Introduction
The classical enzymatic function of AChE is the
precise temporary hydrolysis of ACh at the cholinergic synapse.
However, the biological role of AChE is not limited to cholinergic
transmission (1,2). The aforementioned findings are
supported by the high levels of AChE activity estimated in
nonneuronal tissues, such as blood cells, erythrocytes, lymphocytes
and megakaryocytes (3). The
structure and functions of AChE have been the subject of various
investigations worldwide. However, a number of unresolved questions
remain, both in terms of its mechanism of action, synthesis
processes and transport through cellular compartments. The
physiological meaning of AChE molecular polymorphisms is also
unknown, although their roles in apoptosis, proliferation, cell
differentiation, cell adhesion, neurogenesis and hematopoiesis and
possible participation in other biological activities have begun to
be elucidated (1,4-8).
The structural complexity of human AChE is due to a
gene with a size of 7 kb located in the q22 region of chromosome 7
(9,10). The ACHE gene produces three
different mRNAs (T, H and R) through an alternative splicing
process (11). The generated mRNAs
share exons E2, E3 and E4, which encode the catalytic domain of
AChE (12,13). The AChE-T-type mRNA is assembled
with exons E2-E3-E4 and E6; it encodes amphiphilic monomers, dimers
and tetramers (G1A, G2A
and G4A), hydrophilic tetramers
(G4H) and hetero-oligomeric associations with
a collagen-like stem Q, structurally conforming to the asymmetric
forms (A4, A8 and A12, which
contain one, two and three tetramers, respectively), or with the
transmembrane protein proline-rich membrane anchor (PRiMA),
constituting membrane-bound tetramers
PRiMA-G4A (14-16).
The AChE-H-type mRNA contains exons E2-E3-E4-E5, which encode Type
I amphiphilic monomers and dimers (G1A and
G2A) in which each subunit has a covalently
linked glycophosphatidylinositol residue (11,16,17).
The AChE-R-type mRNA, also called read-through, does not undergo
splicing after the last exon encoding the catalytic domain and,
therefore, has the arrangement E2-E3-E4-I4-E5. The C-terminus of
the R subunit consists of 30 amino acids and lacks Cys and thus the
subunits remain as hydrophilic monomers (G1H)
(18,19). Abnormalities in the ACHE gene have
been observed in various types of cancer, such as ovarian, breast,
prostate, liver and some types of myeloid leukemia (5,20-22).
In general, both brain and nonbrain tumors present
an alteration in AChE activity; in a number of cases, a decrease in
specific AChE activity is observed (23,24).
Lung cancer shows a decrease in AChE-T mRNA levels (25), whereas colon cancer shows decreases
in AChE-T, AChE-H and AChE-R mRNA levels (26,27).
The decrease in mRNA levels is related to a decrease in AChE
activity (25,28). Furthermore, alterations in the ACHE
gene in breast cancer decrease AChE enzymatic activity in axillary
lymph nodes (29). In other types
of cancer, such as in gliomas (30), liver tumors (5,31),
gastric cancer (32) and head and
neck carcinomas, a reduction in the enzymatic activity of AChE has
been observed, where the low activity of AChE is associated with
worse survival (33,34). In patients with prostate cancer, a
decrease in AChE activity in the blood has been reported compared
with that in a control group of healthy subjects; this finding
opens the possibility of proposing AChE as a cancer biomarker
(35). All these reported events,
where a decrease in both the expression and enzymatic activity of
AChE occur, cause an increase in the amount of ACh that leads to
cholinergic overstimulation and an increase in cell proliferation
(36-40).
Antisense polynucleotides against AChE produce
essential changes in hematopoiesis, decreasing apoptosis and
increasing the proliferation of blood cells (41,42).
Organophosphate pesticides are potent inhibitors of AChE and their
use in agricultural activities is associated with the development
of non-Hodgkin lymphoma and leukemia (21,43-46).
Other studies of rat mammary glands have demonstrated that the
inhibition of AChE with physostigmine induces cancer development,
revealing a relationship between a decrease in AChE activity and
carcinogenesis (47).
A clear relationship exists between low AChE
activity and a poor prognosis for cancer patients. However,
determining why reducing AChE activity and protein content induces
tumor progression and aggressiveness is important. With respect to
the information available regarding AChE gene and protein
expression in cancerous tissues, research still needs to be
performed to clarify the molecular mechanisms by which AChE could
participate in types of cancer. Exploring study models that reveal
new information about the relationship of this enzyme with the
processes of altered cell proliferation is essential (5). Based on the aforementioned findings,
hematopoietic cells, mainly T lymphocytes, have a well-established
nonneuronal cholinergic system called the lymphocytic cholinergic
system (3,48-50),
which is why the present study proposed it as a study model to
obtain an improved understanding of the differences in AChE
expression between normal cells and cancer cells.
Based on the reported evidence that AChE expression
is altered in cancer cells compared with that in their normal
counterparts, the present study aimed to compare AChE expression at
the mRNA and protein levels, as well as the glycosylation and
enzymatic activity of the AChE protein between normal T lymphocytes
and the human T lymphoblast cell line Jurkat E6-1.
Materials and methods
Reagents and general materials
Fetal bovine serum (FBS), penicillin, streptomycin,
L-glutamine and sodium pyruvate solutions were purchased from
Invitrogen (Thermo Fisher Scientific, Inc.). The other reagents
mentioned were acquired from MilliporeSigma.
ethylenediaminetetraacetic acid (EDTA)-Vacutainer tubes for blood
collection from healthy volunteers were acquired from Becton,
Dickinson and Company.
Cell culture
The human T lymphoblast cell line Jurkat E6-1
(TIB-152) was obtained from the American Type Culture Collection.
Jurkat E6-1 cells were cultured in RPMI-1640 medium
(MilliporeSigma) supplemented with 10% (v/v) FBS, 2 mM L-glutamine,
1 mM sodium pyruvate, 100 U/ml penicillin and 100 mg/ml
streptomycin. Jurkat cells from passages 2 to 5 were grown at 37˚C
in a humidified 5% CO2 atmosphere for all
experiments.
The human liver cancer cell line HepG2 (HB-8065) was
obtained from the American Type Culture Collection. HepG2 cells
were cultured in DMEM (MilliporeSigma) supplemented with 10% (v/v)
FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, 100 U/ml penicillin
and 100 mg/ml streptomycin at 37˚C in a humidified atmosphere
containing 5% CO2.
Obtaining T lymphocytes from healthy
adults
The use of lymphocytes from healthy donors was
included to perform comparative tests with leukemic cells. The
sample consisted of 10 young adults (aged 18-22 years) selected
from the period between January, 2023 and April, 2023, according to
the following inclusion and exclusion criteria.
The inclusion criteria were: Age between 18-22
years, weight between 50-80 kg, systolic blood pressure between
90-160 mm Hg, diastolic blood pressure between 60-90 mm Hg, pulse
between 50-100 beats and fasting for 6-8 h. The volunteers could
not consume foods with fat, eggs, milk, or their derivatives 24 h
before donation.
The exclusion criteria were as follows: Pregnant or
breastfeeding women; individuals who had suffered from flu, cough,
diarrhea or dental infection in the last 14 days; individuals who
had taken medications in the last five days; individuals who had
undergone endodontic treatment, acupuncture or had tattoos or
piercings in the last 12 months; those who had undergone surgery in
the last six months; those who had been vaccinated in the last 30
days; and those who consumed alcoholic beverages in the 72 h prior
to donation.
Informed consent was obtained from all individual
participants included in the study to the collection and use of
their cells strictly for research purposes. The research ethics
committee approved the use of blood samples donated by healthy
adults with a registration in the National Bioethics Commission
with the number CONBIOETICA-09-CEI-025-20161215 of the National
Institute of Pediatrics, Mexico.
Density gradient separation was performed from 20 ml
of blood, which was generated via density gradient separation using
Lymphoprep density gradient medium (STEMCELL Technologies) and
centrifuged at 800 x g for 20 min at room temperature (20-25˚C) to
extract the mononuclear cells (including T lymphocytes), which were
subsequently resuspended in 1 ml of PBS.
Magnetic beads conjugated with an anti-CD3 antibody
(specific T lymphocyte marker) MAC (Miltenyi Biotec, Inc.) were
added to the suspension of mononuclear cells. T lymphocytes were
isolated by magnetic separation with MiniMACS Starting Kit
(Miltenyi Biotec, Inc.) according to the manufacturer's protocol.
