Introduction
Certain papillomavirus (PV) genotypes are associated
with development of benign lesions while others can be associated
with malignancy. Cancer-related PV types include 13 high risk HPVs
(HPV-16, -18, -31, -33, -39, -45, -51, -52, -56, -58, -59, -66 and
-68) and bovine papillomavirus (BPV) genotypes BPV-1, -2 and -4
(1,2). By contrast, low risk PVs usually cause
benign lesions or warts. For example, HPV-6 and -11 cause genital
warts while BPV-3 causes epithelial papillomas.
The infectious life cycle of PVs is tightly linked
to epithelial cell differentiation (3). Synthesis of PV capsid proteins L1 and
L2 encoded by the late genes is restricted to differentiated
epithelial cells (2,4,5). PV
late gene expression in undifferentiated keratinocytes is inhibited
by post-transcriptional mechanisms. In this way PVs avoid immune
detection (4,6). A number of studies have revealed the
presence of cis-acting RNA elements in the late 3'
untranslated regions (3'UTR) of PV late mRNAs which control this
differentiation-specific expression (7-11).
The best characterized element is that of BPV-1 (8,12).
This 53-nt negative-regulatory element contains one motif with
extremely good sequence homology to a consensus 5' splice site,
which is the binding site of U1 snRNP, one of the splicing
complexes that comprise the spliceosome. Binding of U1 snRNP to the
element leads to inhibition of gene expression by its component U1
70K protein, which inhibits poly A polymerase activity in poly A
tail addition, thereby destabilizing viral late mRNAs (12). BPV-1 is the only BPV genotype where
an inhibitory element in the late 3'UTR has been identified so
far.
Similar to BPV-1, regulatory elements and the
proteins that they bind, have also been characterized in the late
3'UTRs of cutaneous-infective HPV-1 and in the mucosal-infective
high risk HPVs-16 and -31 (7,9-11).
HPV-1 late regulatory element has previously been shown to bind
HuR, hnRNPC and poly(A) binding protein (PABPC) in vitro
(10,13,14).
It has been suggested that HuR destabilizes late mRNAs while PABPC
binding interferes with mRNA translation. The HPV-16 and -31 3'UTR
regulatory elements bind auxiliary splicing factor U2AF,
polyadenylation factor CstF-64 and HuR (11,15-18).
The HPV-16 element is also shown to bind U1A (a component of U1
snRNP) splicing regulatory factors SRSF1, hnRNPA1 and CUG-BP
(19-21).
These factors may control late mRNA splicing and polyadenylation
and/or stability. There is evidence that some of these splicing
factors, e.g. SRSF1, can also regulate gene expression at a
translational level (22-24).
Thus, PV 3'UTR elements could regulate various facets of both RNA
processing and translation.
A previous study sought to compare activities of
3'UTR elements of low (HPV-1, -2, -6b, -41 and -61) and high risk
HPVs (HPV-16, -18 and -31) (25).
All eight elements inhibited gene expression in HeLa cells but to
varying degrees. The present study chose to compare the activity of
the late 3'UTR elements from five HPV and three BPV genotypes
representing different genera and species, as well as pathological
properties, to determine how widespread the inhibitory elements
were in papillomavirus evolution. Every 3'UTR element inhibited
gene expression in keratinocytes. The data suggested that 3'UTR
elements are conserved in BPVs. Bioinformatics investigation of
RNA-binding protein motifs at high stringency level revealed
binding sites of cellular proteins relevant to RNA metabolism
specific to genus and species. These predictions were verified
using HPV-1a as a testable model and it showed PABC4 to be a host
protein capable of controlling the HPV-1a regulatory element.
Materials and methods
Plasmids
A pSV-beta-galactosidase report gene system
(Promega Corporation) lacking a 3'UTR was used to determine
efficiency of each PV late 3'UTR in gene expression regulation.
pSV-beta-galactosidase control vector, containing the
beta-galactosidase reporter gene and 3'UTR (Promega
Corporation) was used as positive control. The 8 PV genomes used in
this study (kindly provided by Professor S. Campo, University of
Glasgow) included four non-cancer related types (HPV-1a, -6b, -11
and BPV-3) and four cancer-related types (HPV-16, -31, BPV-1 and
-4). Virus genome sequences [Genbank accession nos. U06714.1
(HPV-1a), AF092932.1 (HPV-6b), FN907963.1 (HPV-11), AF125673.1
(HPV-16), J04353.1 (HPV-31), AF486184.1 (BPV-3), X05817.1 (BPV-4)
and NC_001522.1 (BPV-1)] were used in designing primers (Table I) to PCR-amplify the late 3'UTR
fragment of each PV. PCR products were ligated into the
pSV-beta-galactosidase vector in place of the 3'UTR of the
beta-galactosidase gene. The cloning strategy for the HPV1a
3'UTR used a PCR-based strategy. The late 3'UTR from HPV-1a was
cloned into the pSV-beta-galactosidase vector using an In
Fusion cloning kit from Takara Bio, Inc. and following the
manufacturer's the instructions therein. Primers pSVfw
5'-CTGCAGGCATGCAAGCTGG-3' and pSVrv 5'-GATCCAGACATGATAAGATACATTG-3'
were used to linearize the vector by inverse PCR prior to
cloning.
 | Table IDetails of primer sequences, product
sizes, genomic location of late 3'UTR of each PV generated by PCR.
Underlined sequences represent restriction sites. |
Table I
Details of primer sequences, product
sizes, genomic location of late 3'UTR of each PV generated by PCR.
Underlined sequences represent restriction sites.
| PV type | 5'-3' Sequences
(Tm) | Restriction
site | Genomic location
(nt) | Product size
(bp) |
|---|
| BPV-1 |
| Forward |
GGATCCGCTTTCTTTGGACTTAG (67.9˚C) | BamHI | 6960-7305 | 345 |
| Reverse |
CTGCAGCGCTGAGAAGGCAGGATTCGG (78.4˚C) | PstI | | |
| BPV-3 |
| Forward |
GGATCCGAGGACCCTTATGCAAAGTACAC
(73.9˚C) | BamHI | 6668-7190 | 522 |
| Reverse |
CTGCAGGCGTACTGCCAGTATGTGC (75.6˚C) | PstI | | |
| BPV-4 |
| Forward |
GGATCCCACTCAGGTTAAAGAAGACCCC (73.9˚C) | BamHI | 6906-7254 | 348 |
| Reverse |
GTCGACCACACAAGTGCCGGAGATG (75.2˚C) | SalI | | |
| HPV-1aa |
| Forward |
TATCATGTCTGGATCCATCCTAGTCTTAGAAAGCG
(71.0˚C) | - | 5380-7087 | 1,707 |
| Reverse |
CTTGCATGCCTGCAGCCAAGACTATTTTATGTAAATACA
(72˚C) | | | |
| HPV-6b |
| Forward |
GGATCCCAAAGTGGATATAGGGGACGG (73.8˚C) | BamHI | 7185-7673 | 488 |
| Reverse |
CTGCAGGTGGAAAGTGTATGCCAAGGGC (78.2˚C) | PstI | | |
| HPV-11 |
| Forward |
GGATCCCCTCTACAGCCCCCAAACG (76.6˚C) | BamHI | 7233-7622 | 389 |
| Reverse |
CTGCAGTAAGTGTATGTAAGGGCAACCG (74.3˚C) | PstI | | |
| HPV-16 |
| Forward |
GGATCCCTATGAATTCCACTATTTTGGAGG ACTGG
(74.2˚C) | BamHI | 6814-7460 | 646 |
| Reverse |
GTCGACCGAATTCGGTTGAAGCAC (71.6˚C) | SalI | | |
| HPV-31 |
| Forward |
GGATCCCAAAAGCCCAAGGAAGATC (72.0˚C) | BamHI | 6854-7374 | 520 |
| Reverse |
GTCGACGGCAGAAATAAGTACATGAC (69.7˚C) | SalI | | |
Transient transfections
Recombinant plasmids or the pSV positive control
(Promega Corporation) were transiently co-transfected with pEGFP as
an internal transfection control (a gift from Dr Tungkeangsirisin,
Silpakorn University, Nakhon Pathom, Thailand) or pmaxGFP (Lonza
Group, Ltd.), into HeLa cells using Lipofectamine®
reagent 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) with a
ratio of pSV + PV late 3'UTR:pEGFP at 2:0.5 µg/µl and incubated at
37˚C for 6 h. The medium was then removed before adding DMEM
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS.
