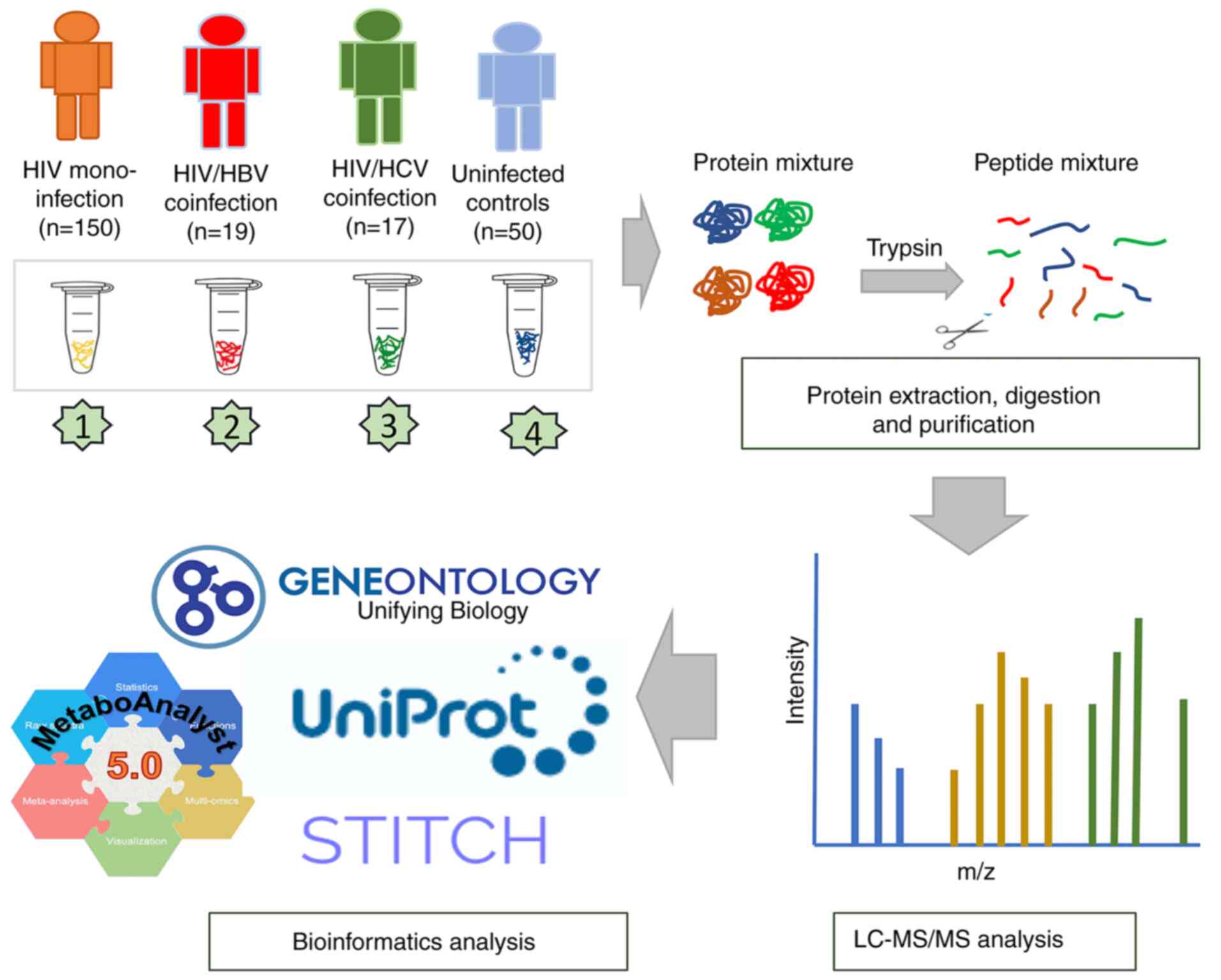
Introduction
Human immunodeficiency virus (HIV) infection is a
major health issue globally, with ~39.0 million patients living
with HIV (PLWH) and ~29.8 million PLWH accessing anti-retroviral
therapy (ART) in 2022(1). Effective
ART has notably reduced the rates of acquired immunodeficiency
syndrome (AIDS)-associated morbidity and mortality. HIV infection
is considered to be a chronic disease in countries where ART is
available and PLWH have longer life expectancy (2,3).
However, since the introduction of combination ART, chronic liver
disease (CLD) has become a key concern in PLWH. The persistently
low levels of viremia during ART can cause chronic hepatic injury
and inflammation, potentially leading to hepatocellular carcinoma
(HCC) development (3,4). To the best of our knowledge, there
have only been three cohort studies that investigated HCC in PLWH
populations (5-7).
Clinical vigilance of CLD and HCC in PLWH is suggested (8).
Coinfection with hepatotropic hepatitis B and C
virus (HBV and HCV, respectively) is a major cause of CLD in PLWH
receiving ART (9,10). HIV infection alone can cause liver
steatosis and fibrosis (11-13),
which aggravate the damage caused by HBV and HCV to promote liver
fibrosis and cirrhosis in patients coinfected with HBV and HCV
(5,14-16).
Different mechanisms underlying acceleration of this form of
disease progression have been proposed: Hepatotropic virus
infection has been reported to induce persistent immune activation
and inflammation, which participate in progressive liver disease in
PLWH receiving ART (3,9). In patients coinfected with HIV/HCV,
elevated levels of certain immune activation and inflammatory
biomarkers such as soluble CD14 and CD163, IL-6 and -8 and
IFN-γ-inducible protein 10, which are associated with
hepatocellular injury and severity of liver fibrosis, have been
previously found (14,17). Distinct apoptosis and inflammatory
marker profiles have also been observed in patients with HBV and
HIV/HBV, indicating different mechanisms of immune activation and
disease progression between these patients (18). Previous studies have also suggested
that HIV potentiates HCV-induced fibrogenesis by activating the
profibrogenic TGF-β pathways in hepatocyte and hepatic stellate
cells (HSCs), resulting in excessive production of extracellular
matrix protein type I collagen by HSCs (19,20).
To the best of our knowledge, there is no evidence of TGF-β
activation following HIV/HBV coinfection; however, TGF-β serum
levels are elevated in patients with chronic hepatitis B and
hepatitis B with liver cirrhosis compared with those in normal
control individuals, potentially linking chronic hepatitis B with
the severity of liver cirrhosis (21). While mechanisms for hepatic
inflammation and fibrosis caused by HIV infection and coinfection
with HBV and HCV have been previously documented (4,9), the
precise molecular pathways underlying progression to advanced liver
disease and HCC require further clarification.
Previous studies have reported relatively high
prevalence of HBV (11.4%) and HCV (7.6%) coinfection and liver
fibrosis in PLWH populations receiving suppressive ART (22,23).
This suggests that PLWH may progress to advanced chronic liver
disease and HCC, warranting investigations into the molecular
changes that occur in PLWH receiving long-term ART. The present
study aimed to explore proteomic profiles associated with HIV
infection and coinfection with HBV and HCV in PLWH using shotgun
proteomics. Patients infected with HIV and uninfected control
individuals were recruited and non-invasively assessed for the
degree of liver fibrosis. Plasma samples were subjected to
proteomics analysis. The plasma protein profiles may be used to
predict disease progression and provide potential biomarkers for
CLD caused by HIV and hepatotropic HBV and/or HCV.
Materials and methods
Study population, clinical data and
laboratory investigation
A total of 186 patients with HIV (age, 18-86 years;
61.8% male and 38.2% female) attending the Antiretroviral Therapy
Clinic in Nakorn Nayok Hospital, Nakorn Nayok (Thailand) from April
2014 to October 2020 were recruited into the present study. The
inclusion criteria were as follows: i) Patients aged >18 years
with documented HIV infection; ii) receiving suppressive ART for
>6 months and iii) available blood samples and clinical data.