This device collects the entire cell suspension and separates the
cells into a positive fraction (T lymphocytes) and a negative
fraction (the remainder of the cells). The magnetic field of the
MACS column allows for minimal labeling of target cells with small
microbeads. This means that there are plenty of surface epitopes
available for fluorescent staining and flow cytometry analysis. The
positive fraction was recovered and the purity of the T lymphocytes
was determined via flow cytometry with a FACSCalibur 2 Laser, 4
Color flow cytometer (Becton, Dickinson and Company); the CD3-PerCP
antibody was used incubated for 30 min at room temperature
(20-25˚C). The samples were processed with CellQuest Pro v 5.2.1.
Software (BD Biosciences) via the SSC and the association with
CD3-PerCP.
Trypan blue (MicroLab-Mexico) was used to determine
the viability and density of total leukocytes, purified T
lymphocytes and Jurkat cells. Cells were incubated with equal
volume of 0.4% trypan blue solution for 3 min at room temperature
(20-25˚C). Viable cells at a density of 1x107 cells/ml
were used for the assays.
Total RNA extraction
Total RNA was extracted from 10x107 cell
samples with 1 ml of TRIzol (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. The isolated RNA was
partially dissolved in 100 µl of RNase-free water and incubated at
55˚C for 5 min. The RNA concentration and purity were determined
with a Nanodrop spectrophotometer (Thermo Fisher Scientific, Inc.)
by calculating the optical density ratio at wavelengths of 260/280
nm and 260/230 nm.
Reverse transcription (RT)-PCR
amplification
The RT-PCR process was performed using the one-step
RT-PCR kit (Qiagen, Inc.) and the following primer pairs: AChE-H
(487 bp), forward 5'-TCTCGAAACTACACGGCAGA-3' and reverse
5'-TGAGGAGGAAGGGAGCACTA-3'; AChE-T (444 bp), forward
5'-TCTCGAAACTACACGGCAGA-3' and reverse 5'-GCCCAGCCCTGAAATAAATAG-3';
AChE-R (333 bp), forward 5'-TCTCGAAACTACACGGCAGA-3' and reverse
5'-GGGGAGAAGAGAGGGGTTAC-3'; and β-actin (500 bp) as a housekeeping
gene, forward 5'-CACTGGCATCGTGATGGACT-3' and reverse
5'-AATGCCAGGGTACATGGTGG-3'. A total of 1 µM of each primer, 1 µl of
a 40 mM dNTP mixture, 5 µl of 5X Q solution, 1 µl of enzyme mix and
1.5 µg of RNA sample were used (titrations were performed using
different amounts of RNA, 0.5-2.0 µg), for a final volume of 25 µl
that was completed with RNase-free water.
The samples were placed in a Mastercycler Gradient
Thermal Cycler (Eppendorf SE) and cycled with the following
program: 50˚C for 30 min and 95˚C for 15 min for reverse
transcription, followed by 39 cycles (1 min at 94˚C, 1 min at 50˚C
and 1 min at 72˚C) for amplification. After the final cycle, the
temperature was maintained at 72˚C for 10 min. The amplified DNA
fragments were visualized on a 2% (w/v) agarose gel containing 0.5
µg/ml ethidium bromide. Image acquisition was performed using a
Molecular Imager Gel Doc XR + system (Bio-Rad Laboratories, Inc.).
The optical density of each band was calculated after background
subtraction and normalization to β-actin for semiquantitative
analysis using Image Studio 4.0 software (LI-COR Biosciences).
Solubilization of the AChE
protein
For AChE solubilization from isolated T lymphocytes
and Jurkat cells, 1x107 cells were used per sample. Each
sample was homogenized with HEPES saline buffer [15 mM HEPES, 1 M
NaCl, 50 mM MgCl2, 1 mM ethylene
glycol-bis(2-aminoethylether)-N,N,N',N'-tetraacetic acid and 3 mM
EDTA, pH 7.5] with a mixture of protease inhibitors at 10% cell
weight/buffer volume (w/v). The homogenate was centrifuged at
200,000 x g in a Beckman Ti 60 rotor (Beckman Coulter, Inc.) for 1
h at 4˚C to separate soluble AChE from AChE weakly bound to the
membranes. After the first supernatants (S1) were
collected, the precipitates were homogenized with HEPES saline
buffer supplemented with 1% (w/v) Triton X-100. These suspensions
were subsequently centrifuged at 200,000 x g for 1 h at 4˚C to
obtain the enzyme bound to the membranes in the second supernatant
(S2). Centrifugation was performed using a Beckman XL-90
Optima centrifuge (Beckman Coulter, Inc.). The enzymatic activity
of AChE was subsequently determined in both fractions
(S1 and S2).
Estimation of AChE enzymatic
activity
The measurement of AChE activity was performed via
spectrophotometry with a Cary 50 spectrophotometer (Agilent
Technologies, Inc.) using the Ellman method (51). The reaction medium contained 100 mM
phosphate buffer, pH 8.0, with 0.33 mM
5,5'-dithiobis-(2-nitrobenzoic) acid, 1 mM acetylthiocholine iodide
and 50 µM Iso-OMPA, butyrylcholinesterase inhibitor. One AChE unit
(U) represents the enzyme that hydrolyzes 1 µmol of substrate per
min at 37˚C.
AChE activity in the fractions collected from the
sucrose gradients was determined using the Ellman method adapted to
a microassay method using a microplate spectrophotometer Epoch
(BioTek; Agilent Technologies, Inc.), for which transparent plastic
plates were used (Nunc), with 96 wells of 400 µl and a flat bottom.
Enzymatic activity was expressed in arbitrary units (AUs) such that
one AU represented an increase in A405 of 0.001 per min and for
each µl of the sample at room temperature. The Bradford method was
used to determine the protein content with bovine serum albumin as
a standard protein (52).
Profile of the molecular forms of the
AChE protein
The molecular forms of AChE were determined by
gradient centrifugation using two sucrose solutions of different
concentrations (5 and 20% w/v) prepared in 10 mM Tris-HCl buffer,
pH 7.0, with 1 M NaCl, 50 mM MgCl2 and Triton X-100
(0.5%, W/V). The proteins used as sedimentation standards were
bovine liver catalase (11.4S) and bovine intestine phosphatase
(6.1S). The gradients were centrifuged at 200,000 x g for 18 h at
4˚C in an SW41Ti tilting rotor (Beckman Coulter, Inc.). Following
centrifugation, 37-40 fractions were collected using the
peristaltic pump Econo Gradient Pump and a fraction collector model
2110 (both from Bio-Rad Laboratories, Inc.). Sedimentation
coefficients were determined according to Martin and Ames's method
(53), comparing the distance
traveled by the AChE protein with that of standard proteins.
AChE protein glycosylation
profile
Lectins from Concanavalia ensiformis (ConA),
Lens culinaris (LCA), Triticum vulgaris (WGA) and
Ricinus communis (RCA) were used to determine possible
differences in the binding of AChE from T lymphocytes and Jurkat
cells to glycans. ConA recognizes mannose residues, LCA recognizes
D-mannose bound to fucosylation centers, WGA recognizes
N-acetyl-glucosamine and RCA recognizes galactose or sialic
acid.
Aliquots of the mixture of
S1+S2 extracts (0.5 ml) of T lymphocytes and
Jurkat cells were incubated with 0.25 ml of Sepharose-4B
(nonspecific binding enzyme control) or ConA, LCA, RCA or WGA
overnight at 4˚C with constant stirring. AChE activity in the
supernatants was assessed via the microassay method to calculate
the percentage of protein interacting with lectin and the
supernatant corresponding to Sepharose-4B, which represented 100%
of the non-retained proteins, was used as a reference.
Statistical analysis
The values of the means and variances for
solubilized enzyme activity, protein content, interaction with
lectins and the proportion of each molecular form were obtained for
normal lymphocytes and leukemic cells for statistical analysis. The
results were compared using the Mann-Whitney U test to establish a
significant difference. Data were analyzed using NCSS 2007
Statistical Software (NCSS, LLC). P<0.05 was considered to
indicate a statistically significant difference.