After incubating at 37˚C for 48 h, cells were harvested and
immediately subjected to preparation of cell lysate for
beta-galactosidase activity assay or RNA extraction. For siRNA
depletion experiments, pSV-HPV1a-3'UTR plasmid was co-transfected
with 10 nM siRNA pools of duplexes against either PABPC1
(sc-108012), PAPBC4 (sc-106347) or HuR (sc-35619) provided by Santa
Cruz Biotechnology, Inc. in 6-well dishes according to the protocol
for Lipofectamine® plasmid/siRNA co-transfection. siGLO
(D-001630-01-20; Horizon Discovery Ltd.) was used as a non-target
siRNA control and to monitor transfection efficiency. The cells
were then incubated in DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS, at 37˚C, 5% CO2 for 24 h (RNA
preparation) or 48 h (protein preparation or beta-galactosidase
assays). The cells were harvested and used for reverse
transcription PCR and beta-galactosidase activity assay. The
experiments were conducted in triplicate.
HaCaT spontaneously transformed keratinocytes were
purchased from CLS Cell Lines Service GmbH (accession number
CVCL_0038) (26). Cells were grown
in serum-free Keratinocyte Growth Medium (KGM Gold; Lonza Group,
Ltd.) in 6-well plates. Triplicate transfections were performed.
pMaxGFP (1 µg; Lonza Group, Ltd.) was transfected as a positive
control for transfection and the pSV-beta-galactosidase
plasmids with the HPV and BPV 3'UTRs or a
pSV-beta-galactosidase vector lacking a 3'UTR as a control
using Effectene Transfection Reagent (Qiagen GmbH). Cells were
incubated at 37˚C in 5% CO2. At 24 h post transfection,
cells at 80% confluence were harvested into 500 µl of QIAzol Lysis
Reagent (Qiagen GmbH) and lysates were stored at -80˚C. Total RNA
was prepared from samples using the RNeasy kit (Qiagen GmbH) in
accordance with the manufacturer's protocol. RNA was eluted with 60
µl of RNase-free water. The concentration of RNA in the eluates was
determined using a Nanodrop One/One Microvolume UV-Vis
Spectrophotometer (Thermo Fisher Scientific, Inc.). cDNA was
synthesized from total RNA (250 ng) using the Maxima First Strand
cDNA Synthesis Kit according to manufacturer's instructions (Thermo
Fisher Scientific, Inc.) for reverse transcription-quantitative
(RT-q) PCR with dsDNase as per the manufacturer's instructions.
RT-qPCR
RT-qPCR was performed using a 7500 Real Time PCR
System (Thermo Fisher Scientific, Inc.). For each RT-qPCR reaction
(total volume of 20 µl), 10 µl of Takyon ROX Probe 2X MasterMix
dTTP blue (Eurogentec), 4 µl of Primer Probe Mix (final
concentrations 900 nM and 100 nM for primers and probes,
respectively) and 1 µl of water were used. Primer and probe sets
for beta galactosidase were forward primer:
5'-GCGATTACCGTTGATGTTGAAG-3', reverse primer
5'-CCCTAATCCGAGCCAGTTTAC-3', probe: 5'-CAGCTGGCAGTTCAGGCCAATC-3'
and the housekeeping gene β-actin forward primer:
5'-AGCGCGGCTACAGCTTCA-3', reverse primer:
5'-CGTAGCACAGCTTCTCCTTAATGT C-3', probe: 5'-ATTTCCCGCTCGGCCGTGGT-3'
were included. A total of 5 µl of cDNA was included for each
reaction. Reaction conditions were one cycle at 50˚C for 2 min, one
cycle of 95˚C for 3 min followed by 40 cycles of 95˚C for 10 sec,
followed by 60˚C for 1 min. Each sample was run in triplicate.
Data produced in each qPCR reaction was analyzed on
the 7500 Real-Time SDS Software (Thermo Fisher Scientific, Inc.).
The threshold line for Cq determination was assigned automatically
and was always within the exponential phase. Relative
quantification of viral mRNA was performed using the Livak method
(2-ΔΔCq) (27) using
β-actin as a reference and comparing the increase in
expression in relation to the pSV-beta-galactosidase plasmid
lacking a 3'UTR.
Reverse transcription PCR and band
intensity assay
Total RNA was extracted by TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. The RNA was treated with RQ RNase-free
DNase (Promega Corporation) prior to performing end point RT-PCR
(Qiagen GmbH). The reaction mixture was 100 ng RNA, 1x Onestep
RT-PCR buffer, 10 µM forward primer, 10 µM reverse primer, 10 mM
dNTP, 2 µl One-step RT-PCR enzyme mix, in a total volume of 50 µl.
The primer pairs used in the study were Lac Z forward primer:
5'-GTTGCAGTGCACGGCAGATACACTTGCTGA-3'; Lac Z reverse primer:
5'-GCCACTGGTGTGGGCCATAATTCAATTCGC-3'; GFP forward primer:
5'-GCCCATCCTGGTCGAGCTGG-3'; GFP reverse primer:
5'-GTGCCCCAGGATGTTGCCGTC-3'. Reverse transcription was carried out
at 50˚C, 60 min. PCR conditions were 95˚C for 15 min, then 35
cycles of 95˚C for 30 sec, 58˚C for 30 sec, 72˚C for 1 min,
followed by a final extension at 72˚C for 7 min and stored at 4˚C.
The PCR products were separated by 1% agarose gel electrophoresis
and visualized by ethidium bromide staining. The band intensity of
PCR products was measured using gel and image analysis and
quantitation software, GelQuantNET provided by BiochemLabSolutions.com.
Cell lysate preparation and
beta-galactosidase activity assay
After transient co-transfection for 24 h, HeLa cells
were washed with PBS 2-3 times before incubation in 1X Reporter
Lysis Buffer (Promega Corporation) at room temperature for 15 min.
The lysate was centrifuged at 16,128 x g for 2 min, at 4˚C to
collect supernatant. To assay the enzyme activity, the supernatant
was mixed with 2X assay buffer (Promega Corporation) at a ratio of
1:1. The reaction was kept in the dark at 37˚C for 3-4 h or until
the solution turned a yellow color. The reaction was stopped by
adding 500 µl of 1 M sodium carbonate (Promega Corporation). The
enzyme activity was analyzed by measuring absorbance at 420 nm
using spectrophotometry (T70 UV/VIS Spectrometer; PG Instruments
Ltd.).
Protein extract preparation and
western blotting
Cells (1x106) were washed twice in PBS at
4˚C and lysed in 2X BOLT protein loading buffer (Invitrogen; Thermo
Fisher Scientific, Inc.). Protein extracts were syringe-passaged
through a 22-gauge needle 15 times then sonicated in an ultrasonic
bath (Guyson-Kerry) for 3x30 sec pulses at room temperature. The
samples were boiled at 100˚C for 5 min before loading on a 12%
NuPAGE gel (Invitrogen; Thermo Fisher Scientific, Inc.) and
electrophoresed at 150 V for 1 h in 1X MES buffer (Invitrogen;
Thermo Fisher Scientific, Inc.). Proteins were transferred to a
nitrocellulose membrane using the iBlot transfer kit and iBlot Gel
Transfer Stacks (Invitrogen; Thermo Fisher Scientific, Inc.) as per
the manufacturer's instructions. Membranes were blocked in 5% milk
powder in PBST (PBS + 0.1%Tween-20) at room temperature for 1 h.
Membranes were washed 3 times in PBST for 5 min each then incubated
with primary antibody. PABPC1 and PABPC4 antibodies were used at
dilutions of 1:10,000 and 1:1,500, respectively (28). HuR antibody 3A2 (Santa Crux
Biotechnologies) was used at a dilution of 1:500. The GAPDH
antibody clone 6C5 (Thermo Fisher Scientific) and the
beta-galactosidase antibody GAL-40 (MilliporeSigma) were used at
1:1,000 dilution. The blots were incubated in their respective
antibody for 1 h at room temperature or overnight at 4˚C. After 1
h, the blots were washed 3 times in PBS-T for 5 min. They were then
placed in secondary antibody for 1 h at room temperature
(HRP-linked goat anti-mouse; Pierce, Thermo Fisher Scientific used
at a 1:2,000 dilution). Blots were washed 3 times in PBST for 5 min
before incubation with ECL western blot substrate. The blots were
exposed to X-ray film (Thermo Fisher Scientific, Inc.) and
processed in a Kodak X-Omat processor (Kodak). It is impossible to
determine mass of protein when BOLT buffer is used for the lysate
since Bromo Phenol Blue alters the spectrophotometry values in a
traditional protein concentration assay such as Bradford's assay.
To ensure that protein quantities in each western blot could be
compared to another, every experiment used the same number of cells
(i.e. a very similar amount of protein) and this was verified in
all western blots by running a GAPDH loading control. All
quantification was carried out relative to the GAPDH loading
control and densitometry analysis was carried out using ImageJ.