Patients who consumed alcohol, herbal medicine and steroidal drugs,
in addition to patients with active opportunistic infections, were
excluded. The patients were divided into 150 HIV-moninfected
patients, aged 18-86 years, 58.7% male and 41.3% female, 19
HBV-coinfected HIV patients, 29-58 years, 73.7% male and 26.3%
female, and 17 HCV-coinfected HIV patients, aged 20-62 years, 76.5%
male and 23.5% female. In total, 50 uninfected controls, aged 20-60
years, 40% male and 60% female, were recruited from Police General
Hospital, Bangkok (Thailand) in November 2022. Inclusion criteria
for controls were individuals aged >18 years with available
blood samples and clinical data who were seronegative for HIV, HBV
and HCV infection. Individuals with notable liver fibrosis,
determined by fibrosis-4 (FIB-4) score >1.45 or aspartate
aminotransferase (AST)-to-platelet ratio index (APRI) >0.5, were
excluded (24-26).
The present study protocol was approved by the Human Ethics
Committee No. 3, Thammasat University, Pathumthani (Thailand;
approval no. 116/2565) and the Certificated Biological Safety
Committee of Thammasat University, Pathumthani, (Thailand, approve
no. 079/2565).
Clinical and laboratory data of the HIV group were
obtained and characterized as described in previous studies
(22,23). Briefly, the data collected from the
medical records of the patients were age, sex, CD4+ cell
count, HBV/HCV coinfection, levels of AST and alanine
aminotransferases (ALT), platelet count, HIV viral load and ART
regimen and duration. Anti-HIV, HBV surface antigen and anti-HCV
tests were performed to recruit seronegative HIV, HBV and
HCV-uninfected controls. The extent of liver fibrosis in
HIV-infected patients and uninfected controls was evaluated by
measuring non-invasive markers FIB-4 and APRI, which are commonly
used scoring systems recommended for assessing liver fibrosis in
the presenceof chronic HCV by the World Health Organization
(27). FIB-4 score >1.45 or APRI
>0.5 was considered to be significant liver fibrosis (24-26).
Additionally, levels of fibrotic markers, namely laminin (LN),
procollagen type III N-terminal peptide (PIIINP), hyaluronic acid
(HA) and type IV collagen (IVC), in addition to tumor marker
α-fetoprotein (AFP), were measured using electrochemiluminescence
immunoassay according to the maufacturer's instructions (Mindray
CL-900i using LN, PIIINP, HA, IVC and AFP test kits (cat. nos.
LV111, PIIINP111, HA111, C IV111 and AFP111, respectively; all
Mindray Medical International Co., Ltd.).
EDTA-blood samples left over from routine
examination were collected from the clinical laboratory of Nakorn
Nayok Hospital. Plasma was separated within 8 h of collection by
centrifuging at 2,000 x g for 10 min at room temperature and stored
at -80˚C until further use. Plasma samples from 186 patients
infected with HIV were classified into the following three groups:
HIV-monoinfection (n=150), HIV/HBV coinfection (n=19) and HIV/HCV
coinfection (n=17). Blood samples from seronegative HIV, HBV and
HCV individuals (n=50) with FIB-4 score ≤1.45 and APRI ≤0.5 were
designated as uninfected controls. All samples were then subjected
to proteomic analysis (Fig. 1).
Sample preparation and liquid
chromatography-tandem mass spectrometry (LC-MS/MS)
Total protein concentration in plasma samples was
measured using the Lowry method, with bovine serum albumin applied
as the standard (28). Protein
samples were subjected to in-solution digestion. The samples, which
were dissolved in 10 mM ammonium bicarbonate (AMBIC), underwent
disulfide bond reduction using 5 mM dithiothreitol in 10 mM AMBIC
at 60˚C for 1 h and alkylation of sulfhydryl groups using 15 mM
iodoacetamide in 10 mM AMBIC at room temperature for 45 min in the
dark, followed by digestion in porcine trypsin (Promega
Corporation) in a 1:20 ratio at 37˚C overnight. Peptides were dried
and resuspended with 0.1% formic acid prior to nano-LC-MS/MS
analysis. The LC-MS/MS analysis was performed using an Ultimate
3000 Nano/Capillary LC System (Thermo Fisher Scientific, Inc.)
coupled to a ZenoTOF 7600 mass spectrometer (SCIEX). Peptide
separation was performed using a 75 µm x 15 cm column, packed with
Acclaim PepMap RSLC C18 resin (2 µm, 100Å, nanoViper, Thermo
Scientific, UK). The C18 column was maintained at 60˚C within a
thermostatted column oven. Solvent A (0.1% formic acid in water)
and solvent B (0.1% formic acid in 80% acetonitrile) were delivered
to the analytical column. A gradient elution of 5-55% solvent B was
employed to separate peptides at a constant flow rate of 0.30
µl/min over a 30-min period.
The electrospray ionization source was set in
positive ion mode with an ion source temperature of 200˚C, spray
voltage of 3300 V, curtain gas (N2) of 35 psi, nebulizer
gas of 50 psi, auxiliary heating gas of 55 psi, and declustering
potential of 80 V. Top 50 most abundant precursor ions per survey
MS1 for MS/MS with an intensity threshold exceeding 150 cps were
selected. Precursor ions were dynamically excluded for 8 sec after
two incidences of MS/MS sampling (with dynamic collision energy
enabled). The MS2 spectra were collected in the range 350-1,800 m/z
with a 250-ms accumulation time and Zeno trap enabled. The
collision energy parameters included a declustering potential of 80
V, no DP spread, and a CE spread of 0 V. The time bins were summed
(with all channels enabled) using a 150,000 cps Zeno trap
threshold. The cycle time for the Top 50 DDA method was 3.0 sec. To
minimize the effect of experimental variation, three independent
LC-MS runs were performed for each sample.
Protein identification and label-free
quantification
Proteins were quantified using MaxQuant 2.1.4.0
(Max-Planck Institute for Biochemistry) using the Andromeda search
engine to match MS/MS spectra to the Uniprot Homo sapiens
database (uniprot.org/) (29). Label-free quantification with
MaxQuant's standard settings was performed as follows: i) Maximum
of two missing cleavages; ii) mass tolerance of 0.6 Da for main
search; iii) trypsin as a digesting enzyme; iv)
carbamidomethylation of cystein as fixed modification and v)
oxidation of methionine and acetylation of the protein N-terminus
as variable modifications. Peptides with ≥7 amino acids and ≥1
unique peptide were chosen for protein identification. Only
proteins with ≥2 two peptides and ≥1 unique peptide were considered
as being identified and subjected to further analysis. The false
discovery rate (FDR) was set at 1% and estimated using the reversed
search sequences. The maximal number of modifications/peptide was
set to 5. The proteins present in the H. sapiens proteome
were then downloaded from Uniprot as a search FASTA file. Potential
contaminants in the contaminant FASTA files were added to the
search space using the MaxQuant software.
Data processing and bioinformatics
analysis
MaxQuant ProteinGroups.txt file was loaded into
Perseus software version 1.6.6.0(29) and potential contaminants that did
not correspond to any Universal Proteomics Standard Set 1 (UPS1)
protein were removed. Mass intensities were log2
transformed before pairwise comparisons using unpaired t tests.
Missing values were imputed with a constant value (0) using the
Perseus software. Intensity values of the MS/MS spectra were
analyzed using partial least squares-discriminant analysis (PLS-DA)
model, one-way ANOVA (P-value and FDR <0.05) and Kruskal-Wallis
test using web-based tool MetaboAnalyst version 6.0 (metaboanalyst.ca/) (30,31).
Heatmap analysis was performed by using the Morpheus online tool
(software.broadinstitute.org/morpheus/) (32). Molecular function and subcellular
localization of identified proteins were obtained from Gene
Ontology (GO; geneontology.org/) and Uniprot databases (uniprot.org/). A protein-protein interaction network
was constructed according to the STITCH 5.0 database (stitch.embl.de/).