Results
Collection of T lymphocytes and
analysis of purity using flow cytometry
The average number of leukocytes isolated from 10
blood samples of 20 ml was 7.13x107±0.89. The average
number of T lymphocytes isolated from the total leukocyte
population by magnetic separation was 1.02x107±1.12 and
their viability was 100% (Table I).
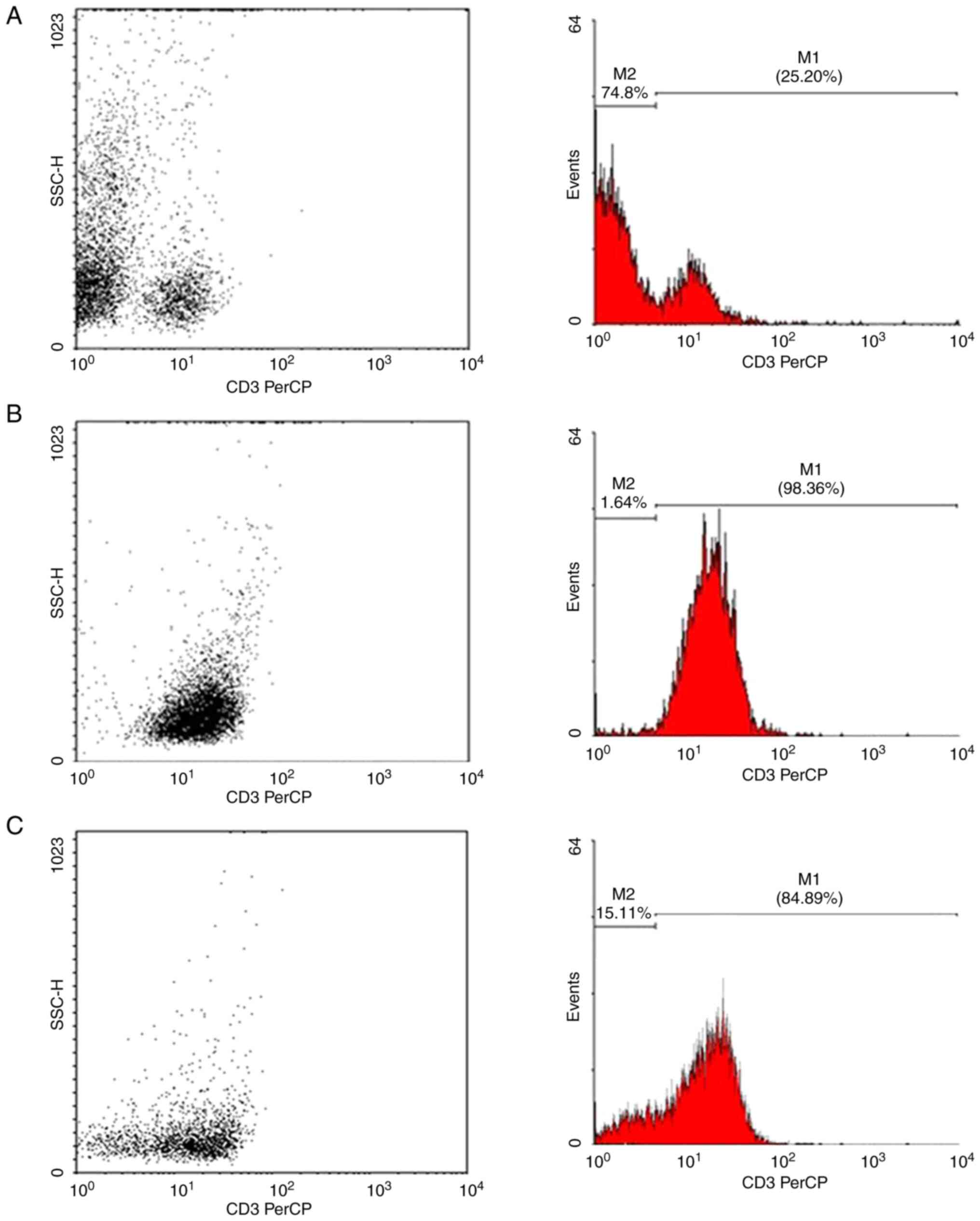
Fig. 1 shows the magnetic
separation of T lymphocytes by flow cytometry. Of the total
leukocytes, ~25% were positive for the CD3 surface marker,
indicating the presence of T lymphocytes. The average percentage of
the 10 samples analyzed using flow cytometry was 98.36%, indicating
that the cell purification process provided high performance.
 | Table IViability and density of total
leukocytes and purified T lymphocytes. |
Table I
Viability and density of total
leukocytes and purified T lymphocytes.
| Source | Cell number/ml
(n=10) | Viability
(n=10) |
|---|
| Leukocytes |
7.13x107±0.89 | 100% |
| T lymphocytes |
1.02x107±1.12 | 100% |
Jurkat cells were also labeled with CD3-PerCP to
determine the percentage of CD3+ cells. The results
revealed that 84.89% of these cells were positive for the CD3
surface marker. Similarly, Fig. 1C
shows an analysis performed using flow cytometry on a sample of
Jurkat cells labeled with CD3-PerCP.
One-step RT-PCR of the ACHE mRNA
Normal human T lymphocytes were used for mRNA
titration with different primers. The results revealed that the
optimal concentration for amplification assays was 1.5 µg of
RNA.
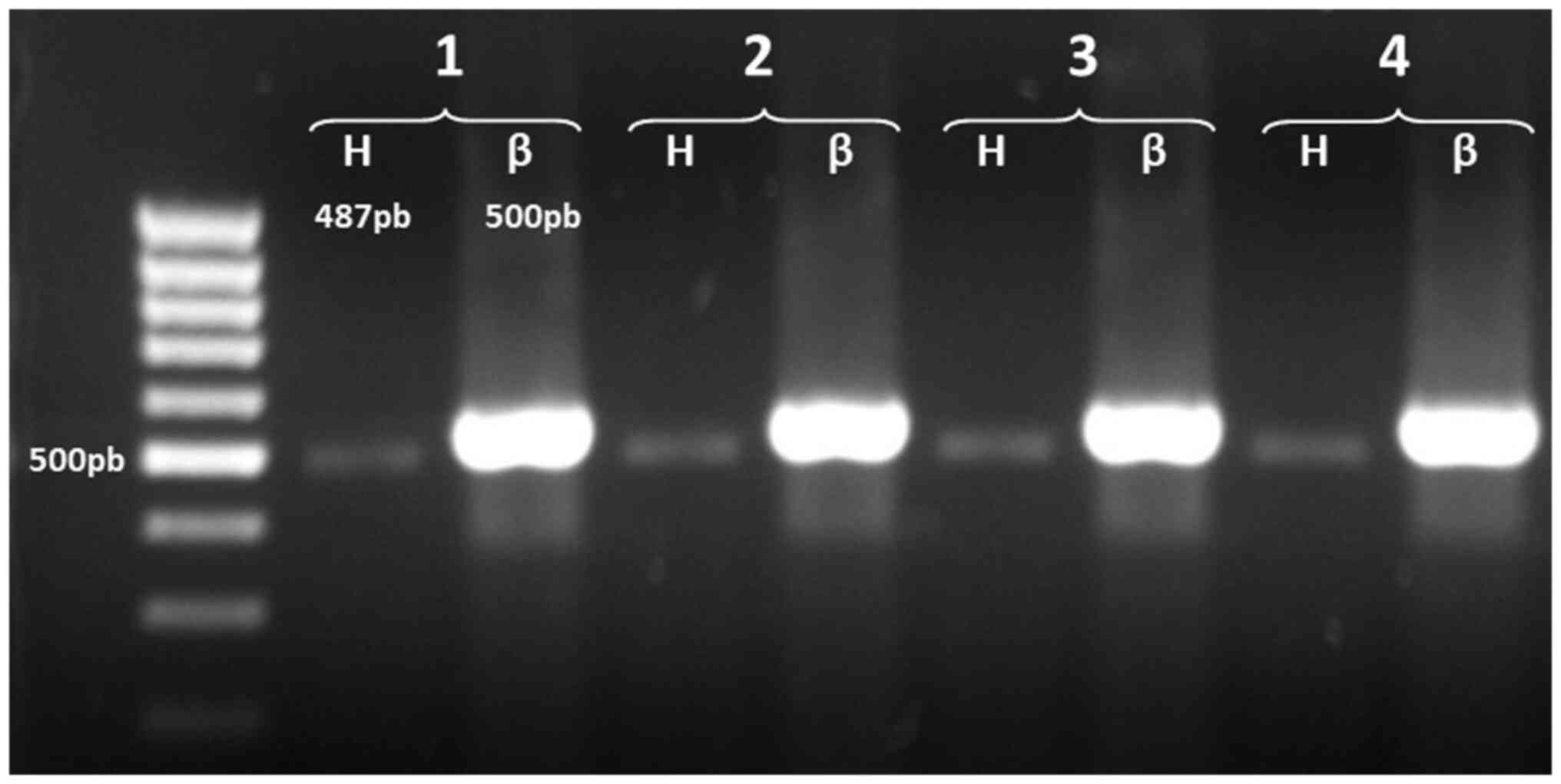
The detection of AChE mRNA variants in normal T
cells revealed only one amplified product, corresponding to the
AChE-H transcript; the products of the amplifications revealed a
band in the gel of ~487 bp (Fig.