Phylogenetic tree, bioinformatics
sequence alignment and RNA-binding protein motif analysis of late
3'UTR of PVs
The molecular phylogenetic tree of 8 PV late 3'UTRs
was analyzed using the MEGA X 10.0.5 version win 32(29) and the maximum-likelihood method
based on the Kimura two-parameter model (30) with 1,000 bootstrap replicates. The
multiple sequence alignment was carried out by using Clustal Omega
(31) and visualized with Jalview
(32). RNA-binding protein motifs
in the late 3'UTR of 8 PV types were analyzed by using RBPmap
version 1.1 (http://rbpmap.technion.ac.il/) (33). The input was set up to Human Genome
Database (Dec 2013; GRCh38/hg38) and searching for Human/Mouse
motifs. For advanced options, P-value (significant) <0.001 and
P-value (suboptimal) <0.01 was chosen for high stringency
level.
Statistical analysis
Statistical analyses were performed on all data
using SPSS (version 21; IBM Corp.). Fold inhibition was calculated
as the ratio of the average band intensity of the PCR product (for
mRNA level) or average absorbance of cell lysates from the
beta-galactosidase activity assays (for protein level) obtained
with the pSV positive control compared with the average band
intensity or average absorbance of cell lysates from pSV vectors
containing each PV late 3'UTR. Significant differences of fold
inhibition on beta-galactosidase expression in mRNA and protein
levels between different PV fragments were tested using a one-way
ANOVA followed by Tukey's post hoc test. P<0.05 was considered
to indicate a statistically significant difference.
Results
The late 3' untranslated region of
eight PV genomes
The present study chose to study four HPV genotypes
in the Alphapapillomavirus genus, where the cancer-related
types (HPV-16 and -31) are in species 9 (alpha-9) and the
non-cancer related types (HPV-6b and -11) in species 10 (alpha-10).
HPV-1a was included as a member of the Mupapillomavirus
genus. For BPVs, BPV-1, a Deltapapillomavirus species, was
included and BPV-3 and -4 which are in the Xipapillomavirus
genus. The well-mapped late regulatory element of HPV-16 spans the
3' end of last exon and extends into the late 3'UTR (7,8),
whereas the equivalent elements of HPV-1a and BPV-1 start
immediately after the L1 stop codon and for HPV-31 are entirely in
the late 3'UTR (Fig. 1A) (9-11).
Accordingly, because the regulatory elements of HPV-6b, -11, BPV-3
and -4 were unmapped, gene fragments were cloned starting near the
3' end of the last exon (at least >40 nts from the L1 stop
codon) and extending past all the predicted polyadenylation sites
(PAS) to ensure that the fragments would cover all potential
regulatory sequences, which can occasionally lie downstream of the
PAS (11). All primers used to
generate PV fragments were designed to have annealing temperature
at >58˚C in order to avoid non-specific binding.
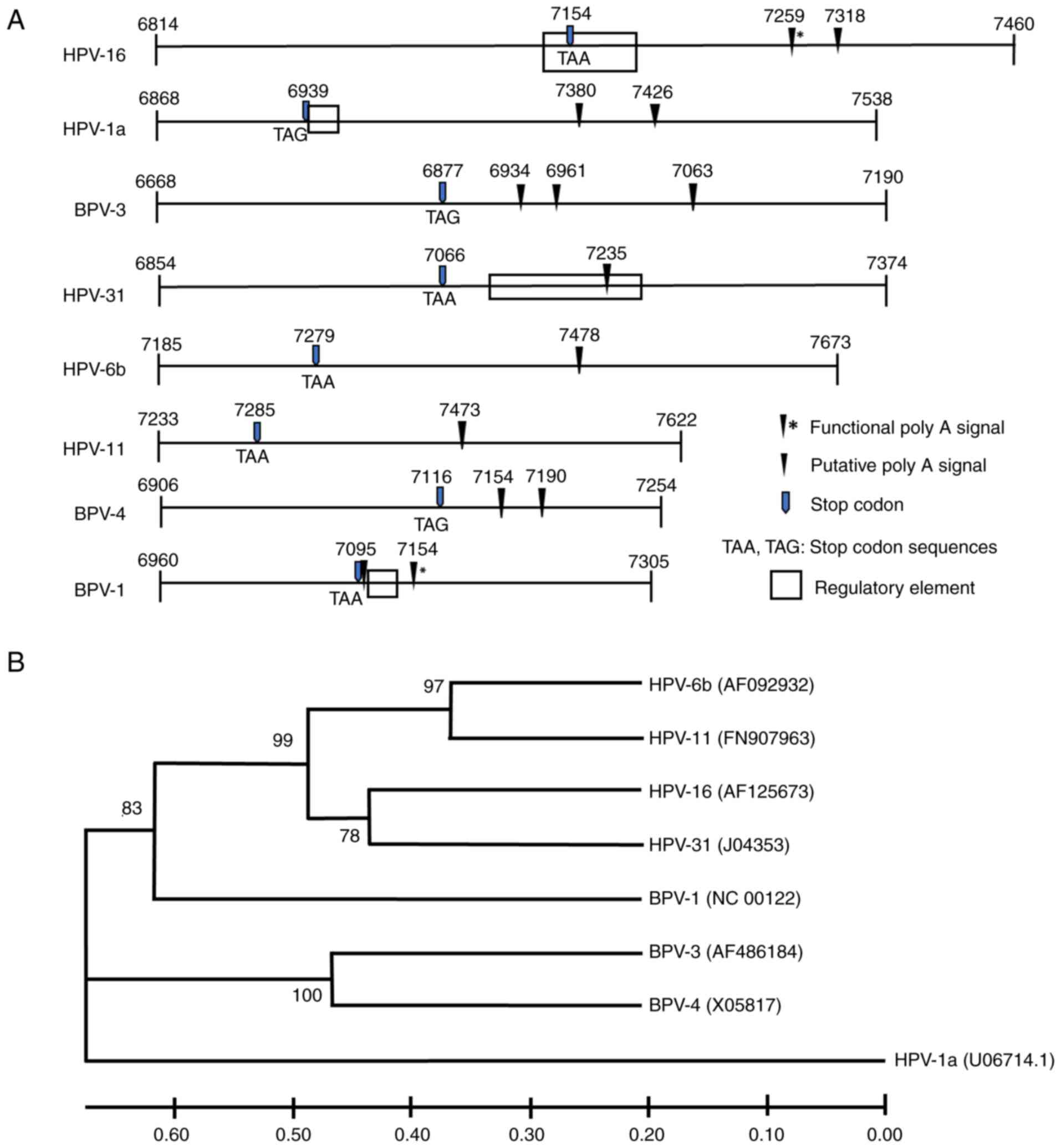 | Figure 1Relatedness of the 3'UTRs of selected
PVs. (A) Diagram of the region of cloned fragments, starting from
the 3' end of last exon and expanding into late 3' UTR, of eight
PVs analyzed in this study. Numbers indicate genomic nucleotide
positions. Regulatory elements have been functionally defined for
HPV-1a, -16, 31 and BPV-1 (7-11)
but not for the other PVs. (B) Phylogenetic tree of eight PV late
3'UTRs, the PV late 3'UTR sequences were retrieved from Genbank.
Accession numbers appear in parentheses. Bootstrap (1,000
replicates) values in percentage are shown. Bar at 0.1
substitutions per nucleotide. 3'UTRs, 3' untranslated region; PV,
papillomaviruses; HPV, human papillomavirus; BPV, bovine
papillomavirus. |
Table II and
Fig. 1A show details of the lengths
of the 3'UTRs and numbers of predicted PAS for each PV. The length
of the 3'UTR, between the L1 stop codon and the last poly A signal
(PAS), of each PV was variable. HPV-1a had the longest 3'UTR (487
nts) whilst those for BPV-4 and -1 were considerable shorter at 71
and 59 nts, respectively (Table
II). BPV-3 had three predicted late PAS, whilst the HPV-1a,
-16, BPV-1 and -4 sequences contained two PAS. There was only one
late PAS detected for HPV-31, -6b and -11. For BPV-4, there was no
available information on the stop codon of the L1 ORF. Based on the
nucleotide alignment results (Fig.
S1), the stop codon was presumed to share a similar sequence
and position on the genome as BPV-3. Phylogenetic analysis was then
performed to compare the evolutionary relatedness of the 3'UTR
sequences among HPV-1a, -6b, -11, 16, -31, BPV-1, -3 and -4.
