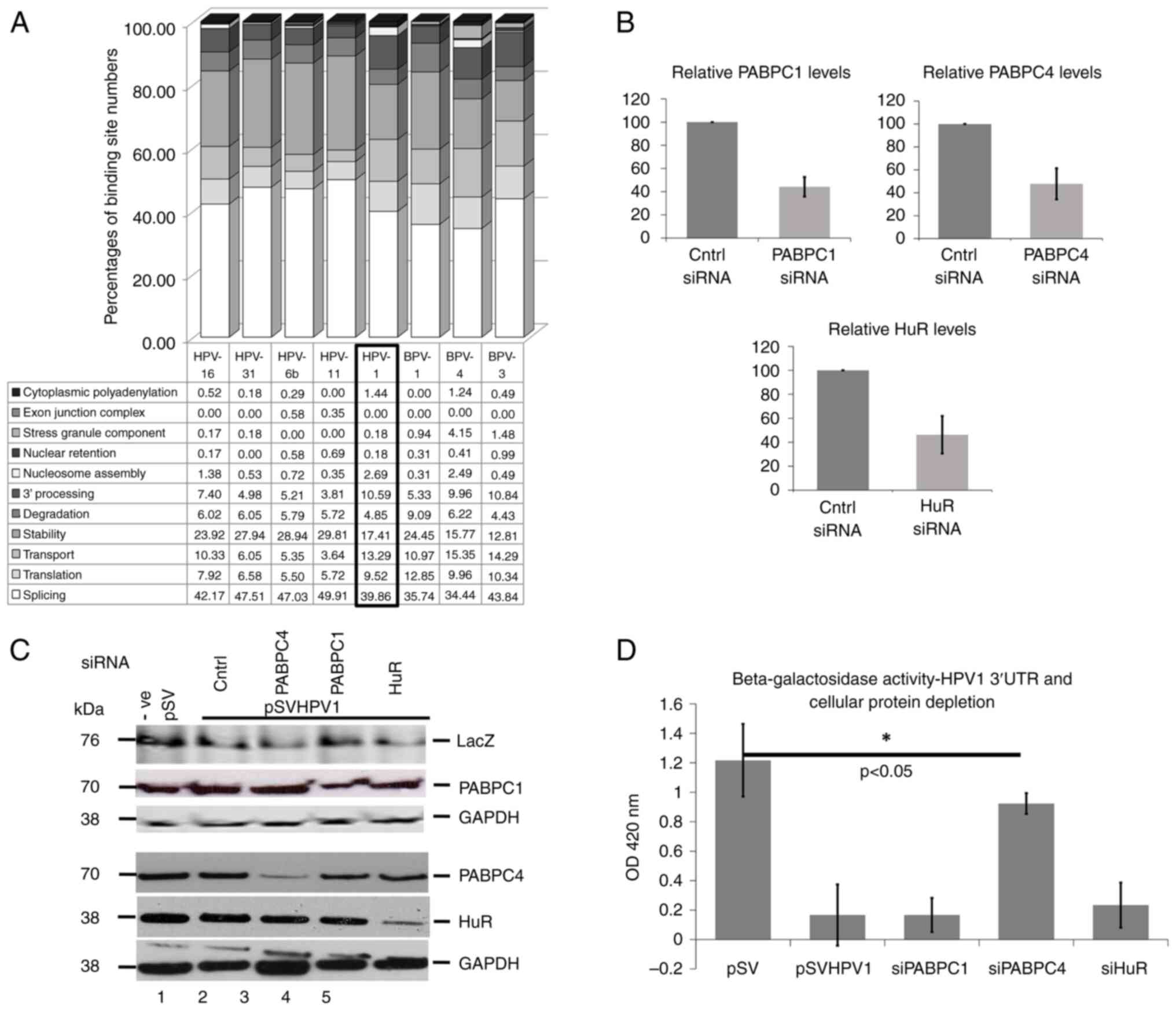
|
1
|
Nasir L and Campo MS: Bovine
papillomaviruses: Their role in the aetiology of cutaneous tumours
of bovids and equids. Vet Dermatol. 19:243–254. 2008.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Graham SV: The human papillomavirus
replication cycle, and its links to cancer progression: A
comprehensive review. Clin Sci (Lond). 131:2201–2221.
2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Peh WL, Middleton K, Christensen N,
Nicholls P, Egawa K, Sotlar K, Brandsma J, Percival A, Lewis J, Liu
WJ and Doorbar J: Life cycle heterogeneity in animal models of
human papillomavirus-associated disease. J Virol. 76:10401–10416.