2). These results indicated that human T lymphocytes express
only the AChE-H transcript, which encodes membrane-anchored
amphiphilic dimers (17,54).
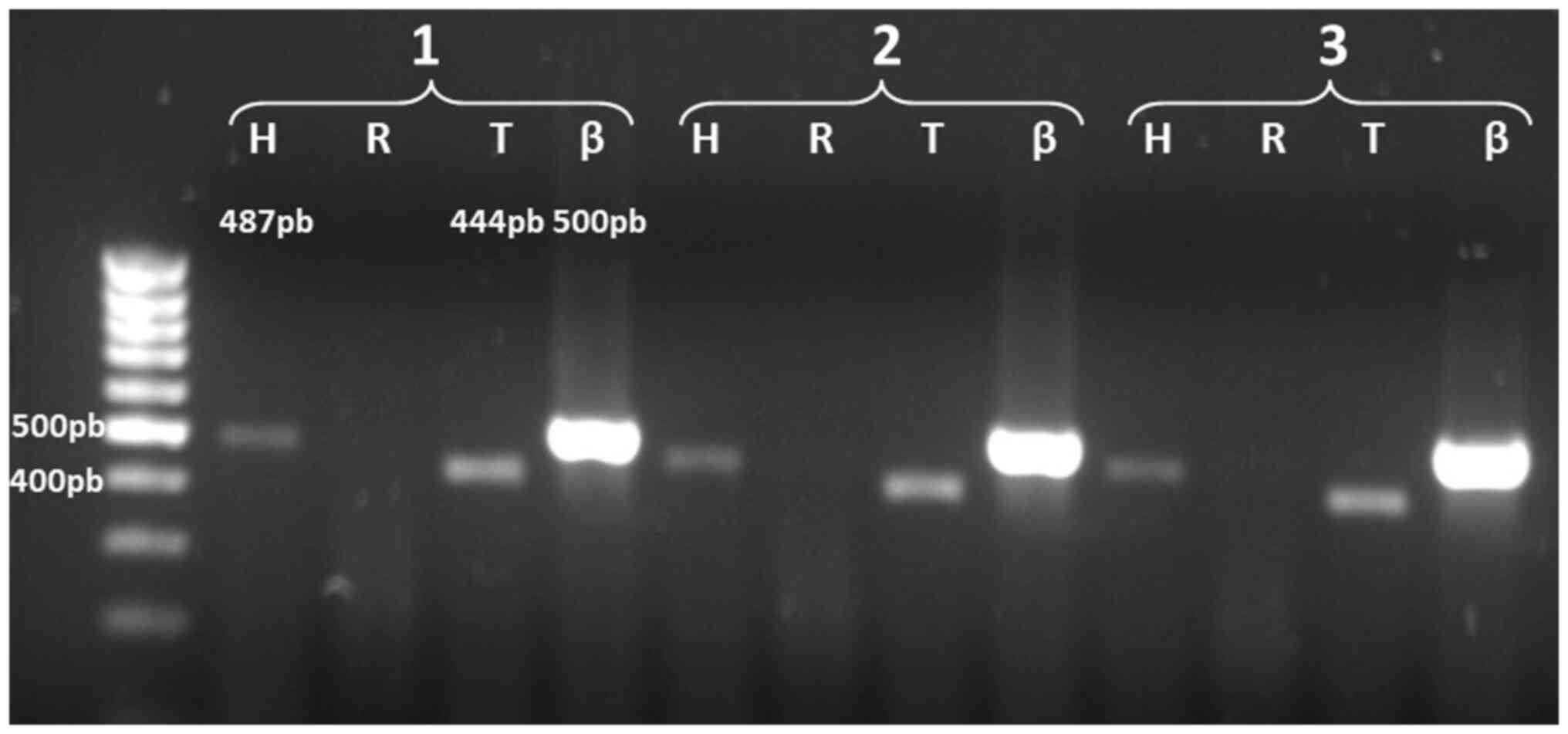
In the analysis of the expression of the ACHE gene
transcripts in Jurkat leukemia cells, the results revealed that the
transcripts detected corresponded to AChE-H (487 bp) and AChE-T
(444 bp); the latter was not detected in normal T lymphocytes
(Fig. 3). With these data, a
comparative analysis of the expression of AChE in T lymphocytes and
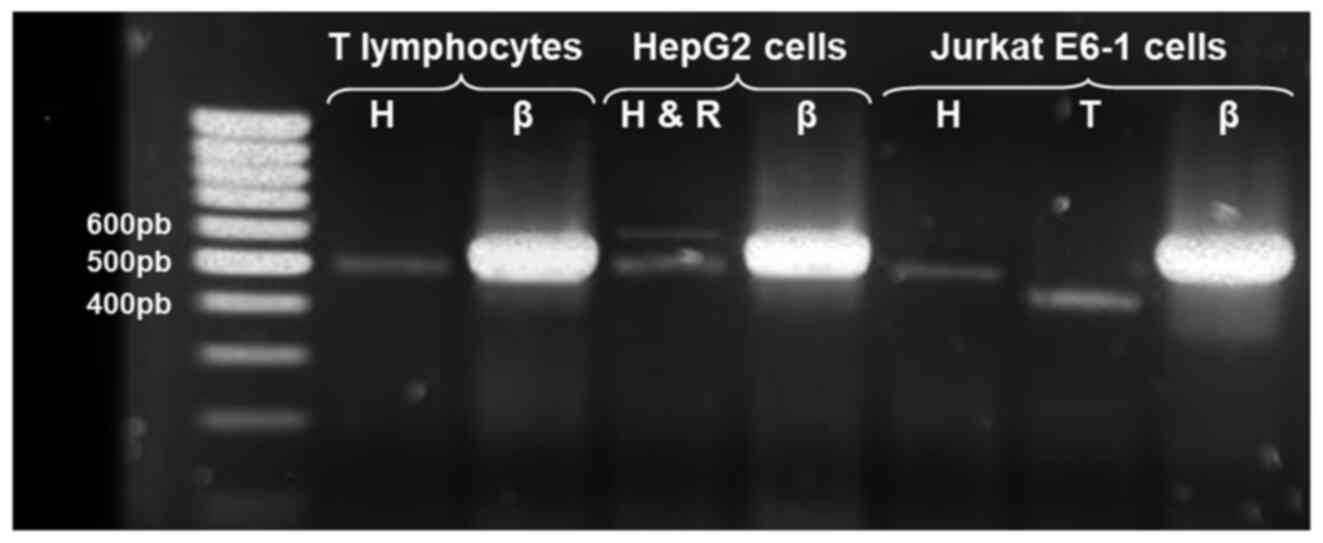
leukemic cells could be later conducted. Fig. 4 shows the amplification products of
the alternative AChE transcripts detected in normal T cells and
Jurkat cells; as a positive control for AChE-R, a 574 bp product
was obtained using the same set of primers for AChE-H and mRNA
extracted from HepG2 cells.