Although the late 3'UTRs of the eight PVs showed a high diversity
of length and sequence, a phylogenetic tree revealed relationships
of all PV fragments in accordance with their taxonomic
classification (Fig. 1B).
 | Table IILength of late 3'UTR, numbers of PAS
and sequences of L1 stop codon in each HPV and BPV. |
Table II
Length of late 3'UTR, numbers of PAS
and sequences of L1 stop codon in each HPV and BPV.
| PV type | Length (L1 stop
codon-the last PAS) | Number of PAS | Stop codon |
|---|
| HPV-1a | 487 nt | 2 | TAG |
| HPV-6b | 199 nt | 1 | TAA |
| HPV-11 | 188 nt | 1 | TAA |
| HPV-16 | 164 nt | 2 | TAA |
| HPV-31 | 169 nt | 1 | TAA |
| BPV-1 | 59 nt | 2 | TAA |
| BPV-3 | 186 nt | 3 | TAG |
| BPV-4 | 74 nt | 2 | TAG |
Relative inhibitory activity of the
seven PV late 3'UTRs
Investigation of the influence of the late 3'UTRs on
gene expression was performed using a beta-galactosidase
gene reporter system and transient co-transfection with pEGFP as a
transfection efficiency control. Following transient transfection
in HeLa cells, cellular RNA and protein lysates were prepared for
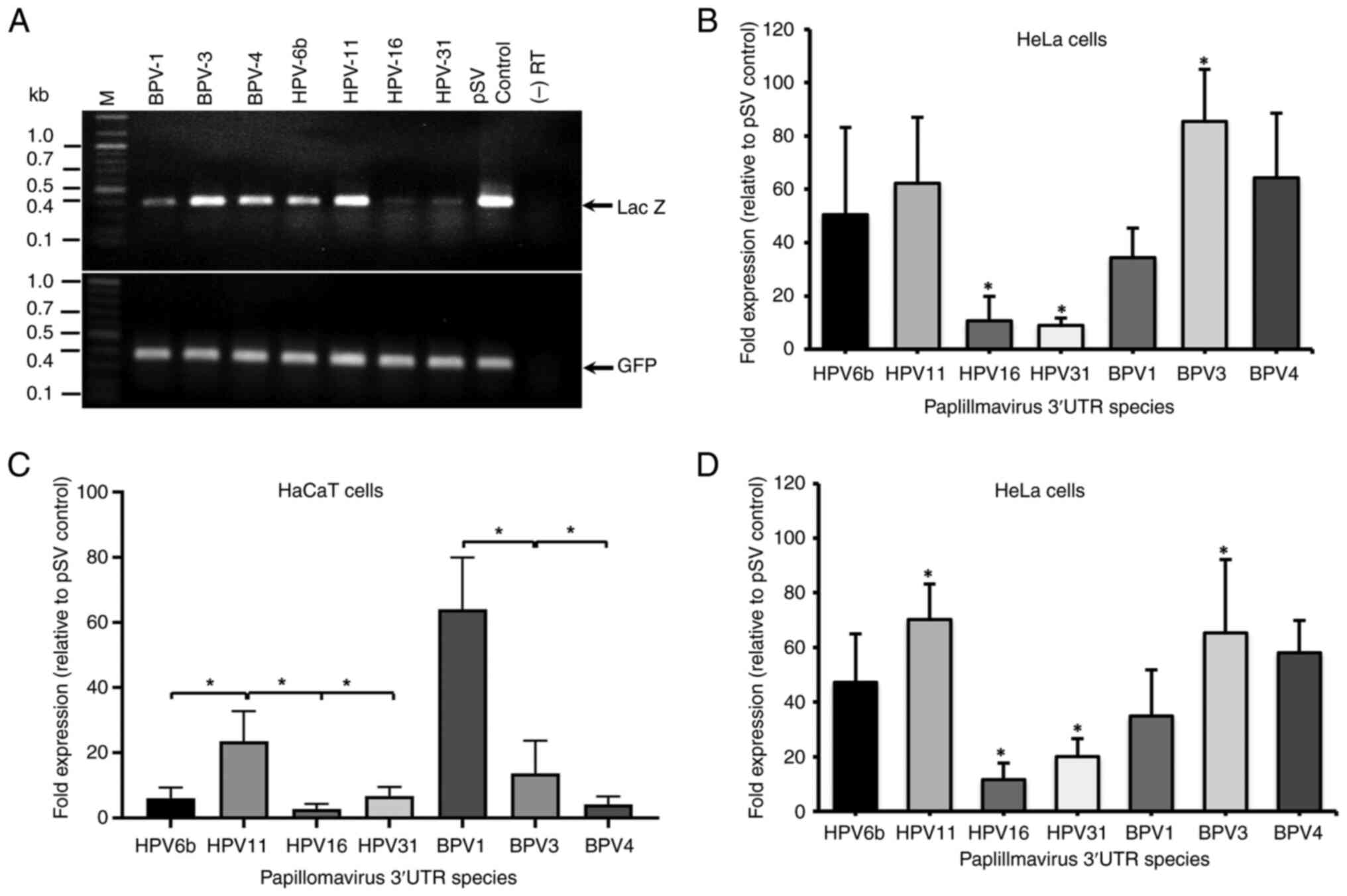
end point PCR and enzyme activity assays. Fig. 2A shows PCR products size-separated
on a 1% agarose gel. Compared with the internal control, GFP
expression, differences in Lac Z band intensity were
observed among cDNAs containing the different PV 3'UTRs (Fig. 2B), implying that they could control
mRNA production or stability. As expected, due to transfection into
undifferentiated cells, all seven 3'UTRs inhibited gene expression.
The inhibitory activity had no relationship to the length of the
cloned fragments, e.g., the BPV-1 region (345 nts) had greater
inhibitory activity than the BPV-3 region (522 nts). The inhibitory
efficiency of late 3'UTRs on mRNA levels can be ranked as HPV-16
and -31 (high activity); BPV-1 and HPV-6b (medium activity); BPV-3,
-4 and HPV-11 (low activity; Table
III). If the observed inhibitory activities were specific to
the HeLa cancer cell line, expression of each reporter construct
was determined in HaCaT spontaneously immortalized keratinocytes
(Fig. 2C). Values are shown
relative to pSV control lacking a 3'UTR. BPV-1 3'UTR had the least
inhibitory activity, followed by HPV-11 and BPV-3. BPV-4, HPV-16
and HPV-31 each displayed high levels of inhibition. These data
revealed that HPV-16 and HPV-31 display the greatest inhibitory
activity in keratinocytes, the natural host of PV infection.
 | Table IIIFold inhibition of
beta-galactosidase (lac Z) expression at mRNA and
protein levels observed in HeLa cells. |
Table III
Fold inhibition of
beta-galactosidase (lac Z) expression at mRNA and
protein levels observed in HeLa cells.
| PV type | Genus | Species | Cancer-related | Fold inhibition
lac Z mRNA mean (SD)a | Inhibition fold of
lac Z activity (SD)a |
|---|
| HPV-16 | Alpha | 9 | Yes | 14.45 (0.14) | 11.63 (8.43) |
| HPV-31 | Alpha | 9 | Yes | 11.81 (0.07) | 5.42 (2.11) |
| HPV-6b | Alpha | 10 | No | 3.16 (0.56) | 2.33 (0.88) |
| HPV-11 | Alpha | 10 | No | 1.83 (0.44) | 1.46 (0.30) |
| BPV-1 | Delta | 4 | Yes | 3.10 (0.91) | 3.57 (2.21) |
| BPV-4 | Xi | - | Yes | 1.74 (0.76) | 1.78 (0.41) |
| BPV-3 | Xi | - | No | 1.21 (0.28) | 1.77 (0.87) |
Regulation of beta-galactosidase expression
by the late 3' UTRs of the seven PVs was also determined at the
protein level (Fig. 2D). Similar to
mRNA expression levels, enzyme activity in cell lysates transfected
with pSV vectors containing different PV 3'UTRs was lower than that
of the pSV positive control. The order of inhibitory efficiencies
was similar to those found for mRNA expression levels. However,
fold inhibition of lacZ enzyme activity in all BPV 3'UTRs was
always higher than fold inhibition at the level of mRNA expression
and vice versa for HPVs (Table
III). The inhibitory efficiency of HPV-11 and BPV-3 late 3'UTRs
are significantly different from HPV-16 (P=0.007 and 0.013,
respectively) and HPV-31 (P=0.022 and 0.043, respectively).