Statistical analysis
Descriptive statistics were conducted to analyze
characteristics of the study groups. Continuous variables are
presented as median (range); categorical variables are reported as
n (%). χ2 and Fisher's exact tests were used to
determine the association between viral infection status and
categorical variables, whereas the Kruskal-Wallis and Dunn's post
hoc tests were performed to compare the median of continuous
variables between study groups. P<0.05 was considered to
indicate a statistically significant difference. SPSS for Windows
version 18.0 (SPSS, Inc.) and GraphPad Prism 9.1.1 (Dotmatics) were
used for the statistical analysis.
Results
Characteristics of the study
population
Table I demonstrates
general and clinical characteristics of each group. All patients
infected with HIV received suppressive ART for >6 months and had
no detectable HIV viral load (data not shown), whereas no specific
medications for HBV and HCV infection were used. All study groups
had similar median ages, infected groups showed the higher
male-to-female sex ratio than the uninfected control group.
Although platelet counts of the three HIV-infected groups and
uninfected group were not significantly different, medians of liver
enzymes AST and ALT, in addition to the fibrosis markers FIB-4 and
APRI, were significantly different. The additional analysis also
indicated that the median values of AST, ALT, FIB-4 score and APRI
in the three HIV-infected groups compared with the uninfected group
differed significantly, excluding the FIB-4 score of
HIV-mono-infection group. Differences in immune status and ART
between the HIV-infected groups were also tested. Median levels of
CD4+ cell count and duration of ART in the
HIV-monifected, HIV/HBV and HIV/HCV coinfected groups, were
compared, yielding no statistical difference. ART drug regimens
used in these three infected groups were not found to be
significantly different (Table
I).
 | Table IGeneral and clinical characteristics
of the patients infected with HIV and the uninfected control group
recruited into the present study. |
Table I
General and clinical characteristics
of the patients infected with HIV and the uninfected control group
recruited into the present study.
| Characteristic | Uninfected
(n=50) | HIV (n=150) |
P-valuec | HIV/HBV (n=19) |
P-valuec | HIV/HCV (n=17) |
P-valuec |
P-valued |
|---|
| Age
(years)a | 44.5
(20.0-77.0) | 44.0
(18.0-86.0) | | 46.0
(29.0-58.0) | | 43.0
(20.0-62.0) | 0.965 | |
| Sexb | | | | | | | | 0.012 |
|
Male | 20(40) | 88 (58.7) | | 14 (73.7) | | 13 (76.5) | | |
|
Female | 30(60) | 62 (41.3) | | 5 (26.3) | | 4 (23.5) | | |
| Platelet count
(x103 cells/µl)a | 274 (194-435) | 275.5 (34-922) | | 276 (110-387) | | 279 (133-447) | 0.816 | |
| Aspartate
aminotransferase (U/l)a | 19 (6-52) | 26 (13-161) | <0.001 | 25 (17-168) | <0.001 | 37 (16-162) | <0.001 | |
| Alanine
aminotransferase (U/l)a | 16 (4-167) | 24 (6-169) | 0.016 | 21 (9-111) | <0.001 | 32 (7-178) | <0.001 | |
| Fibrosis-4
scorea | 0.74
(0.20-1.38) | 0.83
(0.23-9.57) | 0.057 | 0.99
(0.31-3.01) | 0.044 | 1.30
(0.28-3.94) | 0.001 | |
| Alanine
aminotransferase to platelet ratio indexa | 0.18
(0.06-0.38) | 0.23
(0.05-5.09) | <0.001 | 0.23
(0.13-1.77) | 0.004 | 0.53
(0.13-1.34) | <0.001 | |
| CD4+
cell count (cells/µl)a | - | 550 (1-1,644) | | 365 (30-1,016) | | 421 (13-896) | 0.326 | |
| Duration of
anti-retroviral therapy (months)a | - | 62.2
(6.0-169.0) | | 69.3
(6.1-308.4) | | 75.0
(6.7-156.5) | 0.351 | |
| Anti-retroviral
drugsb | | | | | | | | 0.418 |
|
Lamivudine/zidovudine/nevirapine | - | 34 (22.7) | | 2 (10.5) | | 1 (5.9) | | |
|
Tenofovir-based | - | 86 (57.3) | | 13 (68.4) | | 12 (70.6) | | |
|
Other
regimens | - | 30 (20.00) | | 4 (21.1) | | 4 (23.5) | | |
| Nevirapine
experienceb | | | | | | | | 0.205 |
|
Non-nevirapine-based
regimen | - | 112 (75.6) | | 14 (73.7) | | 16 (94.1) | | |
|
Nevirapine-based
regimen | - | 33 (25.33) | | 5 (26.3) | | 1 (5.9) | | |
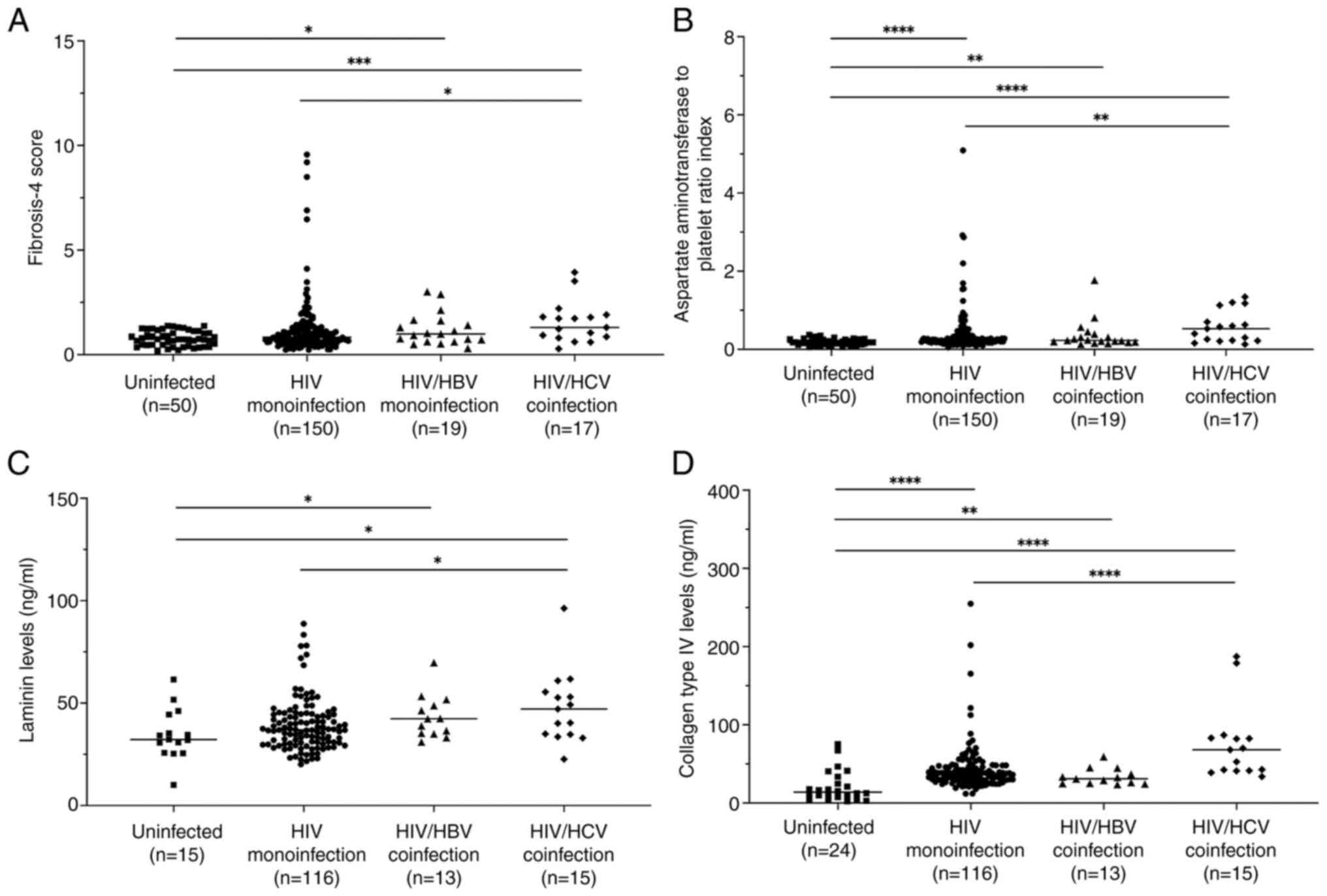
There were higher FIB-4 scores and APRI in the three
HIV-infected groups compared with the uninfected control group,
except for the FIB-4 score in the HIV-monoinfected group (Fig. 2A and B). In addition, significantly higher FIB-4
score and APRI in the HIV/HCV group compared with the
HIV-monoinfection group were observed. Key components of the
extracellular matrix that typically accumulate during liver
fibrosis, namely LN, IVC, PIIINP and HA, coupled with the tumor
marker AFP (33), were examined in
the plasma samples. HIV groups also had significantly higher levels
of LN and IVC compared with the uninfected control group, except
for LN levels in the HIV-monoinfection group (Fig. 2C and D). Consistent with FIB-4 score and APRI,
LN and IVC levels were found to be significantly higher in the
HIV/HCV group compared with HIV monoinfection. Notably, elevated
plasma PIIINP, HA and AFP levels in the infected groups was not
observed, whereas the HIV/HCV group had significantly higher levels
of all of these markers compared with the HIV-monoinfection group
(data not shown).