Finally, densitometry analyses were performed to
obtain the intensity of each band, thus obtaining an average of the
intensity of the β-actin control, which facilitated establishing
the relative intensity index of each band of the amplicons
corresponding to AChE in each sample (Table II).
 | Table IIRelative intensity of mRNA splicing
isoforms for the ACHE gene. |
Table II
Relative intensity of mRNA splicing
isoforms for the ACHE gene.
| | Expressed AChE
transcript |
|---|
| Cell type | H | R | T |
|---|
| Normal T
lymphocytes | 0.2326±0.0102 | ND | ND |
| Jurkat E6-1
cells | 0.1909±0.0050 | ND | 0.4638±0.0058 |
| HepG2 cells | 0.2615±0.0308 | 0.0657±0.0128 | - |
Estimation of AChE activity in normal
T lymphocytes and Jurkat cells
The AChE activity in the enzyme extracts of T
lymphocytes solubilized by ionic force (S1) or detergent
(S2) did not change. These results indicated that in
human T lymphocytes, a similar proportion of AChE is weakly bound
and that AChE is strongly bound to the membrane.
On the other hand, the results obtained from the
solubilization of the AChE protein in Jurkat cells revealed higher
AChE activity in the weakly membrane-bound fraction (S1)
than in the strongly membrane-bound fraction (S2).
Table III shows the protein
content, AChE activity and AChE-specific activity obtained from the
S1 and S2 extracts of normal T lymphocytes
and Jurkat cells. Specifically, Jurkat cells had lower activity
than normal T lymphocytes. These data were thought-provoking as,
although the Jurkat cells expressed a transcript, their activity
levels decreased.
 | Table IIISolubilized acetylcholinesterase
activity of normal T lymphocytes and Jurkat cells. |
Table III
Solubilized acetylcholinesterase
activity of normal T lymphocytes and Jurkat cells.
| | Protein
(mg/ml) | AChE (U/ml) | A.E. AChE
(U/mg) |
|---|
| T-lymphocytes | | | |
|
Fraction
S1 | 1.96±0.0035 | 0.216±0.0849 | 0.1101±0.0433 |
|
Fraction
S2 | 2.22±0.0049 | 0.294±0.0255 | 0.1323±0.0115 |
| Jurkat Cells | | | |
|
Fraction
S1 | 4.86±0.0045 | 0.287±0.0312 | 0.0590±0.0064 |
|
Fraction
S2 | 2.50±0.0164 | 0.088±0.0000 | 0.0353±0.0000 |
Glycosylation profile of the AChE
protein in T lymphocytes and Jurkat cells
An analysis of the interaction with lectins revealed
the posttranslational maturation process that glycoproteins
undergo. In the case of T lymphocytes, the AChE protein had a weak
interaction with the ConA lectin (33%), the interaction with LCA
was 66%, the interaction level observed with WGA was 63% and the
interaction with RCA was 75% (Fig.
5). Meanwhile, the glycosylation profile of the AChE protein
differed in Jurkat leukemic cells. The interactions of the AChE
protein with the ConA, LCA, WGA and RCA lectins were 66, 72, 67 and
67%, respectively. The difference in the level of AChE
glycosylation between T lymphocytes and leukemic T cells indicated
that the posttranslational maturation process of AChE is modified
in the neoplastic state.
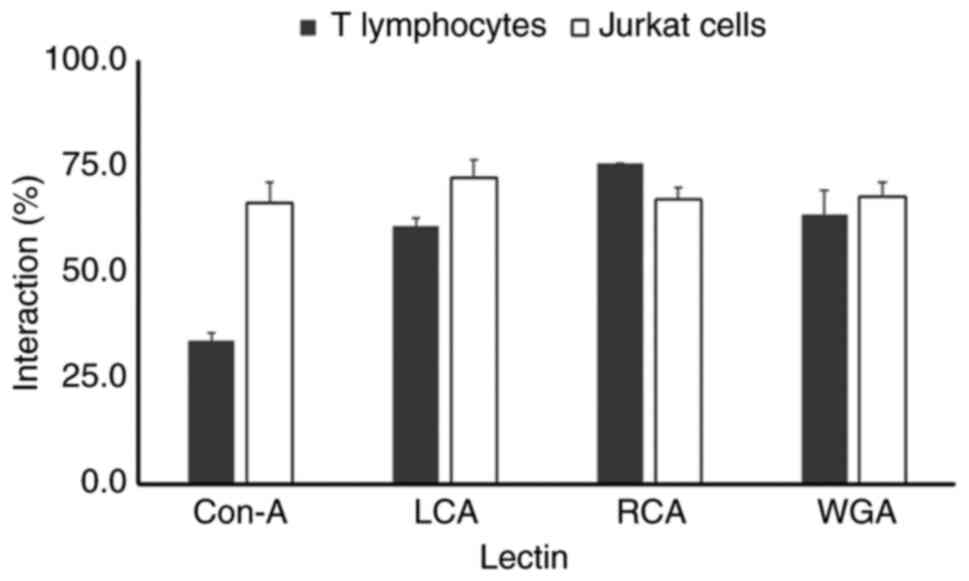 | Figure 5Lectin affinity chromatography
analysis of AChE glycosylation in human T lymphocytes and Jurkat
cells. Solubilized AChE protein extracts
(S1+S2 fractions) from normal human T
lymphocytes and Jurkat E6-1 leukemia cells were incubated overnight
at 4˚C with Sepharose-4B beads (control for nonspecific binding) or
lectin-conjugated Sepharose-4B beads specific for ConA, recognizing
mannose residues, LCA recognizing D-mannose bound to fucose
residues, RCA, recognizing galactose or terminal sialic acid
residues and WGA recognizing N-acetyl-glucosamine residues. Unbound
AChE activity in the supernatants was measured via a microplate
assay and expressed as a percentage of the total AChE activity in
the Sepharose-4B control (100%). T lymphocytes: AChE displayed
decreased binding to ConA (33%), moderate binding to LCA (66%) and
WGA (63%) and increased binding to RCA (75%). Jurkat cells: AChE
exhibited increased binding to ConA (66%) and LCA (72%) compared
with T-lymphocytes, suggesting increased levels of mannose and
fucose-associated mannose residues. Conversely, Jurkat cells showed
decreased binding to RCA (67%) compared with T lymphocytes,
indicating potentially reduced levels of galactose or sialic acid
residues. Binding to WGA (67%) remained statistically similar
(P<0.05) between both cell lines. AChE, acetylcholinesterase;
ConA, Concanavalin A; LCA, Lens culinaris agglutinin; RCA,
Ricinus communis agglutinin; WGA, wheat germ agglutinin. |
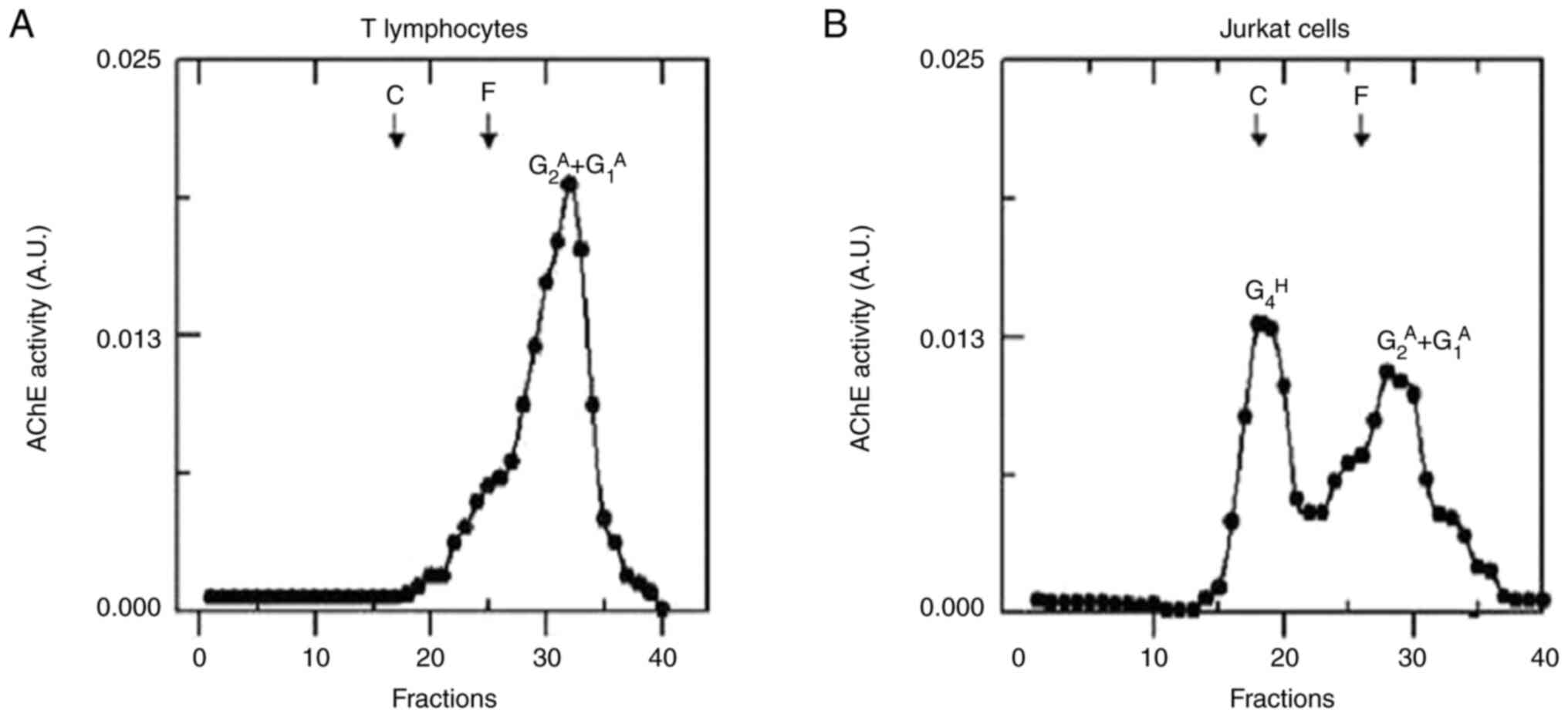
Profile of the molecular forms of AChE
in T lymphocytes and Jurkat cells
The results for the molecular forms of AChE with
sedimentation coefficients of 5.2S and 3.5S were consistent with
previously reported values for amphiphilic dimers
(G2A, 5.2S) and amphiphilic monomers
(G1A, 3.5S) found in T-cell extracts
(55,56). In the sedimentation analysis of the
Jurkat cell extracts, amphiphilic dimers
(G2A, 5.2S), amphiphilic monomers
(G1A, 3.5S) and hydrophilic tetramers
(G4H, 10.6) were the AChE molecular forms
identified. The latter is encoded by the AChE-T transcript, thus
confirming the presence of this alternative splicing variant in the
studies conducted via RT-PCR (57).