Bioinformatics analysis of RNA-binding
protein motifs in the late 3'UTRs
PV late gene expression can be controlled at
multiple post-transcriptional and translational steps (34). Post-transcriptional regulation
requires recognition of specific RNA sequences on late viral RNAs
by cellular regulatory proteins. Such RNA-protein interactions are
very well defined in several open-access databases. The present
study therefore identified protein binding sites in the PV 3'UTRs
and used their known functions to infer which cellular RNA
metabolic pathways could be affected as a result of interaction
between RNA-binding proteins and viral sequences. Using a
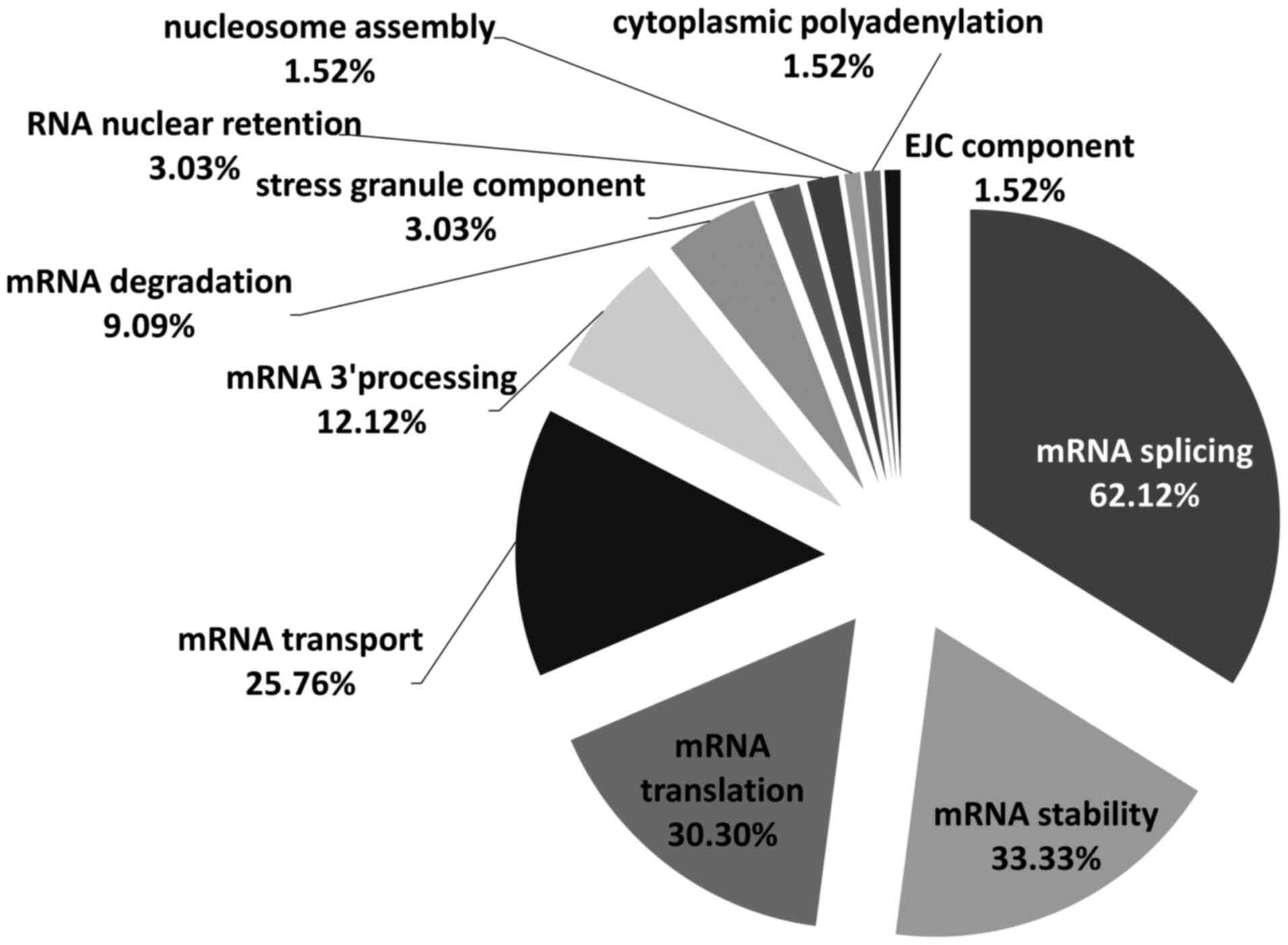
computational tool RBPmap version 1.1(33), a total of 66 RNA-binding proteins
were identified as having highly statistically significant
(P=0.001) putative binding site(s) in at least one PV late 3'UTR
(Fig. 3). These proteins are all
known to be involved in key RNA-related pathways. Of the total 66
proteins, 62.12% were involved in mRNA splicing, following by mRNA
stability (33.33%), translation (30.30%), transport (25.76%), mRNA
3' end processing (12.12%) and degradation (9.09%) (Fig. 3). In addition, 3.03% functioned as
stress granule components and RNA nuclear retention and 1.52% were
found to be involved in nucleosome assembly, cytoplasmic
polyadenylation and exon junction complex.
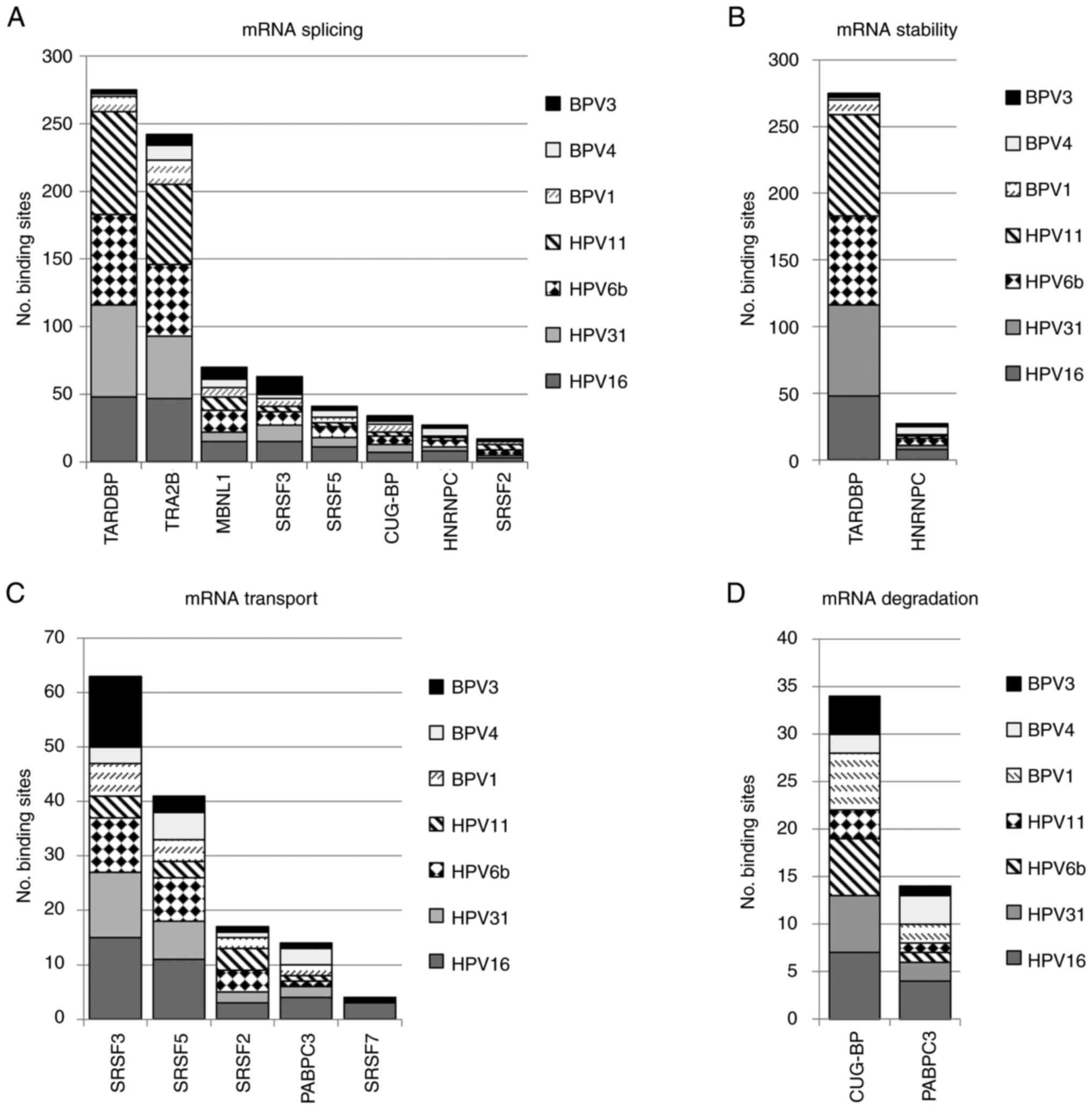
Among 11 different mRNA processing pathways, mRNA
splicing proteins (42 proteins) dominated the list (Fig. 4A). In all, there were eight splicing
factors (CUG-BP, HNRNPC, MBNL1, SRSF2, SRSF3, SRSF5, TARDBP and
TRA2B) where binding sites were found in every PV sequence
(Fig. S2). Of these, CUG-BP,
HNRNPC and SRSF3 have already been proven to bind PV inhibitory
elements (13,21,35).
The RNA processing pathway that contained the second highest number
of predicted-binding site proteins (22 proteins) was mRNA
stability. From a total of 22 proteins, binding sites for two,
TARDBP and HNRNPC, appeared in all seven PVs (Fig. 4B). SRSF2, SRSF3 and SRSF5 had
predicted binding sites for all PV 3'UTRs and together with PABPC3
were identified as involved in mRNA transport in all PV 3'UTR
sequence sets (Fig. 4C). Likewise,
the mRNA degradation set contained two proteins (CUG-BP and PABPC3)
that had binding sites in every PV sequence. (Fig. 4D). There was no protein involved in
mRNA translation and stress granules where binding sites appeared
in all PV sequences (Fig. S3).