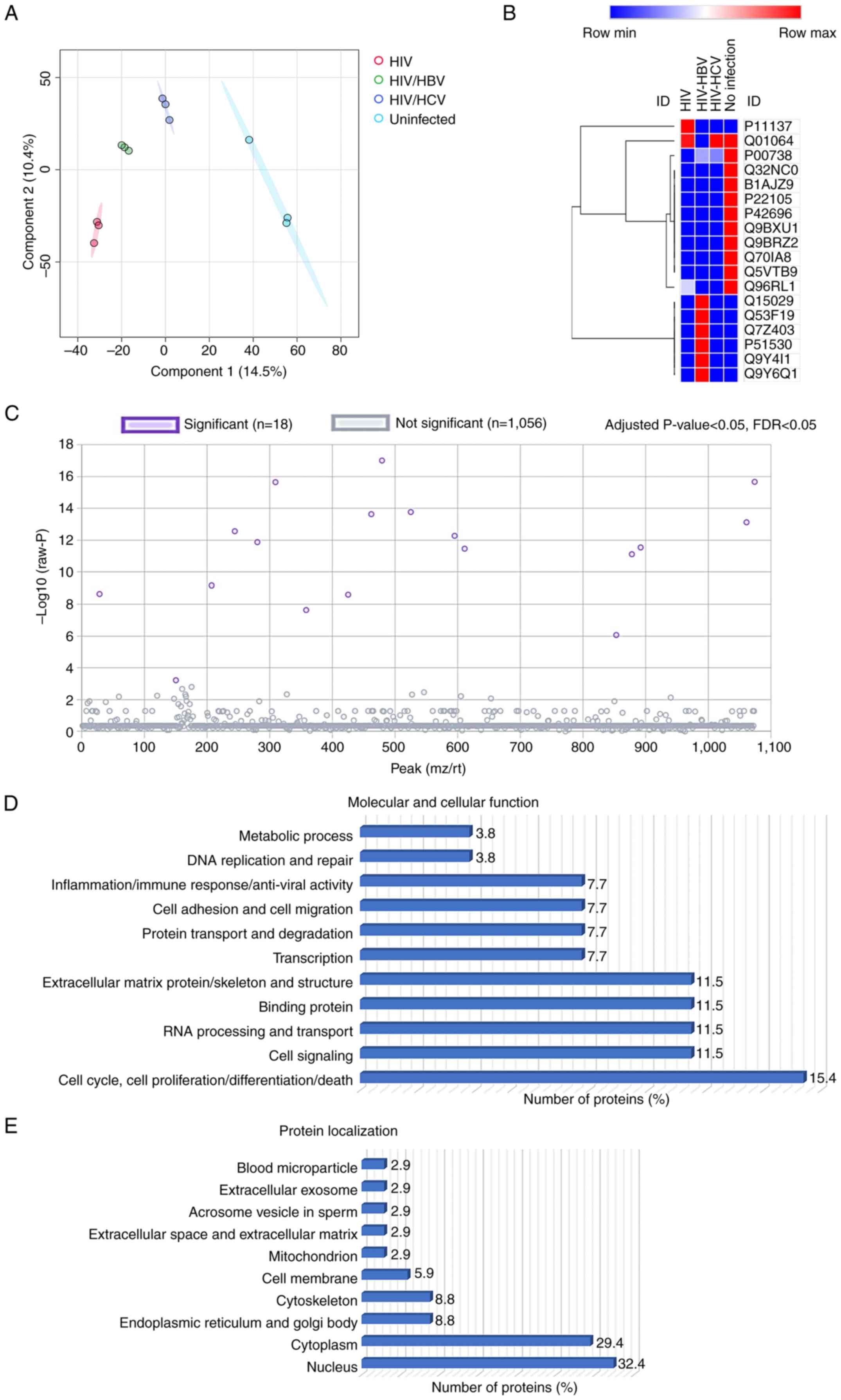
Plasma proteomic profiles
Shotgun proteomics was conducted to explore the
plasma protein profiles. A total of 1,074 proteins were
differentially expressed. PLS-DA model demonstrated clear
separation of the proteome between sample groups, indicating
significant differences in the plasma protein profiles (Fig. 3A). Of these, 18 were significantly
differentially expressed (Fig. 3C;
Table II). PLS-DA model of 18
selected proteins also indicated a clear clustering of proteins
from the four sample groups (data not shown). Analysis for
localization and function of the 18 proteins was conducted using GO
and Uniprot databases (Fig. 3D and
E). These proteins were primarily
located in the nucleus (32.4%) and cytoplasm (29.4%) and in other
intracellular and extracellular compartments (Fig. 3E). The cellular and molecular
functions were enriched in molecular processes, including ‘cell
cycle, cell proliferation, cell differentiation and cell death’
(15.4%), ‘cell signaling’ (11.5%), ‘RNA processing and transport’
(11.5%), ‘binding protein’ (11.5%), ‘extracellular matrix protein
and cytoskeleton’ (11.5%), ‘transcription’ (7.7%), ‘protein
transport and degradation’ (7.7%), ‘cell adhesion and cell
migration’ (7.7%), ‘inflammation, immune response and anti-viral
activity’ (7.7%) ‘DNA replication and repair’ (3.8%) and ‘metabolic
process’ (3.8%; Fig. 3D).
 | Table IISignificantly differentially
expressed proteins between HIV-monoinfected, HIV/hepatitis B/C
virus-coinfected and uninfected controls. |
Table II
Significantly differentially
expressed proteins between HIV-monoinfected, HIV/hepatitis B/C
virus-coinfected and uninfected controls.
