Clin Lab
Invest. 66:607–621. 2006.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Sáenz JB and Mills JC: Acid and the basis
for cellular plasticity and reprogramming in gastric repair and
cancer. Nat Rev Gastroenterol Hepatol. 15:257–273. 2018.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Vinasco K, Mitchell HM, Kaakoush NO and
Castaño-Rodríguez N: Microbial carcinogenesis: Lactic acid bacteria
in gastric cancer. Biochim Biophys Acta Rev Cancer.
1872(188309)2019.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Ferreira RM, Pereira-Marques J,
Pinto-Ribeiro I, Costa JL, Carneiro F, Machado JC and Figueiredo C:
Gastric microbial community profiling reveals a dysbiotic
cancer-associated microbiota. Gut. 67:226–236. 2018.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Wang LL, Liu JX, Yu XJ, Si JL, Zhai YX and
Dong QJ: Microbial community reshaped in gastric cancer. Eur Rev
Med Pharmacol Sci. 22:7257–7264. 2018.PubMed/NCBI View Article : Google Scholar
|