Protein binding site prediction can
infer PV late gene regulatory mechanisms
Since modulation of gene expression mostly occurs
through interaction between regulatory proteins and specific
binding sequences, the nature of the binding motifs on viral late
3'UTRs may reflect the post-transcriptional and/or translational
mechanisms that control viral late gene expression. To test the
hypothesis that RNA binding protein motif analysis could predict
inhibitory mechanisms, the present study chose the late 3'UTR of
HPV-1a (Mu papillomavirus) as a model because there is good in
vitro evidence that inhibitory sequences in the HPV-1 late
3'UTR can bind proteins that act at post-transcriptional but also
at translational levels (9,14). RNA-binding protein motifs in the
HPV-1a late 3'UTR were analyzed using RBPmap version 1.1 and
compared with the seven PVs already analyzed (Fig. 5). Fig.
5A shows comparisons of percentages of binding sites for
proteins representing the 11 mRNA metabolic pathways in all eight
late 3'UTRs. HPV1a had fewer predicted binding sites for proteins
involved in mRNA splicing and mRNA stability than HPV-6b, -11, -16,
-31 whereas there was a reverse situation for protein binding sites
related to mRNA 3'end processing. Notably, HPV-1a contained a
higher percentages of binding sites for proteins involved in mRNA
transport (P=0.0057) and translation (P=0.0038). The pattern of
protein binding motifs in HPV-1a late 3'UTR appeared to be closer
to BPV-3 and -4 rather than any HPV. Taken together, the data
showed that predicted protein binding motifs on the HPV-1a late
3'UTRs were closely associated with the known mechanisms that
regulate HPV-1 late gene expression.
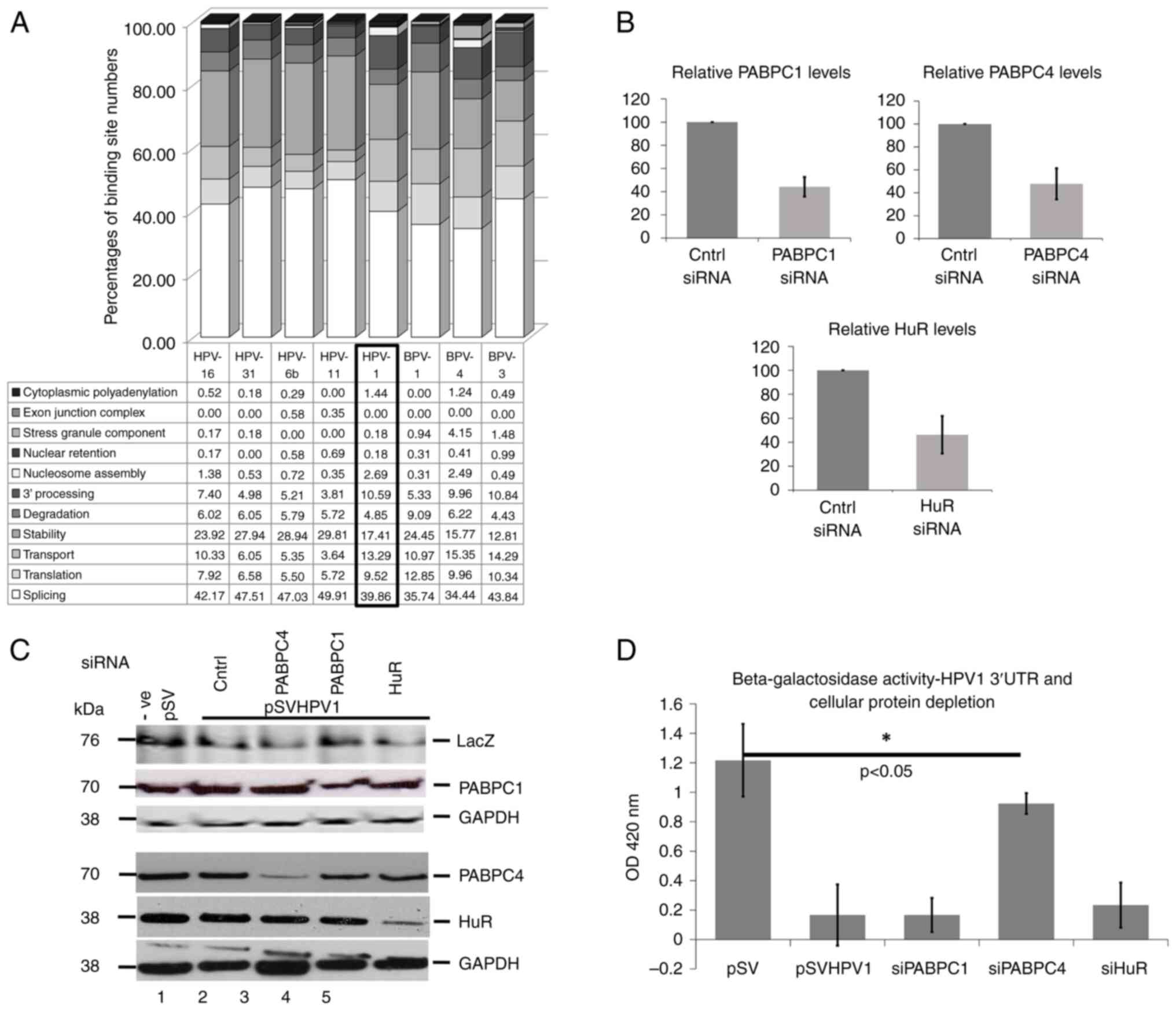 | Figure 5Comparisons of predicted RNA binding
protein sites in HPV-1a late 3'UTR and the 7 PVs (A) Percentage of
RNA binding protein binding sites found in eight PV 3'UTRs
categorised into 11 mRNA metabolic pathways/events analyzed using
RBPmap version 1.1(31) at high
stringency level, P-value (significant) <0.001 and P-value
(suboptimal) <0.01, (B) Graphs showing relative levels of siRNA
knockdown of PABPC1, PABPC4 and HuR with siRNA control. (C) Western
blot analysis of HeLa cell lysate after siRNA depletion using
primary antibodies against PABPC1, PABPC4, HuR and GAPDH. The GADPH
control for PABPC4 and HuR shows faint bands above the main band.
This is due to the X-ray film moving during exposure. (D) Average
absorbance from beta-galactosidase activity assays measured in HeLa
cell lysates transiently co-transfected with pSV-HPV-1a late 3'UTR
and siRNA for PABPC1, PABPC4 and HuR depletion. Bar charts show the
mean and standard error of the mean from three separate
experiments. *P<0.05. HPV, human papillomavirus;
3'UTR, 3' untranslated region; PV, papillomavirus; si, small
interfering; PABPC, poly(A) binding protein; HuR, human antigen R;
pSV, pSV-beta-galactosidase positive control. |
Next, the predictions were tested using a genetic
approach. Translation factor, poly(A) binding protein (PABPC) was
previously shown to bind the HPV-1 3'UTR in vitro (14), although the present study could not
differentiate between different PABPC family members. The HPV-1a
RNA binding protein dataset predicted that PABPC family members,
PABPC1 and PABPC4, could bind the 3'UTR. In addition, the
stability/translation factor HuR, detected in the dataset of the
present study, had also been shown to bind the HPV-1 3'UTR in
vitro (10). The present study
examined the role of these proteins in regulating the HPV-1a 3'UTR
activity by transiently transfecting the pSV-HPV-1a-3'UTR together
with siRNAs to deplete each of these three proteins. Depletion and
level of each protein after siRNA knockdown is shown in Fig. 5B and C. Using an antibody that reacts against
E. coli beta-galactosidase, increased protein levels were
seen when PABPC4 was depleted. Finally, beta-galactosidase assays
showed that only PABPC4 depletion increased levels of the enzyme
significantly (Fig. 5D). These data
confirm that bioinformatics predictions can be important indicators
of genetic factors controlling PV late gene expression.
Discussion
The papillomavirus family is large and infects a
wide range of host species; however, the study of regulatory
elements and RNA-binding proteins in the late 3' untranslated
regions has previously mostly been carried out in HPVs and BPV-1
(7-11).
This is because these are the best characterized cancer-causing PVs
and understanding regulation of their gene expression could lead to
novel antiviral approaches. As aforementioned, the best
characterized 3'UTR regulatory elements are those of BPV-1 and
HPV16. The present study aimed to determine similarities and
differences in RNA binding protein profiles for other HPVs,
particularly low risk HPVs and other BPVs. Future studies will
clone and examine a range other animal PV 3'UTR regulatory
elements.
HeLa cells were used as a model for the present
study because they are a cervical adenocarcinoma cell line that
provides a model for undifferentiated epithelial cells in which the
3'UTR regulatory elements actively repress viral late gene
expression. Moreover, this cell line has been commonly used in the
study of regulatory elements and RNA-binding proteins in the late
3' untranslated regions of HPVs and BPV-1 (7,9-11,13,14,21,25).