Fig. 6 shows the density gradient
and the molecular forms of AChE present in normal T lymphocytes and
Jurkat cells.
Discussion
The expression of the ACHE gene in normal T
lymphocytes and Jurkat E6-1 leukemic cells was analyzed using
RT-PCR. AChE-H transcripts were detected in the first cell type,
but AChE-T or AChE-R transcripts were not. These results agreed
with a previous study of peripheral blood cells, including
lymphocytes and erythrocytes, where the exclusive presence of the
AChE-H transcript has been reported. This transcript encodes the
AChE molecular forms of amphiphilic dimers and monomers
(G2A and G1A) anchored
to the plasma membrane by a glycosylphosphatidylinositol bond
(17).
By contrast, the analysis of AChE transcripts in
Jurkat leukemic cells using RT-PCR allowed the detection of AChE-H
transcripts and the AChE-T variant, which demonstrated an
alteration in the expression of the AChE gene in this cell line.
This finding coincided with the results observed by Liao et
al (58) regarding the
expression of these transcripts in the Jurkat line.
In a previous study of P19 semidifferentiated
neuronal cells via microarrays, overexpression of AChE-T
transcripts was observed, which is associated with high expression
levels of antiapoptotic genes and genes related to the splicing
process (59). In Jurkat cells, the
AChE-T transcript is probably involved in cell proliferation by
regulating the expression of antiapoptotic agents and allowing the
cell to survive.
The data obtained in the present study revealed that
Jurkat cells presented a notable decrease in AChE activity compared
with that of normal T lymphocytes. Other studies have reported
lower AChE activity in cancer cells compared with normal cells
(5,26,29).
In the case of lung tumors, AChE activity decreases with increasing
malignancy, which is related to an increase in ACh levels in these
cells (25). The increase in AChE
activity may be related to the development of hematological
diseases and the regulation of immune function (35).
The function of lymphocytes is regulated not only by
the cytokine system but also by the lymphoid cholinergic system
independent of cholinergic neurons (3). A number of the components found in the
nervous system, including ACh, choline acetyltransferase (ChAT),
high-affinity choline transporter, muscarinic and nicotinic ACh
receptors (mAChR and nAChR, respectively) and AChE, which together
are known as the nonneuronal cholinergic system (NNCS), are
expressed in lymphocytes (49,50,60).
On the other hand, ACh and agonists of mAChRs and nAChRs have been
reported to induce a variety of effects on lymphocytes, such as
increases in the fluidity of the membrane, the synthesis of
inositol triphosphate, the expression of the c-fos gene, the
activation of DNA and RNA synthesis and cell proliferation. These
findings support the hypothesis that ACh may regulate the functions
of T lymphocytes (49,50,60).
A decrease in AChE activity can lead to an increase
in the content of ACh present in the cell when it is not
hydrolyzed. In the case of leukemic cells, the present study found
that the specific activity of AChE was much lower than that of
normal T lymphocytes. Similar results have been reported by Fujii
et al (60), who documented
that the levels of ACh in T lymphocytes are much lower than those
in Jurkat cells. Thus, an increase in ACh levels in leukemic cells,
produced by a lack of AChE activity, leads to an overregulation of
the functions controlled by the NNCS, including the activation of
cell proliferation (3). Fig. 7 shows a model that attempts to
explain the induction of cell proliferation by ACh levels.
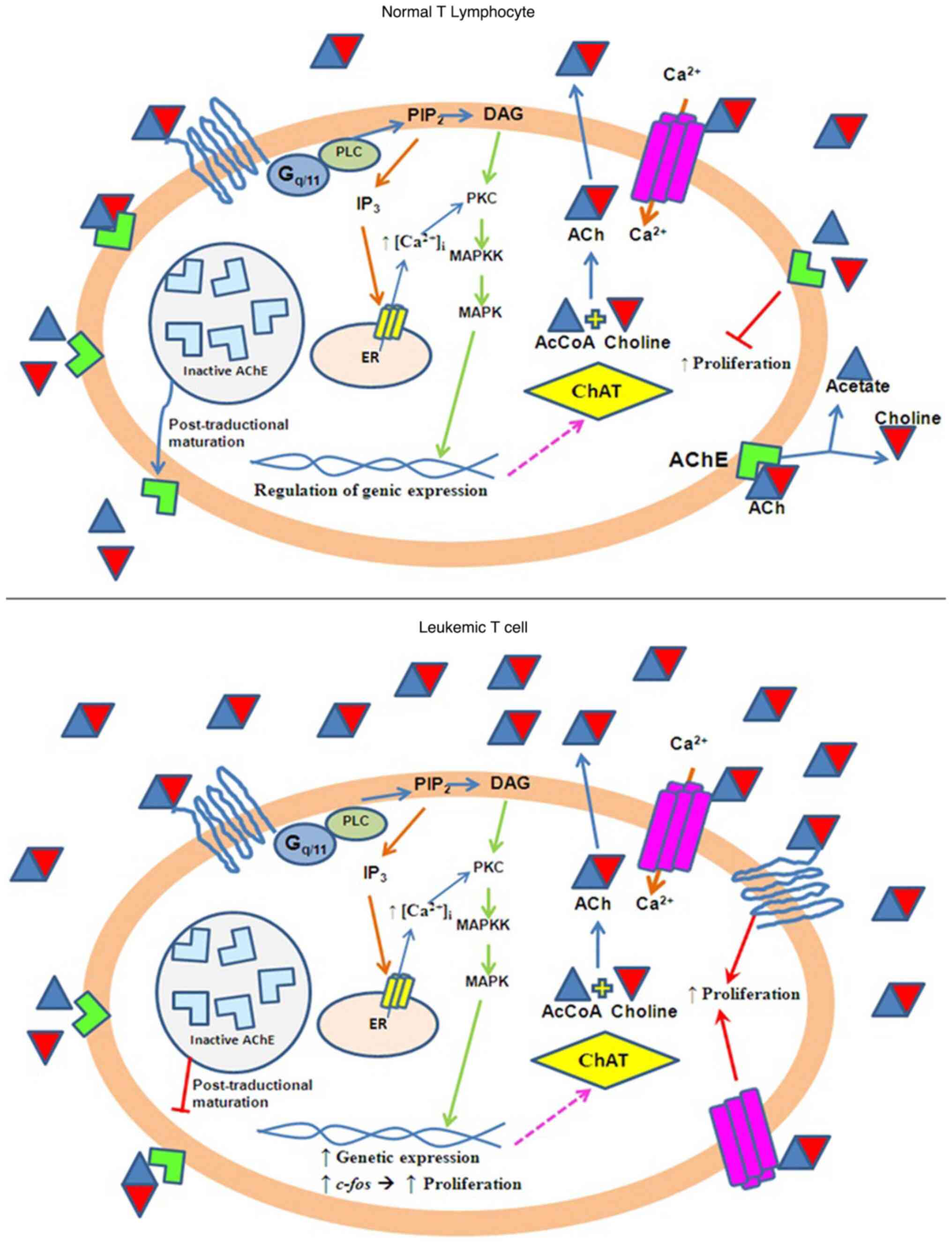 | Figure 7Schematic model of ACh-mediated
induction of cell proliferation in Jurkat leukemia cells compared
with normal cells. Normal cells: Functional AChE maintains
steady-state levels of ACh via hydrolysis. ACh interacts nAChRs and
mAChRs receptors at physiological levels, leading to balanced
regulation of intracellular signaling pathways. Jurkat cells:
Deficient AChE activity leads to increased ACh accumulation.