| No. | Accession no. | Peptide
sequence | Protein | False discovery
rate | P-value |
|---|
| 1 | Q53F19 |
AGSFITGIDVTSKEAIEK | Nuclear cap-binding
protein subunit 3 |
1.06x10-14 |
9.89x10-18 |
| 2 | Q9Y6Q1 | DLRTYRR | Calpain-6 |
8.12x10-14 |
2.16x10-16 |
| 3 | P51530 | AVLSETFR | DNA replication
ATP-dependent helicase/nuclease DNA 2 |
8.12x10-14 |
2.27x10-16 |
| 4 | Q5VTB9 | FEEYEWCGQKR | E3
ubiquitin-protein ligase RNF220 |
4.59x10-12 |
1.71x10-14 |
| 5 | Q32NC0 | GLHDSCPGQAR | UPF0711 protein
C18orf21 (HBV X-transactivated gene 13 protein) |
5.01x10-12 |
2.33x10-14 |
| 6 | Q9Y4I1 | AACIRIQK | Unconventional
myosin-Va |
1.35x10-11 |
7.56x10-14 |
| 7 | P22105 |
DLRSGTLYSLTLYGLRGPHK | Tenascin-X |
4.23x10-11 |
2.75x10-13 |
| 8 | Q70IA8 | ALCLKQVFAKDK | MOB kinase
activator 3C |
7.25x10-11 |
5.40x10-13 |
| 9 | P42696 |
ESALASADLEEEIHQKQGQKR | RNA-binding protein
34 |
1.60x10-10 |
1.34x10-12 |
| 10 | Q9BXU1 | AATYHRAWR |
Serine/threonine-protein kinase 31 |
3.09x10-10 |
2.87x10-12 |
| 11 | Q7Z403 | ELLAEWQLR | Transmembrane
channel-like protein 6 |
3.39x10-10 |
3.46x10-12 |
| 12 | Q9BRZ2 | AAAAFAR | E3
ubiquitin-protein ligase TRIM56 |
6.87x10-10 |
7.67x10-12 |
| 13 | P11137 |
AEKGLSSVPEIAEVEPSK |
Microtubule-associated protein 2 |
5.70x10-8 |
6.90x10-10 |
| 14 | B1AJZ9 | AAGASGR | Forkhead-associated
domain-containing protein 1 |
1.77x10-7 |
2.30x10-9 |
| 15 | Q15029 |
AFIPAIDSFGFETDLR | 116 kDa U5 small
nuclear ribonucleoprotein component |
1.81x10-7 |
2.53x10-9 |
| 16 | Q01064 |
DWLASTFTQQARAKGR | Dual specificity
calcium/calmodulin-dependent 3', 5'-cyclic nucleotide
phosphodiesterase |
1.57x10-6 |
2.34x10-8 |
| 17 | Q96RL1 |
ADQGDGPEGSGRACSTVEGK | BRCA1-A complex
subunit RAP80 |
5.55x10-5 |
8.73x10-7 |
| 18 | P00738 |
AVGDKLPECEADDGCPKPPEIAHGYVEHSVR | Haptoglobin
(zonulin) |
3.62x10-3 |
6.07x10-4 |
Heatmap analysis demonstrated differential
expression of the 18 selected proteins (Fig. 3B). A total of 10 proteins was
downregulated in the HIV-monoinfected, HIV/HBV and HIV/HCV groups
compared with those in the uninfected control group, included
haptoglobin (HP), HBV X-transactivated gene 13 protein,
forkhead-associated domain-containing protein 1, tenascin-X,
RNA-binding protein 34, serine/threonine-protein kinase 31, E3
ubiquitin-protein ligase tripartite motif-containing 56, Mps one
binder kinase activator 3C, E3 ubiquitin-protein ligase ring finger
protein 220 and BRCA1-A complex subunit RAP80; six proteins were
upregulated in the HIV/HBV group compared with the uninfected
control group (116 kDa U5 small nuclear ribonucleoprotein
component, nuclear cap-binding protein subunit 3, transmembrane
channel-like protein 6, DNA replication ATP-dependent
helicase/nuclease DNA 2, unconventional myosin-V and calpain-6). In
addition, dual specificity calcium/calmodulin-dependent
3',5'-cyclic nucleotide phosphodiesterase was downregulated only in
the HIV/HBV group and microtubule-associated protein 2 was
upregulated only in the HIV-monoinfection group. The differential
expression of these proteins suggests their potential roles in HIV
monoinfection and coinfection with HBV and HCV.
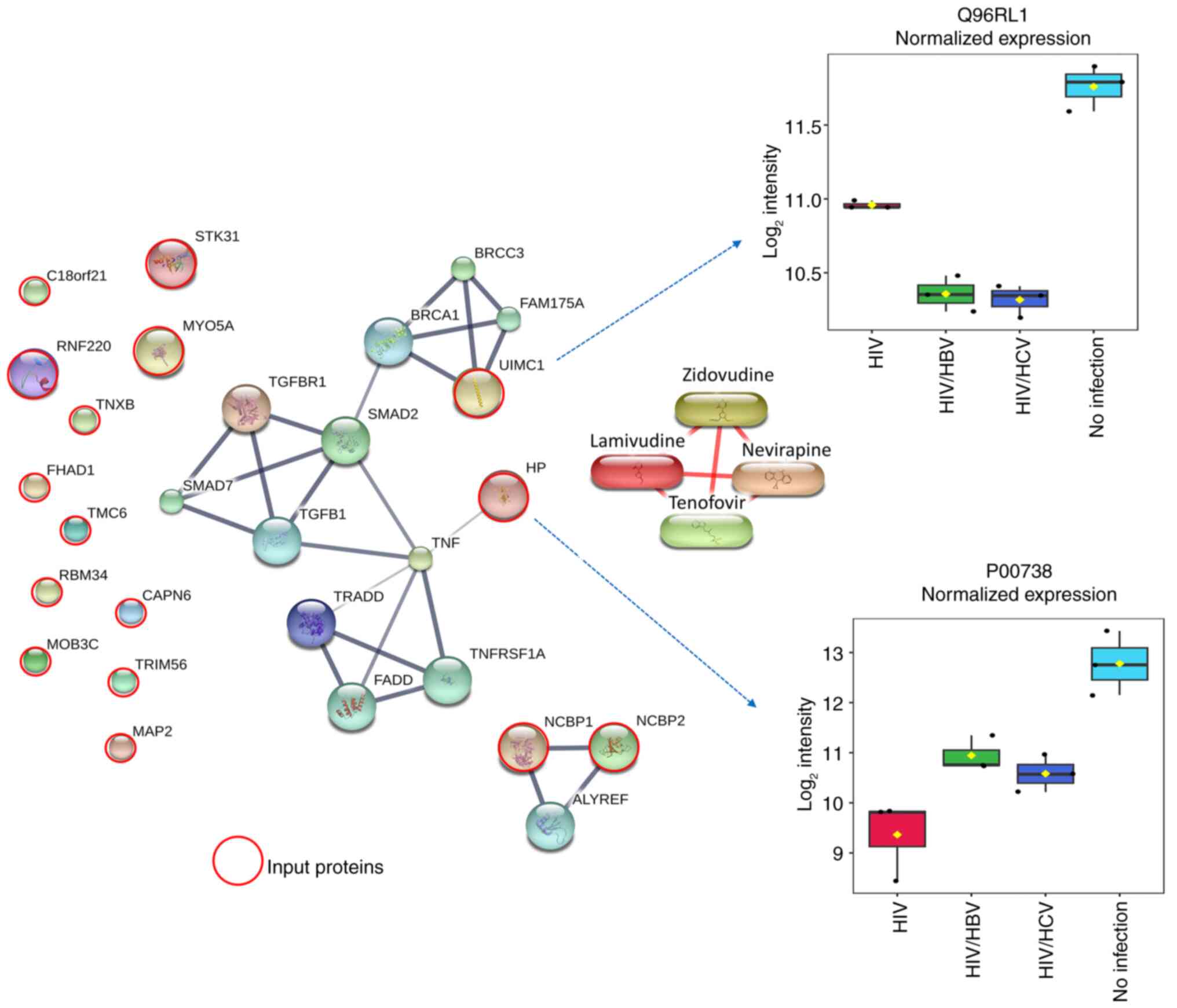
Association between 18 candidate
proteins with profibrogenic, inflammatory and tumorigenic
pathways
To identify the molecular function of plasma protein
profiles potentially associated with HIV infection and coinfection
with HBV and HCV under long-term ART, the 18 selected proteins were
analyzed for their interaction with proteins and chemicals in the
STITCH 5.0 database (Fig. 4). Since
accumulating evidence has indicated key roles of the profibrogenic
TGF-β/SMAD and inflammatory TNF pathways in liver fibrogenesis and
inflammation, especially in HIV and hepatotropic HBV and HCV
(18,21,34,35),
the proteins TGF-β, SMAD, TNF and ART drugs used in the patient
groups (zidovudine, lamivudine, nevirapine and tenofovir) were
included in the STITCH model. Input proteins serine/threonine
kinase 31, chromosome 18 open reading frame 21, ring finger protein
220, myosin VA, tenascin XB, forkhead-associated, phosphopeptide
binding domain 1, transmembrane channel-like 6, RNA binding motif
protein 34, calpain 6, MOB kinase activator 3C, tripartite motif
containing 56, microtubule-associated protein 2, nuclear cap
binding protein subunit 1 and nuclear cap binding protein subunit 2
did not form any interaction with the protein networks. However,
the model predicted close interactions between ubiquitin
interaction motif containing 1 (UIMC1), also named BRCA1-A complex
subunit RAP80, and BRCA1 and TGF-β/SMAD pathways. In addition, an
interaction between haptoglobin (HP) and the TNF pathway was found.