Using HeLa cells in the present study allowed the analyze of data
in comparison with previous studies. HeLa cells contain integrated
portions of the HPV-18 genome including the long control region,
the E6, E7, E1 genes and partial coding regions of the E2 and L1
genes. Subsequently, HPV-18 was not included in the present study
because of a concern that the endogenous HPV-18 genome sequences
and any viral proteins expressed might confuse the results of the
experiments leading into mis-interpretations.
The present study revealed inhibitory activity in
keratinocytes of the late 3'UTRs of papillomavirus genomes from
different genera and species, as well as pathological properties.
This suggested that such elements are conserved in papillomavirus
evolution and thus important for viral replication. The present
study demonstrated for the first time regulatory sequences in the
3'UTRs of non-oncogenic HPV-11 (non-oncogenic
Alphapapillomavirus) and, in the Xipapillomaviruses,
non-oncogenic BPV-3 and oncogenic BPV-4. In 2007, Zhao et al
(25) showed inhibitory activity in
HeLa cells of eight HPV late 3'UTRs, including HPV-1, -6b, -16 and
-31 that were analyzed in the present study. The present study,
although using a different experimental approach, including
analysis in keratinocytes, the target cells for PV infection, gave
similar results showing that the highest inhibitory efficiency was
found in HPV-16, following by HPV-31 and -6b. Taken together with
the demonstration of regulatory elements in HPV-1, -2, -6b, -16,
-18, -31, -41 and -61 (7,9-11,25),
it is probable that the presence of regulatory sequences in the 3'
end of the late regions is a conserved property among HPVs and
BPVs.
Bioinformatics analysis of protein binding motifs
revealed binding sites of high statistical significance for a total
of 66 RNA-binding proteins. Of the 66 proteins, there were seven
that had previously been shown to directly bind to papillomavirus
regulatory elements, CUG-BP (CELF1), U2AF2, HuR, HNRNPA1, SNRPA
(U1A; a component of U1 snRNP), HNRNPC, PABPC (9,10,13-18,20,21)
and one for indirect interaction, SRSF1(19). Comparison of percentages of
predicted protein binding sites among 11 mRNA metabolic pathways
showed that the highest percentage of binding motifs in all PV
sequences were binding sites for proteins implicated in mRNA
splicing (Fig. 3). This is
consistent with previous findings in BPV-1, HPV-16 and -31, where
the interaction of splicing factors e.g. U1A, U1 70K, U2AF2, SRSF1,
CUG-BP and HNRNPA1 with the regulatory sequences may contribute to
repression of viral late gene expression via inhibition of terminal
exon definition and 3' end formation (12,16,17,19-21).
Considering Table
III, HPV late 3'UTRs predominantly inhibited lac Z gene
expression at an mRNA level. Conversely, fold inhibition of BPV
late 3'UTRs appeared to be greater at the protein level. Analysis
of protein binding motifs revealed that all BPVs contained higher
percentages of binding sites for factors involved in mRNA transport
and translation and, except for BPV-3, lower percentages of binding
motifs for proteins involved in mRNA splicing, when compared with
HPVs (Fig. 5A). Notably, analysis
of protein binding motifs in the HPV-1a late 3'UTR showed more
similarity to BPV protein binding motifs than other HPVs and it
will be interesting to determine if regulation of gene expression
by the late 3'UTRs of these BPV types may also be predominantly at
the protein synthesis level.
It was observed that patterns of protein binding
motifs on 3'UTRs corresponded with viral taxonomy. For example,
binding sites for members of the CUG binding protein Elav-like
family (CELF), BRUNOL4, BRUNOL5 and BRUNOL6, which control splicing
and mRNA stability (36-38),
were predicted in HPV late 3'UTRs but not in BPVs. On the other
hand, binding sites of RBM46, an mRNA decay factor (39), were identified in only BPV late
3'UTRs. The present study also found binding sites of RBM8A only in
the alpha-10 types, HPV-6b and -11. RBM8A participates in formation
of the exon-junction complex and nonsense-mediated mRNA decay (NMD)
and mRNA splicing (40-42).
For TARDBP and TRA2B, their binding motifs were identified in all
PV 3'UTRs but a larger number of binding sites were found in HPV
sequences (46-76 sites) compared with BPV sequences (2-19 sites;
Figs. 4A and S2). Finally, HNRNPF binding motifs were
specific to HPV 3'UTRs and a higher number of binding sites were
found in non-cancer-related HPVs (10-13 sites) compared with
cancer-related HPVs (2-5 sites). Therefore, although there may be
some commonalities, it is possible that different PVs use different
strategies to regulate late gene expression. This may result from
adaptation of the virus to different targets of infection e.g.
cutaneous and mucosal epithelial cells and fibroblasts, reflecting
important divergence during virus evolution (43).
Results from protein binding motif analysis revealed
binding sites of PABPC family members in all PV late 3'UTRs
including HPV-1a. Functions of PABPCs involve mRNA translation and
stability of mRNA (44). Consistent
with the observation of the present study, PAPBCs have been
demonstrated to bind directly to the HPV-1 late 3'UTR (14). Furthermore, using genetic methods,
it was demonstrated that depletion of PABPC4, a PAPBC family
member, in HeLa cells transiently transfected with pSV-HPV-1a 3'UTR
resulted in significantly increased gene expression. Based on the
results from the present study, bioinformatics analysis may be
considered as one approach for protein binding site prediction used
as a primary step for analysis of PV late gene regulatory
mechanisms.
The present study generated new hypotheses and
identified potential new RNA/protein interactions which may
regulate papillomavirus gene expression. The presence of regulatory
sequences in the 3' end of the late regions is most probably a
conserved property among papillomaviruses. The results from the
bioinformatics analyses revealed binding motifs of nine proteins
common to all investigated PV 3'UTRs, as well as patterns of
protein binding motifs on 3'UTRs which corresponded with viral
taxonomy. It is possible that adaptation of the virus to different
targets of infection resulted in different PVs developing different
strategies to regulate late gene expression.
Supplementary Material
Multiple alignments of DNA sequences
in late 3'UTR fragment. Sequence alignments of 8 PVs were performed
using Clustal Omega (29) and
visualized with Jalview (30).
3'UTR, 3' untranslated region; PV, papillomavirus.
Diagram of RNA binding protein sites
commonly found in eight PV late 3'UTRs predicted by using RBPmap
version 1.1 at high stringency level, P-value (significant)
<0.001 and P-value (suboptimal) <0.01. PV, papillomavirus;
3'UTR, 3' untranslated region.
Predicted RNA binding proteins and
binding site numbers that have functions related to mRNA splicing,
translation and stress granules by using RBPmap version 1.1
(http://rbpmap.technion.ac.il/) (31), at high stringency level [P-value
(significant) <0.001 and P-value (suboptimal) <0.01].
Acknowledgements
The authors thank Professor Saveria Campo
(University of Glasgow, UK) for kindly providing virus DNA samples
and Dr Wisit Tungkeangsirisin (Silpakorn University, Nakhon Pathom,
Thailand) for pEGFP preparation.
Funding
Funding: This research was funded in the fiscal year 2017, The
Silpakorn University Research, Innovation and Creativity
Administration Office (SURIC), grant no. SURDI 60/01/05. We
acknowledge funding from the Medical Research Council (MC_
UU_12014) as core funding for the MRC University of Glasgow Centre
for Virus Research.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
NI, AS and AK were responsible for investigation and
formal analysis. NK and SVG were responsible for conceptualization
and writing and editing the manuscript. TC was responsible for
conceptualization, methodology, investigation, data curation,
formal analysis and writing the original draft. TC and SVG confirm
the authenticity of all the raw data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Authors' information
Nakarin Kitkumthorn ORCID iD:
0000-0003-0616-6039
Anna Kirk ORCID iD: 0000-0001-8175-942X
Thanaporn Chuen-im ORCID iD: 0000-0003-0101-9419
Sheila V Graham ORCID iD: 0000-0002-7140-8279
References
|
1
|
Nasir L and Campo MS: Bovine
papillomaviruses: Their role in the aetiology of cutaneous tumours
of bovids and equids. Vet Dermatol. 19:243–254. 2008.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Graham SV: The human papillomavirus
replication cycle, and its links to cancer progression: A
comprehensive review. Clin Sci (Lond). 131:2201–2221.
2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Peh WL, Middleton K, Christensen N,
Nicholls P, Egawa K, Sotlar K, Brandsma J, Percival A, Lewis J, Liu
WJ and Doorbar J: Life cycle heterogeneity in animal models of
human papillomavirus-associated disease. J Virol. 76:10401–10416.
2002.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Barksdale SK and Baker CC:
Differentiation-specific alternative splicing of bovine
papillomavirus late mRNAs. J Virol. 69:6553–6556. 1995.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Egawa N, Egawa K, Griffin H and Doorbar J:
Human papillomaviruses; epithelial tropisms and the development of
neoplasia. Viruses. 7:3863–3890. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Stoler MH, Wolinsky SM, Whitbeck A, Broker
TR and Chow LT: Differentiation-linked human papillomavirus types 6
and 11 transcription in genital condylomata revealed by in situ
hybridization with message-specific RNA probes. Virology.