2002.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Barksdale SK and Baker CC:
Differentiation-specific alternative splicing of bovine
papillomavirus late mRNAs. J Virol. 69:6553–6556. 1995.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Egawa N, Egawa K, Griffin H and Doorbar J:
Human papillomaviruses; epithelial tropisms and the development of
neoplasia. Viruses. 7:3863–3890. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Stoler MH, Wolinsky SM, Whitbeck A, Broker
TR and Chow LT: Differentiation-linked human papillomavirus types 6
and 11 transcription in genital condylomata revealed by in situ
hybridization with message-specific RNA probes. Virology.
172:331–340. 1989.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Kennedy IM, Haddow JK and Clements JB:
Analysis of human papillomavirus type 16 late mRNA 3' processing
signals in vitro and in vivo. J Virol. 64:1825–1829.
1990.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Furth PA and Baker CC: An element in the
bovine papillomavirus late 3'untranslated region reduces
polyadenylated cytoplasmic RNA levels. J Virol. 65:5806–5812.
1991.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Sokolowski M, Zhao C, Tan W and Schwartz
S: AU-rich mRNA instability elements on human papillomavirus type 1
late mRNAs and c-fos mRNAs interact with the same cellular factors.
Oncogene. 15:2303–2319. 1997.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Sokolowski M, Furneaux H and Schwartz S:
The inhibitory activity of the AU-rich RNA element in the human
papillomavirus type 1 late 3' untranslated region correlates with
its affinity for the elav-like HuR protein. J Virol. 73:1080–1091.
1999.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Cumming SA, Repellin CE, McPhilips M,
Redford JC, Clements JB and Graham SV: The human papillomavirus
type 31 late 3' untranslated region contains a complex bipartite
negative regulatory element. J Virol. 76:5993–6003. 2002.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Gunderson SI, Polycarpou-Schwarz M and
Mattaj IW: U1 snRNP inhibits pre-mRNA polyadenylation through a
direct interaction between U1 70K and poly(A) polymerase. Mol Cell.
1:255–264. 1998.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Sokolowski M and Schwartz S: Heterogeneous
nuclear ribonucleoprotein C binds exclusively to the functionally
important UUUUU-motifs in the human papillomavirus type-1 AU-rich
inhibitory element. Virus Res. 73:163–175. 2001.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wiklund L, Sokolowski M, Carlsson A, Rush
M and Schwartz S: Inhibition of translation by UAUUUAU and
UAUUUUUAU motifs of the AU-rich RNA instability element in the
HPV-1 late 3' untranslated region. J Biol Chem. 277:40462–40471.
2002.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Dietrich-Goetz W, Kennedy IM, Levins B,
Stanley MA and Clements JB: A cellular 65-kDa protein recognizes
the negative regulatory element of human papillomavirus late mRNA.
Proc Natl Acad Sci USA. 94:163–168. 1997.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Koffa MD, Graham SV, Takagaki Y, Manley JL
and Clements JB: The human papillomavirus type 16 negative
regulatory RNA element interacts with three proteins that act at
different posttranscriptional levels. Proc Natl Acad Sci USA.
97:4677–4682. 2000.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Cumming SA, McPhillips MG, Veerapraditsin
T, Milligan SG and Graham SV: Activity of the human papillomavirus
type 16 late negative regulatory element is partly due to four weak
consensus 5' splice sites that bind a U1 snRNP-like complex. J
Virol. 77:5167–5177. 2003.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Cumming SA, Chuen-Im T, Zhang J and Graham
SV: The RNA stability regulator HuR regulates L1 protein expression
in vivo in differentiating cervical epithelial cells. Virology.
383:142–149. 2009.PubMed/NCBI View Article : Google Scholar
|
|
19
|
McPhillips MG, Veerapraditsin T, Cumming
SA, Karali D, Milligan SG, Boner W, Morgan IM and Graham SV:
SF2/ASF binds the human papillomavirus type 16 late RNA control
element and is regulated during differentiation of virus-infected
epithelial cells. J Virol. 78:10598–10605. 2004.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Cheunim T, Zhang J, Milligan SG,
McPhillips MG and Graham SV: The alternative splicing factor hnRNP
A1 is up-regulated during virus-infected epithelial cell
differentiation and binds the human papillomavirus type 16 late
regulatory element. Virus Res. 131:189–198. 2008.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Goraczniak R and Gunderson SI: The
regulatory element in the 3'-untranslated region of human
papillomavirus 16 inhibits expression by binding CUG-binding
protein 1. J Biol Chem. 283:2286–2296. 2008.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Moraes KCM, Wilusz CJ and Wilusz J: CUG-BP
binds to RNA substrates and recruits PARN deadenylase. RNA.