Notably, no protein exhibited interactions with ART drugs.
Additionally, box plots indicated that the expression levels of
UIMC1 and HP were 1.6-2.4 and 4.0-5.5 times lower in the three
HIV-infected groups compared with those in the uninfected control
group, who had no significant liver fibrosis, respectively. These
results suggested the potential involvement of BRCA1-linked
TGF-β/SMAD and TNF pathways during HIV-monoinfection and
coinfection with HBV and HCV, in PLWH receiving suppressive
ART.
Discussion
Accumulating evidence suggests an increased burden
of liver inflammation and fibrosis caused by HIV, HBV and HCV
infection in PLWH (3,36,37).
Previous cross-sectional studies in PLWH revealed a relatively high
prevalence of liver fibrosis in patients with HBV or HCV
coinfection (22,23). Due to accumulating evidence
suggesting progression to more advanced CLD and HCC in PLWH with
HBV/HCV (5,14-16),
the present study aimed to examine the molecular changes in PLWH
compared with virus-free uninfected controls. HIV-monoinfection,
HIV/HBV and HIV/HCV groups had similar durations of ART,
anti-retroviral drug regimens and immune status as assessed using
the CD4+ cell count. The uninfected control group had a
similar median age to the HIV-infected groups and no significant
liver fibrosis. The three HIV groups all showed significantly
higher levels of liver fibrosis as evaluated using FIB-4 score and
APRI, coupled with higher levels of extracellular matrix proteins
LN and IVC, compared with the uninfected group; analysis also
revealed higher levels of liver fibrosis in the HIV/HCV group
compared with those in the HIV-monoinfection group. This was
consistent with previous studies, indicating CLD in PLWH receiving
ART and the influence of HCV infection in development of liver
fibrosis in patients infected with HIV/HCV (5,11-16)
The increased levels of liver fibrosis markers in HIV-infected
groups may support potential progression to CLD in PLWH with
HIV-monoinfection, HBV- and HCV-coinfection. Therefore, long-term
monitoring of potential progression to advanced CLD is required in
this study group.
The present study indicate differentially expression
of the 18 candidate proteins suggesting their potential roles in
the disease progrestion of HIV monoinfection and/or coinfection
with HBV and HCV in PLWH and the functions of the candidate
proteins were examined. Molecular functions of plasma proteins
potentially associated with HIV monoinfection and coinfection with
HBV and HCV in PLWH under long-term suppressive ART were predicted
by STITCH 5.0 analysis. The STITCH model indicated no interaction
between any candidate proteins with the four ARV drugs, suggesting
no involvement of the proteins with mechanisms of the ARV drugs
most commonly used in the HIV-moninfection group. However, the
model revealed interactions between UIMC1 protein and BRCA1 and
profibrogenic TGF-β/SMAD pathways, whereas an interaction was also
found between HP and TNF pathways, which serve key roles in liver
fibrogenesis and inflammation in HIV and hepatotropic HBV and HCV
infection (18,21,34,35).
Heatmap analysis and box plots demonstrated that these two proteins
were downregulated in all HIV-infected groups compared with those
in the uninfected control group. The analysis suggested potential
involvement of BRCA1, TGF-β/SMAD and TNF pathways under of
HIV-monoinfection and/or coinfection with HBV and HCV. In addition,
novel protein targets UIMC1 and HP were identified to understand
the molecular mechanisms of liver fibrosis in PLWH receiving
suppressive ART.
UIMC1 or BRCA1-A complex subunit RAP80, a major
contributor in the BRCA1 complex, functions in the DNA damage
repair response (38,39). UIMC1 is a ubiquitin-binding protein
targeting a complex containing BRCA1-BRCA1-associated ring domain
protein 1 E3 ligase at double-strand breaks. This is required for
DNA damage resistance, cell cycle checkpoint and DNA repair
(40,41). In vivo study has indicated
that loss of UIMC1 suppresses recruitment of the BRCA1 complex to
DNA damage sites and abolishes the repair process (42). BRCA1 is a tumor suppressor, the
mutation or epigenetic inactivation of which increases the risk of
various types of cancer, including HCC (43). To the best of our knowledge, there
is a lack of direct evidence for an association between BRCA1
expression and liver disease caused by HIV and hepatotropic
viruses. However, BRCA1/2 is one of the most commonly altered DNA
damage repair genes associated with higher tumor mutation burden in
patients diagnosed with primary liver cancer (44) and BRCA1 expression is associated
with immune cell infiltration and proposed as a prognostic
indicator of HCC (43). Here, the
lower expression of UIMC1 in all three infected groups compared
with that in uninfected groups may have impaired DNA repair
response, potentially supporting tumourigenesis. HP is a secreted
protein that has been reported to be an important regulator of
intestinal barrier function (45,46).
The HP pathway is involved in intestinal innate immunity and is
dysregulated during chronic inflammatory diseases (45,46).
HIV infection causes mucosal disruption in the gut, alteration of
microbial composition and microbial translocation. These processes
lead to chronic immune activation and inflammation, which are
associated with non-AIDS comorbidities, including CLD (3). Consistent with the present study,
previous studies have reported lower levels of serum HP in patients
with chronic hepatitis B compared with controls (47) and an association between decreased
HP levels with mortality in PLWH (48). However, increased serum levels of HP
in more advanced liver disease are also reported (49,50).
Accordingly, the lower expression of UIMC1 and HP proteins in all
three infected groups compared with that in the uninfected control
group, coupled with interactions with the profibrogenic TGF-β/SMAD,
inflammatory TNF and tumor suppressor BRCA1 pathways, suggest
potential roles of these proteins in liver inflammation and the
progression to more advanced liver disease and HCC in PLWH.
The present study had limitations due to
characteristics of the study groups and the cross-sectional design.
The present study contained a relatively small number of subjects
and information on their clinical symptoms was not obtained. In
additon, there were significant differences in the sex distribution
in the uninfected control and HIV-infected groups. However, in a
previous study, multivariate analysis adjusted for different
parameters suggested that sex was not a significant factor for
liver fibrosis in PLWH (23). The
influence of sex differences in the plasma protein profiles in the
present study should be taken into account. There were also no
specific treatments for HBV and HCV coinfection in any of the study
groups, nor was the status of HBV and HCV infection, including HBV
and HCV viral loads, available. Therefore, it remains unclear
whether these parameters are associated with the protein profiles
in the present study. To confirm the reliability of the protein
profiles, the expression of candidate proteins, particularly UIMC1
and HP, should be validated and monitored in longitudinal studies.