172:331–340. 1989.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Kennedy IM, Haddow JK and Clements JB:
Analysis of human papillomavirus type 16 late mRNA 3' processing
signals in vitro and in vivo. J Virol. 64:1825–1829.
1990.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Furth PA and Baker CC: An element in the
bovine papillomavirus late 3'untranslated region reduces
polyadenylated cytoplasmic RNA levels. J Virol. 65:5806–5812.
1991.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Sokolowski M, Zhao C, Tan W and Schwartz
S: AU-rich mRNA instability elements on human papillomavirus type 1
late mRNAs and c-fos mRNAs interact with the same cellular factors.
Oncogene. 15:2303–2319. 1997.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Sokolowski M, Furneaux H and Schwartz S:
The inhibitory activity of the AU-rich RNA element in the human
papillomavirus type 1 late 3' untranslated region correlates with
its affinity for the elav-like HuR protein. J Virol. 73:1080–1091.
1999.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Cumming SA, Repellin CE, McPhilips M,
Redford JC, Clements JB and Graham SV: The human papillomavirus
type 31 late 3' untranslated region contains a complex bipartite
negative regulatory element. J Virol. 76:5993–6003. 2002.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Gunderson SI, Polycarpou-Schwarz M and
Mattaj IW: U1 snRNP inhibits pre-mRNA polyadenylation through a
direct interaction between U1 70K and poly(A) polymerase. Mol Cell.
1:255–264. 1998.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Sokolowski M and Schwartz S: Heterogeneous
nuclear ribonucleoprotein C binds exclusively to the functionally
important UUUUU-motifs in the human papillomavirus type-1 AU-rich
inhibitory element. Virus Res. 73:163–175. 2001.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wiklund L, Sokolowski M, Carlsson A, Rush
M and Schwartz S: Inhibition of translation by UAUUUAU and
UAUUUUUAU motifs of the AU-rich RNA instability element in the
HPV-1 late 3' untranslated region. J Biol Chem. 277:40462–40471.
2002.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Dietrich-Goetz W, Kennedy IM, Levins B,
Stanley MA and Clements JB: A cellular 65-kDa protein recognizes
the negative regulatory element of human papillomavirus late mRNA.
Proc Natl Acad Sci USA. 94:163–168. 1997.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Koffa MD, Graham SV, Takagaki Y, Manley JL
and Clements JB: The human papillomavirus type 16 negative
regulatory RNA element interacts with three proteins that act at
different posttranscriptional levels. Proc Natl Acad Sci USA.
97:4677–4682. 2000.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Cumming SA, McPhillips MG, Veerapraditsin
T, Milligan SG and Graham SV: Activity of the human papillomavirus
type 16 late negative regulatory element is partly due to four weak
consensus 5' splice sites that bind a U1 snRNP-like complex. J
Virol. 77:5167–5177. 2003.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Cumming SA, Chuen-Im T, Zhang J and Graham
SV: The RNA stability regulator HuR regulates L1 protein expression
in vivo in differentiating cervical epithelial cells. Virology.
383:142–149. 2009.PubMed/NCBI View Article : Google Scholar
|
|
19
|
McPhillips MG, Veerapraditsin T, Cumming
SA, Karali D, Milligan SG, Boner W, Morgan IM and Graham SV:
SF2/ASF binds the human papillomavirus type 16 late RNA control
element and is regulated during differentiation of virus-infected
epithelial cells. J Virol. 78:10598–10605. 2004.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Cheunim T, Zhang J, Milligan SG,
McPhillips MG and Graham SV: The alternative splicing factor hnRNP
A1 is up-regulated during virus-infected epithelial cell
differentiation and binds the human papillomavirus type 16 late
regulatory element. Virus Res. 131:189–198. 2008.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Goraczniak R and Gunderson SI: The
regulatory element in the 3'-untranslated region of human
papillomavirus 16 inhibits expression by binding CUG-binding
protein 1. J Biol Chem. 283:2286–2296. 2008.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Moraes KCM, Wilusz CJ and Wilusz J: CUG-BP
binds to RNA substrates and recruits PARN deadenylase. RNA.
12:1084–1091. 2006.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Jean-Philippe J, Paz S and Caputi M: hnRNP
A1: The swiss army knife of gene expression. Int J Mol Sci.
14:18999–19024. 2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Das S and Krainer AR: Emerging functions
of SRSF1, splicing factor and oncoprotein, in RNA metabolism and
cancer. Mol Cancer Res. 12:1195–1204. 2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhao X, Rush M, Carlsson A and Schwartz S:
The presence of inhibitory RNA elements in the late 3'-untranslated
region is a conserved property of human papillomaviruses. Virus
Res. 125:135–144. 2007.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Boukamp P, Petrussevska RT, Breitkreutz D,
Hornung J, Markham A and Fusenig NE: Normal keratinization in a
spontaneously immortalized aneuploid human keratinocyte cell line.
J Cell Biol. 106:761–771. 1988.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Burgess HM, Richardson WA, anderson RC,
Salaun C, Graham SV and Gray NK: Nuclear relocalisation of
cytoplasmic poly(A)-binding proteins PABP1 and PABP4 in response to
UV irradiation reveals mRNA-dependent export of metazoan PABPs. J
Cell Sci. 124:3344–3355. 2011.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Kumar S, Stecher G, Li M, Knyaz C and
Tamura K: MEGA X: Molecular evolutionary genetics analysis across
computing platforms. Mol Biol Evol. 35:1547–1549. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kimura M: A simple method for estimating
evolutionary rates of base substitutions through comparative
studies of nucleotide sequences. J Mol Evol. 16:111–120.
1980.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Madeira F, Park YM, Lee J, Buso N, Gur T,
Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD and
Lopez R: The EMBL-EBI search and sequence analysis tools APIs in
2019. Nucleic Acids Res. 47:W636–W641. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Waterhouse AM, Procter JB, Martin DMA,
Clamp M and Barton GJ: Jalview version 2-a multiple sequence
alignment editor and analysis workbench. Bioinformatics.
25:1189–1191. 2009.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Paz I, Kosti I, Ares M Jr, Cline M and
Mandel-Gutfreund Y: RBPmap: A web server for mapping binding sites
of RNA-binding proteins. Nucleic Acids Res. 42:W361–W367.
2014.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wu C, Kajitani N and Schwartz S: Splicing
and polyadenylation of human papillomavirus type 16 mRNAs. Int J
Mol Sci. 18(366)2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Jia R, Liu X, Tao M, Kruhlak M, Guo M,
Meyers C, Baker CC and Zheng ZM: Control of the papillomavirus
early-to-late switch by differentially expressed SRp20. J Virol.
83:167–180. 2009.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ladd AN, Charlet N and Cooper TA: The CELF
family of RNA binding proteins is implicated in cell-specific and
developmentally regulated alternative splicing. Mol Cell Biol.
21:1285–1296. 2001.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Ladd AN, Nguyen NH, Malhotra K and Cooper
TA: CELF6, a member of the CELF family of RNA-binding proteins,
regulates muscle-specific splicing enhancer-dependent alternative
splicing. J Biol Chem. 279:17756–17764. 2004.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wagnon JL, Briese M, Sun W, Mahaffey CL,
Curk T, Rot G, Ule J and Frankel WN: CELF4 regulates translation
and local abundance of a vast set of mRNAs, including genes
associated with regulation of synaptic function. PLoS Genet.
8(e1003067)2012.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zhai L, Wang C, Chen Y, Zhou S and Li L:
Rbm46 regulates mouse embryonic stem cell differentiation by
targeting β-catenin mRNA for degradation. PLoS One.
12(e0172420)2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Gehring NH, Neu-Yilik G, Schell T, Hentze
MW and Kulozik AE: Y14 and hUpf3b form an NMD-activating complex.
Mol Cell. 11:939–949. 2003.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Michelle L, Cloutier A, Toutant J, Shkreta
L, Thibault P, Mathieu D, Garneau D, Gendron D, Lapointe E, Couture
S, et al: Proteins associated with the exon junction complex also
control the alternative splicing of apoptotic regulators. Mol Cell
Biol. 32:954–967. 2012.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Chuang TW, Chang WL, Lee KM and Tarn WY:
The RNA-binding protein Y14 inhibits mRNA decapping and modulates
processing body formation. Mol Biol Cell. 24:1–13. 2013.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Graham SV: Papillomavirus 3' UTR
regulatory elements. Front Biosci. 13:5646–5663. 2008.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Mangus DA, Evans MC and Jacobson A:
Poly(A)-binding proteins: Multifunctional scaffolds for the
post-transcriptional control of gene expression. Genome Biol.
4(223)2003.PubMed/NCBI View Article : Google Scholar
|