Elevated ACh levels overstimulate nAChRs and mAChRs, resulting in
the excessive activation of ChAT and PLC. PLC activation increases
the Ca²+ release from the endoplasmic reticulum.
Increased Ca²+ signaling promotes the expression of the
proto-oncogene c-fos, which upregulates genes involved in cell
proliferation. ACh, acetylcholine; AChE, acetylcholinesterase;
nAChRs, nicotinic ACh receptor; mAChRs, muscarinic ACh receptor;
ChAT, choline acetyltransferase; PLC, phospholipase C;
PIP2, phosphatidylinositol bisphosphate; DAG,
diacylglycerol; PKC, protein kinase C; IP3, inositol
triphosphate. |
The sedimentation analysis of the molecular forms of
AChE in T lymphocytes revealed the presence of a molecular
structure corresponding to amphiphilic dimers and monomers
(G2A and G1A), which
are anchored to the membrane by GFI and originate through the
expression of AChE-H transcripts (17,57,60).
This finding is evidence of the expression of the AChE-H transcript
detected in this type of cell. On the other hand, the sedimentation
analysis of the AChE molecular forms in Jurkat cells revealed the
presence of amphiphilic monomers and dimers
(G2A and G1A) and
hydrophilic tetramers (G4H). The molecular
forms found in Jurkat cells were confirmed by the presence of the
AChE-H transcripts responsible for the synthesis of the AChE
G2A and G1A forms, as
well as the expression of AChE-T transcripts from which the
molecular form of AChE G4H is derived
(57).
In this context, AChE activity has been proposed as
a generic marker for small extracellular vesicles or exosomes. In
Jurkat cells incubated with an extra depleted medium that can
release AChE protein, a hydrophilic variant is released into the
extracellular medium (58). AChE is
an export protein whose synthesis is restarted in the rough
endoplasmic reticulum, after which it is transported to the Golgi
apparatus for modification and then secreted or deposited on the
cell membrane (61).
Glycosylation variations were detected between both
cell types, which suggested an alteration in the expression of AChE
in a transformed cell compared with a normal cell. The primary
sequence of the AChE protein contains three potential glycosylation
sites linked to asparagine (Asn) residues 265, 350 and 464 through
the glycosylation signal Asn-X-Ser (62). In mammals, these sites are conserved
in all sequenced cholinesterases (63). When all three N-glycosylation
signals of human AChE were suppressed by site-directed mutagenesis,
all the sites were shown to be essential for effective biosynthesis
and secretion. The extracellular AChE levels of mutants defective
at one, two, or all three sites were 20-30%, 2-4% and ~0.5% of the
wild-type level, respectively (62).
In a number of pathological states, the degree of
AChE alteration includes posttranslational maturation, as evidenced
by the modification of the glycosylation pattern of the AChE
protein (25,29,64,65). A
different glycosylation pattern of the AChE proteins contained in
the S1+S2 extracts of normal T lymphocytes
and Jurkat cells was observed when they interacted with lectins of
different specificities. This finding indicated that the
incorporation and/or removal of sugar residues during AChE
maturation in the rough endoplasmic reticulum and the different
lamellae of the Golgi apparatus were altered in the leukemic cell
line, where a decrease in the incorporation of galactose and sialic
acid into the AChE structure was mainly observed. AChE
glycosylation is essential from the point of view of its activity
and circulatory half-life (66).
N-glycosylation may play a specific role in the unresolved
noncholinergic roles of cholinesterases in cellular differentiation
and development (67).
Sialic acid incorporation and its role in the
pharmacokinetic properties of proteins was first reported by Morell
et al (68), who discovered
that sialylation alters the circulatory half-life of proteins by
protecting the penultimate galactose residues, which are recognized
by the asialoglycoprotein receptor (ASGPR). The binding of
glycoproteins to ASGPR initiates protein degradation in the liver
(69-71).
Several studies have been built on the discovery by Morell et
al (68) by increasing the
level of sialic acid in various proteins, including
cholinesterases, to prolong their circulatory retention time
(62,71,72).
The present study was limited, using only one
leukemia cell line (Jurkat). However, it was considered that the
results obtained are a valuable starting point for future studies.
The authors continue to investigate the possible participation of
AChE and other components of the nonneuronal cholinergic system in
cell growth and proliferation, allowing the development of new
proposals for therapeutic targets against acute lymphoblastic
leukemia.
Future research will involve a wider range of
leukemia cell lines and clinical samples. Additionally, it will
include data on activated normal T lymphocytes to gain a
comprehensive understanding of the role of AChE in different
functional states of lymphocytes. Notably, future studies of AChE
expression in samples from patients with leukemia may allow the use
of AChE as a tool for the early detection of leukemia, with the aim
on increasing the effectiveness of current treatments.
The results of the present study showed that the
content and composition of AChE are altered in Jurkat leukemic
cells compared with normal T lymphocytes, from the change in the
expression pattern of the transcripts to the decrease in the
specific activity of AChE, the differences in glycan processing and
the alteration in the assembly of molecular shapes. This opens the
way to explore the possible participation of AChE in the
proliferation and differentiation of lymphoid cells, as well as in
the development of new therapeutic targets against acute
lymphoblastic leukemia and the creation of timely detection
methods. Further studies should be conducted in other leukemia cell
types to elucidate the underlying mechanisms involved in the
generation of the proposed molecular targets.
Acknowledgements
The authors thank Dr María Concepción Gutiérrez
Ruiz, Department of Health Sciences, Metropolitan Autonomous
University-Iztapalapa Campus for providing the RNA samples from the
HepG2 cell line; Dr Rocio Salceda Sacanelles, National Autonomous
University of Mexico and Dr Hector Serrano, Metropolitan Autonomous
University-Iztapalapa Campus for their technical assistance.
Funding
Funding: The present study was funded by the E022 Program:
Recursos Fiscales para Investigación, Instituto Nacional de
Pediatría (grant no. 2022/067 LF-L and 2019/072 SE-F). It was also
supported by Consejo Nacional de Humanidades, Ciencias y
Tecnologías, México (UAM-I: grant nos. 309-0; C/PFPN-2002-35-32)
and Ciencia de Frontera 2023 (grant no. CONAHCYT CF-2023-I-2755 and
CF-2023-I-811).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
Data acquisition, processing and validation was
performed by LF-L, JG-O, SE-F, GL-V, ID-D, RV-R and IG-T; the study
investigation was carried out by LF-L, JG-O and SE-F carried out
project administration; LF-L and JG-O wrote the original draft of
the manuscript; LF-L, JG-O, SE-F, GL-V, ID-D and IG-T reviewed and
edited the manuscript. SE-F, GL-V, ID-D, IG-T, RL-D, RV-R, RV-Q and
MM confirm the authenticity of all the raw data. All authors read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
The research ethics committee of the National
Institute of Pediatrics approved the use of blood samples donated
by healthy adults with a registration in the National Bioethics
Commission with the number CONBIOETICA-09-CEI-025-20161215 of the
National Institute of Pediatrics, Mexico.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Soreq H and Seidman S:
Acetylcholinesterase-new roles for an old actor. Nat Rev Neurosci.