In addition, studies on the molecular and cellular functions of
these proteins in the progression of HIV are warranted.
In conclusion, the present study was conducted on
PLWH with HIV monoinfection or HBV and HCV coinfection receiving
long-term ART. The comparative proteomics analysis revealed plasma
protein profiles potentially associated with HIV infection,
coinfection with HBV and HCV and liver fibrosis. STITCH model
identified UIMC1 and HP proteins, which were associated with the
profibrogenic TGF-β/SMAD, inflammatory TNF and tumor suppressor
BRCA1 pathways, suggesting their potential role in inflammation,
fibrosis and tumorigenesis in the liver. The proteomics data
support the molecular basis of progressive liver disease in PLWH
receiving long-term suppressive ART.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Thailand Science
Research and Innovation Fundamental Fund fiscal year 2022 (grant
no. 2493097) and Thammasat University Research Unit in Diagnostic
Molecular Biology of Chronic Diseases related to Cancer.
Availability of data and materials
The MS/MS raw data and analysis files have been
deposited in the ProteomeXchange Consortium (proteomecentral.proteomexchange.org) via the jPOST
partner repository (jpostdb.org)
with the data set identifier JPST003189 and PXD PXD053504
(proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD053504).
Authors' contributions
CA contributed to funding acquisition resources,
supervision, study design, data analysis, manuscript preparation,
review and editing. CT performed the experiments, data collection
and analysis and manuscript preparation. JK conducted the detection
of ECM fibrosis markers and tumor marker AFP and data analysis. TS
and WS participated in blood samples and clinical data collection.
NP and SR performed LC-MS/MS and data analysis. All authors read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Human Ethics
Committees No. 3, Thammasat University (approval no. 116/2565) and
Certificated Biological Safety Committee, Thammasat University,
Pathumthani, Thailand. Written informed consent was obtained from
the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
UNAIDS: Global HIV & AIDS
statistics-Fact sheet. 2023.
|
|
2
|
Deeks SG, Lewin SR and Havlir DV: The end
of AIDS: HIV infection as a chronic disease. Lancet. 382:1525–1533.
2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Zicari S, Sessa L, Cotugno N, Ruggiero A,
Morrocchi E, Concato C, Rocca S, Zangari P, Manno EC and Palma P:
Immune Activation, Inflammation, and Non-AIDS Co-Morbidities in
HIV-Infected Patients under Long-Term ART. Viruses.
11(200)2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Sherman KE and Thomas DL: HIV and liver
disease: A comprehensive update. Top Antivir Med. 30:547–558.
2022.PubMed/NCBI
|
|
5
|
Han WM, Ueaphongsukkit T, Chattranukulchai
P, Siwamogsatham S, Chaiteerakij R, Sophonphan J, Gatechompol S,
Ubolyam S, Phonphithak S, Ruxrungtham K, et al: Incident liver
cirrhosis, associated factors, and cardiovascular disease risks
among people living with HIV: A longitudinal study. J Acquir Immune
Defic Syndr. 86:463–472. 2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Sun J, Althoff KN, Jing Y, Horberg MA,
Buchacz K, Gill MJ, Justice AC, Rabkin CS, Goedert JJ, Sigel K, et
al: Trends in Hepatocellular Carcinoma Incidence and Risk Among
Persons With HIV in the US and Canada, 1996-2015. JAMA Netw Open.
4(e2037512)2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Torgersen J, Kallan MJ, Carbonari DM, Park
LS, Mehta RL, D'Addeo K, Tate JP, Lim JK, Goetz MB,
Rodriguez-Barradas MC, et al: HIV RNA, CD4+ Percentage, and Risk of
Hepatocellular Carcinoma by Cirrhosis Status. J Natl Cancer Inst.
112:747–755. 2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Chamroonkul N and Bansal MB: HIV and the
liver. Nat Rev Gastroenterol Hepatol. 16:1–2. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ganesan M, Poluektova LY, Kharbanda KK and
Osna NA: Human immunodeficiency virus and hepatotropic viruses
co-morbidities as the inducers of liver injury progression. World J
Gastroenterol. 25:398–410. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Sherman KE, Peters MG and Thomas D: Human
immunodeficiency virus and liver disease: A comprehensive update.
Hepatol Commun. 1:987–1001. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Mohr R, Schierwagen R, Schwarze-Zander C,
Boesecke C, Wasmuth JC, Trebicka J and Rockstroh JK: Liver Fibrosis
in HIV Patients Receiving a Modern cART: Which Factors Play a Role?
Medicine (Baltimore). 94(e2127)2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Rivero-Juarez A, Camacho A, Merchante N,
Pérez-Camacho I, Macias J, Ortiz-Garcia C, Cifuentes C,
Torre-Cisneros J, Peña J, Pineda JA, et al: Incidence of liver
damage of uncertain origin in HIV patients not co-infected with
HCV/HBV. PLoS One. 8(e68953)2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Sulyok M, Ferenci T, Makara M, Horváth G,
Szlávik J, Rupnik Z, Kormos L, Gerlei Z, Sulyok Z and Vályi-Nagy I:
Hepatic fibrosis and factors associated with liver stiffness in HIV
mono-infected individuals. PeerJ. 5(e2867)2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Medrano LM, Garcia-Broncano P, Berenguer
J, González-García J, Jiménez-Sousa MÁ, Guardiola JM, Crespo M,
Quereda C, Sanz J, Canorea I, et al: Elevated liver stiffness is
linked to increased biomarkers of inflammation and immune
activation in HIV/hepatitis C virus-coinfected patients. AIDS.
32:1095–1105. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Portocarrero Nunez JA, Gonzalez-Garcia J,
Berenguer J, Gallego MJV, Loyarte JAI, Metola L, Bernal E, Navarro
G, Del Amo J and Jarrín I: and the Cohort of the Spanish HIV
Research Network (CoRIS). Impact of co-infection by hepatitis C
virus on immunological and virological response to antiretroviral
therapy in HIV-positive patients. Medicine (Baltimore).
97(e12238)2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Kim HN, Newcomb CW, Carbonari DM, Roy JA,
Torgersen J, Althoff KN, Kitahata MM, Reddy KR, Lim JK, Silverberg
MJ, et al: Risk of HCC With Hepatitis B Viremia Among
HIV/HBV-Coinfected Persons in North America. Hepatology.
74:1190–1202. 2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Shmagel KV, Saidakova EV, Shmagel NG,
Korolevskaya LB, Chereshnev VA, Robinson J, Grivel JC, Douek DC,
Margolis L, Anthony DD and Lederman MM: Systemic inflammation and
liver damage in HIV/hepatitis C virus coinfection. HIV Med.
17:581–589. 2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Shata MTM, Abdel-Hameed EA, Rouster SD, Yu
L, Liang M, Song E, Esser MT, Shire N and Sherman KE: HBV and
HIV/HBV infected patients have distinct immune exhaustion and
apoptotic serum biomarker profiles. Pathog Immun. 4:39–65.
2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Lin W, Tsai WL, Shao RX, Wu G, Peng LF,
Barlow LL, Chung WJ, Zhang L, Zhao H, Jang JY and Chung RT:
Hepatitis C virus regulates transforming growth factor beta1
production through the generation of reactive oxygen species in a
nuclear factor kappaB-dependent manner. Gastroenterology.
138:2509–2518, 2518 e1. 2010.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Salloum S, Holmes JA, Jindal R, Bale SS,
Brisac C, Alatrakchi N, Lidofsky A, Kruger AJ, Fusco DN, Luther J,
et al: Exposure to human immunodeficiency virus/hepatitis C virus
in hepatic and stellate cell lines reveals cooperative profibrotic
transcriptional activation between viruses and cell types.