12:1084–1091. 2006.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Jean-Philippe J, Paz S and Caputi M: hnRNP
A1: The swiss army knife of gene expression. Int J Mol Sci.
14:18999–19024. 2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Das S and Krainer AR: Emerging functions
of SRSF1, splicing factor and oncoprotein, in RNA metabolism and
cancer. Mol Cancer Res. 12:1195–1204. 2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhao X, Rush M, Carlsson A and Schwartz S:
The presence of inhibitory RNA elements in the late 3'-untranslated
region is a conserved property of human papillomaviruses. Virus
Res. 125:135–144. 2007.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Boukamp P, Petrussevska RT, Breitkreutz D,
Hornung J, Markham A and Fusenig NE: Normal keratinization in a
spontaneously immortalized aneuploid human keratinocyte cell line.
J Cell Biol. 106:761–771. 1988.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Burgess HM, Richardson WA, anderson RC,
Salaun C, Graham SV and Gray NK: Nuclear relocalisation of
cytoplasmic poly(A)-binding proteins PABP1 and PABP4 in response to
UV irradiation reveals mRNA-dependent export of metazoan PABPs. J
Cell Sci. 124:3344–3355. 2011.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Kumar S, Stecher G, Li M, Knyaz C and
Tamura K: MEGA X: Molecular evolutionary genetics analysis across
computing platforms. Mol Biol Evol. 35:1547–1549. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kimura M: A simple method for estimating
evolutionary rates of base substitutions through comparative
studies of nucleotide sequences. J Mol Evol. 16:111–120.
1980.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Madeira F, Park YM, Lee J, Buso N, Gur T,
Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD and
Lopez R: The EMBL-EBI search and sequence analysis tools APIs in
2019. Nucleic Acids Res. 47:W636–W641. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Waterhouse AM, Procter JB, Martin DMA,
Clamp M and Barton GJ: Jalview version 2-a multiple sequence
alignment editor and analysis workbench. Bioinformatics.
25:1189–1191. 2009.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Paz I, Kosti I, Ares M Jr, Cline M and
Mandel-Gutfreund Y: RBPmap: A web server for mapping binding sites
of RNA-binding proteins. Nucleic Acids Res. 42:W361–W367.
2014.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wu C, Kajitani N and Schwartz S: Splicing
and polyadenylation of human papillomavirus type 16 mRNAs. Int J
Mol Sci. 18(366)2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Jia R, Liu X, Tao M, Kruhlak M, Guo M,
Meyers C, Baker CC and Zheng ZM: Control of the papillomavirus
early-to-late switch by differentially expressed SRp20. J Virol.
83:167–180. 2009.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ladd AN, Charlet N and Cooper TA: The CELF
family of RNA binding proteins is implicated in cell-specific and
developmentally regulated alternative splicing. Mol Cell Biol.
21:1285–1296. 2001.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Ladd AN, Nguyen NH, Malhotra K and Cooper
TA: CELF6, a member of the CELF family of RNA-binding proteins,
regulates muscle-specific splicing enhancer-dependent alternative
splicing. J Biol Chem. 279:17756–17764. 2004.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wagnon JL, Briese M, Sun W, Mahaffey CL,
Curk T, Rot G, Ule J and Frankel WN: CELF4 regulates translation
and local abundance of a vast set of mRNAs, including genes
associated with regulation of synaptic function. PLoS Genet.
8(e1003067)2012.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zhai L, Wang C, Chen Y, Zhou S and Li L:
Rbm46 regulates mouse embryonic stem cell differentiation by
targeting β-catenin mRNA for degradation. PLoS One.
12(e0172420)2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Gehring NH, Neu-Yilik G, Schell T, Hentze
MW and Kulozik AE: Y14 and hUpf3b form an NMD-activating complex.
Mol Cell. 11:939–949. 2003.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Michelle L, Cloutier A, Toutant J, Shkreta
L, Thibault P, Mathieu D, Garneau D, Gendron D, Lapointe E, Couture
S, et al: Proteins associated with the exon junction complex also
control the alternative splicing of apoptotic regulators. Mol Cell
Biol. 32:954–967. 2012.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Chuang TW, Chang WL, Lee KM and Tarn WY:
The RNA-binding protein Y14 inhibits mRNA decapping and modulates
processing body formation. Mol Biol Cell. 24:1–13. 2013.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Graham SV: Papillomavirus 3' UTR
regulatory elements. Front Biosci. 13:5646–5663. 2008.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Mangus DA, Evans MC and Jacobson A:
Poly(A)-binding proteins: Multifunctional scaffolds for the
post-transcriptional control of gene expression. Genome Biol.
4(223)2003.PubMed/NCBI View Article : Google Scholar
|