Hepatology. 64:1951–1968. 2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Ming D, Yu X, Guo R, Deng Y, Li J, Lin C,
Su M, Lin Z and Su Z: Elevated TGF-β1/IL-31 pathway is associated
with the disease severity of hepatitis B virus-related liver
cirrhosis. Viral Immunol. 28:209–216. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Akekawatchai C, Sretapunya W,
Pipatsatitpong D and Chuenchit T: Hepatitis B or C virus
coinfection in and risks for transaminitis in human
immunodeficiency virus-infected Thais on combined antiretroviral
therapy. Asian Biomedicine. 9:353–361. 2015.
|
|
23
|
Chiraunyanann T, Changsri K, Sretapunya W,
Yuenyongchaiwat K and Akekawatchai C: CXCL12 G801A polymorphism is
associated with significant liver fibrosis in HIV-infected Thais: A
cross-sectional study. Asian Pac J Allergy Immunol. 37:162–170.
2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Foca E, Fabbiani M, Prosperi M, Quiros
Roldan E, Castelli F, Maggiolo F, Di Filippo E, Di Giambenedetto S,
Gagliardini R, Saracino A, et al: Liver fibrosis progression and
clinical outcomes are intertwined: Role of CD4+ T-cell count and
NRTI exposure from a large cohort of HIV/HCV-coinfected patients
with detectable HCV-RNA: A MASTER cohort study. Medicine
(Baltimore). 95(e4091)2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sterling RK, Lissen E, Clumeck N, Sola R,
Correa MC, Montaner J, S Sulkowski M, Torriani FJ, Dieterich DT,
Thomas DL, et al: Development of a simple noninvasive index to
predict significant fibrosis in patients with HIV/HCV coinfection.
Hepatology. 43:1317–1325. 2006.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wai CT, Greenson JK, Fontana RJ,
Kalbfleisch JD, Marrero JA, Conjeevaram HS and Lok AS: A simple
noninvasive index can predict both significant fibrosis and
cirrhosis in patients with chronic hepatitis C. Hepatology.
38:518–526. 2003.PubMed/NCBI View Article : Google Scholar
|
|
27
|
World Health Organization: Guidelines for
the Care and Treatment of Persons Diagnosed with Chronic Hepatitis
C Virus Infection. 2018.
|
|
28
|
Lowry OH, Rosebrough NJ, Farr AL and
Randall RJ: Protein measurement with the Folin phenol reagent. J
Biol Chem. 193:265–275. 1951.PubMed/NCBI
|
|
29
|
Tyanova S, Temu T and Cox J: The MaxQuant
computational platform for mass spectrometry-based shotgun
proteomics. Nat Protoc. 11:2301–2319. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Howe EA, Sinha R, Schlauch D and
Quackenbush J: RNA-Seq analysis in MeV. Bioinformatics.
27:3209–3210. 2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Pang Z, Zhou G, Ewald J, Chang L, Hacariz
O, Basu N and Xia J: Using MetaboAnalyst 5.0 for LC-HRMS spectra
processing, multi-omics integration and covariate adjustment of
global metabolomics data. Nat Protoc. 17:1735–1761. 2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Starruss J, de Back W, Brusch L and
Deutsch A: Morpheus: A user-friendly modeling environment for
multiscale and multicellular systems biology. Bioinformatics.
30:1331–1332. 2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Dong H, Xu C, Zhou W, Liao Y, Cao J, Li Z
and Hu B: The combination of 5 serum markers compared to FibroScan
to predict significant liver fibrosis in patients with chronic
hepatitis B virus. Clin Chim Acta. 483:145–150. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Dewidar B, Meyer C, Dooley S and
Meindl-Beinker AN: TGF-β in hepatic stellate cell activation and
liver fibrogenesis-updated 2019. Cells. 8(1419)2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Steele H, Cheng J, Willicut A, Dell G,
Breckenridge J, Culberson E, Ghastine A, Tardif V and Herro R: TNF
superfamily control of tissue remodeling and fibrosis. Front
Immunol. 14(1219907)2023.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Demirkol ME, Aktas G, Bilgin S, Kahveci G,
Kurtkulagi O, Atak BM and Duman TT: C-reactive protein to
lymphocyte count ratio is a promising novel marker in hepatitis C
infection: The clear hep-c study. Rev Assoc Med Bras (1992).
68:838–841. 2022.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Kosekli MA: Mean platelet volume and
platelet to lymphocyte count ratio are associated with hepatitis
B-related liver fibrosis. Eur J Gastroenterol Hepatol. 34:324–327.
2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Anamika Markin CJ, Rout MK and
Spyracopoulos L: Molecular basis for impaired DNA damage response
function associated with the RAP80 ∆E81 defect. J Biol Chem.
289:12852–12862. 2014.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Yan J, Kim YS, Yang XP, Li LP, Liao G, Xia
F and Jetten AM: The ubiquitin-interacting motif containing protein
RAP80 interacts with BRCA1 and functions in DNA damage repair
response. Cancer Res. 67:6647–6656. 2007.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Sobhian B, Shao G, Lilli DR, Culhane AC,
Moreau LA, Xia B, Livingston DM and Greenberg RA: RAP80 targets
BRCA1 to specific ubiquitin structures at DNA damage sites.
Science. 316:1198–1202. 2007.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Wang M, Gong Q, Zhang J, Chen L, Zhang Z,
Lu L, Yu D, Han Y, Zhang D, Chen P, et al: Characterization of gene
expression profiles in HBV-related liver fibrosis patients and
identification of ITGBL1 as a key regulator of fibrogenesis. Sci
Rep. 7(43446)2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Wu J, Liu C, Chen J and Yu X: RAP80
protein is important for genomic stability and is required for
stabilizing BRCA1-A complex at DNA damage sites in vivo. J Biol
Chem. 287:22919–22926. 2012.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Mei J, Wang R, Xia D, Yang X, Zhou W, Wang
H and Liu C: BRCA1 Is a novel prognostic indicator and associates
with immune cell infiltration in hepatocellular carcinoma. DNA Cell
Biol. 39:1838–1849. 2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Lin J, Shi J, Guo H, Yang X, Jiang Y, Long
J, Bai Y, Wang D, Yang X, Wan X, et al: Alterations in DNA damage
repair genes in primary liver cancer. Clin Cancer Res.
25:4701–4711. 2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Sturgeon C and Fasano A: Zonulin, a
regulator of epithelial and endothelial barrier functions, and its
involvement in chronic inflammatory diseases. Tissue Barriers.
4(e1251384)2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Fasano A: Zonulin and its regulation of
intestinal barrier function: The biological door to inflammation,
autoimmunity, and cancer. Physiol Rev. 91:151–175. 2011.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Calgin MK and Cetinkol Y: Decreased levels
of serum zonulin and copeptin in chronic Hepatitis-B patients. Pak
J Med Sci. 35:847–851. 2019.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Hunt PW, Sinclair E, Rodriguez B, Shive C,
Clagett B, Funderburg N, Robinson J, Huang Y, Epling L, Martin JN,
et al: Gut epithelial barrier dysfunction and innate immune
activation predict mortality in treated HIV infection. J Infect
Dis. 210:1228–1238. 2014.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Voulgaris TA, Karagiannakis D, Hadziyannis
E, Manolakopoulos S, Karamanolis GP, Papatheodoridis G and
Vlachogiannakos J: Serum zonulin levels in patients with liver
cirrhosis: Prognostic implications. World J Hepatol. 13:1394–1404.
2021.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Wang X, Li MM, Niu Y, Zhang X, Yin JB,
Zhao CJ and Wang RT: Serum Zonulin in HBV-Associated chronic
hepatitis, liver cirrhosis, and hepatocellular carcinoma. Dis
Markers. 2019(5945721)2019.PubMed/NCBI View Article : Google Scholar
|