2:294–302. 2001.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Grisaru D, Pick M, Perry C, Sklan EH,
Almog R, Goldberg I, Naparstek E, Lessing JB, Soreq H and Deutsch
V: Hydrolytic and nonenzymatic functions of acetylcholinesterase
comodulate hemopoietic stress responses. J Immunol. 176:27–35.
2006.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Fujii T, Mashimo M, Moriwaki Y, Misawa H,
Ono S, Horiguchi K and Kawashima K: Expression and function of the
cholinergic system in immune cells. Front Immunol.
8(1085)2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Onganer PU, Djamgoz MBA, Whyte K and
Greenfield SA: An acetylcholinesterase-derived peptide inhibits
endocytic membrane activity in a human metastatic breast cancer
cell line. Biochim Biophys Acta. 1760:415–420. 2006.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Pérez-Aguilar B, Vidal CJ, Palomec G,
García-Dolores F, Gutiérrez-Ruiz MC, Bucio L, Gómez-Olivares JL and
Gómez-Quiroz LE: Acetylcholinesterase is associated with a decrease
in cell proliferation of hepatocellular carcinoma cells. Biochim
Biophys Acta. 1852:1380–1387. 2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Villeda-González JD, Gómez-Olivares JL,
Baiza-Gutman LA, Manuel-Apolinar L, Damasio-Santana L,
Millán-Pacheco C, Ángeles-Mejía S, Cortés-Ginez MC, Cruz-López M,
Vidal-Moreno CJ and Díaz-Flores M: Nicotinamide reduces
inflammation and oxidative stress via the cholinergic system in
fructose-induced metabolic syndrome in rats. Life Sci.
250(117585)2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zhang JY, Jiang H, Gao W, Wu J, Peng K,
Shi YF and Zhang XJ: The JNK/AP1/ATF2 pathway is involved in
H2O2-induced acetylcholinesterase expression during apoptosis. Cell
Mol Life Sci. 65:1435–1445. 2008.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhang XJ, Yang L, Zhao Q, Caen JP, He HY,
Jin QH, Guo LH, Alemany M, Zhang LY and Shi YF: Induction of
acetylcholinesterase expression during apoptosis in various cell
types. Cell Death Differ. 9:790–800. 2002.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Getman DK, Eubanks JH, Camp S, Evans GA
and Taylor P: The human gene encoding acetylcholinesterase is
located on the long arm of chromosome 7. Am J Hum Genet.
51:170–177. 1992.PubMed/NCBI
|
|
10
|
Taylor P and Radić Z: The cholinesterases:
From genes to proteins. Annu Rev Pharmacol Toxicol. 34:281–320.
1994.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Soreq H, Ben-Aziz R, Prody CA, Seidman S,
Gnatt A, Neville L, Lieman-Hurwitz J, Lev-Lehman E, Ginzberg D,
Lipidot-Lifson Y, et al: Molecular cloning and construction of the
coding region for human acetylcholinesterase reveals a G + C-rich
attenuating structure. Proc Natl Acad Sci USA. 87:9688–9692.
1990.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Massoulié J, Pezzementi L, Bon S, Krejci E
and Vallette FM: Molecular and cellular biology of cholinesterases.
Prog Neurobiol. 41:31–91. 1993.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Massoulié J, Anselmet A, Bon S, Krejci E,
Legay C, Morel N and Simon S: Acetylcholinesterase: C-terminal
domains, molecular forms and functional localization. J Physiol
Paris. 92:183–190. 1998.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Dori A and Soreq H: ARP, the cleavable
C-terminal peptide of ‘readthrough’ acetylcholinesterase, promotes
neuronal development and plasticity. J Mol Neurosci. 28:247–255.
2006.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Perrier NA, Salani M, Falasca C, Bon S,
Augusti-Tocco G and Massoulié J: The readthrough variant of
acetylcholinesterase remains very minor after heat shock,
organophosphate inhibition and stress, in cell culture and in vivo.
J Neurochem. 94:629–638. 2005.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Deutsch VR, Pick M, Perry C, Grisaru D,
Hemo Y, Golan-Hadari D, Grant A, Eldor A and Soreq H: The
stress-associated acetylcholinesterase variant AChE-R is expressed
in human CD34(+) hematopoietic progenitors and its C-terminal
peptide ARP promotes their proliferation. Exp Hematol.
30:1153–1161. 2002.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Pick M, Flores-Flores C, Grisaru D,
Shochat S, Deutsch V and Soreq H: Blood-cell-specific
acetylcholinesterase splice variations under changing stimuli. Int
J Dev Neurosci. 22:523–531. 2004.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Grisaru D, Sternfeld M, Eldor A, Glick D
and Soreq H: Structural roles of acetylcholinesterase variants in
biology and pathology. Eur J Biochem. 264:672–686. 1999.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Sternfeld M, Shoham S, Klein O,
Flores-Flores C, Evron T, Idelson GH, Kitsberg D, Patrick JW and
Soreq H: Excess ‘read-through’ acetylcholinesterase attenuates but
the ‘synaptic’ variant intensifies neurodeterioration correlates.
Proc Natl Acad Sci USA. 97:8647–8652. 2000.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Fischer K, Brown J, Scherer SW, Schramm P,
Stewart J, Fugazza G, Pascheberg U, Peter W, Tsui LC, Lichter P and
Döhner H: Delineation of genomic regions in chromosome band 7q22
commonly deleted in myeloid leukemias. Recent Results Cancer Res.
144:46–52. 1998.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zeng WR, Watson P, Lin J, Jothy S,
Lidereau R, Park M and Nepveu A: Refined mapping of the region of
loss of heterozygosity on the long arm of chromosome 7 in human
breast cancer defines the location of a second tumor suppressor
gene at 7q22 in the region of the CUTL1 gene. Oncogene.
18:2015–2021. 1999.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Neville PJ, Thomas N and Campbell IG: Loss
of heterozygosity at 7q22 and mutation analysis of the CDP gene in
human epithelial ovarian tumors. Int J Cancer. 91:345–349.
2001.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Sáez-Valero J and Vidal CJ: Biochemical
properties of acetyl- and butyrylcholinesterase in human
meningioma. Biochim Biophys Acta. 1317:210–218. 1996.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Vidal CJ: Expression of cholinesterases in
brain and non-brain tumours. Chem Biol Interact. 157-158:227–232.
2005.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Martínez-Moreno P, Nieto-Cerón S,
Torres-Lanzas J, Ruiz-Espejo F, Tovar-Zapata I, Martínez-Hernández
P, Rodríguez-López JN, Vidal CJ and Cabezas-Herrera J:
Cholinesterase activity of human lung tumours varies according to
their histological classification. Carcinogenesis. 27:429–436.
2006.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Montenegro MF, Nieto-Cerón S, Ruiz-Espejo
F, Páez de la Cadena M, Rodríguez-Berrocal FJ and Vidal CJ:
Cholinesterase activity and enzyme components in healthy and
cancerous human colorectal sections. Chem Biol Interact.
157-158:429–430. 2005.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Montenegro MF, Ruiz-Espejo F, Campoy FJ,
Muñoz-Delgado E, de la Cadena MP, Rodríguez-Berrocal FJ and Vidal
CJ: Cholinesterases are down-expressed in human colorectal
carcinoma. Cell Mol Life Sci. 63:2175–2182. 2006.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Martínez-López de Castro A, Nieto-Cerón S,
Aurelio PC, Galbis-Martínez L, Latour-Pérez J, Torres-Lanzas J,
Tovar-Zapata I, Martínez-Hernández P, RodríRodríguez-López JN and
Cabezas-Herrera J: Cancer-associated differences in
acetylcholinesterase activity in bronchial aspirates from patients
with lung cancer. Clin Sci (Lond). 115:245–253. 2008.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ruiz-Espejo F, Cabezas-Herrera J, Illana
J, Campoy FJ, Muñoz-Delgado E and Vidal CJ: Breast cancer
metastasis alters acetylcholinesterase activity and the composition
of enzyme forms in axillary lymph nodes. Breast Cancer Res Treat.
80:105–114. 2003.PubMed/NCBI View Article : Google Scholar
|
|
30
|
García-Ayllón MS, Sáez-Valero J,
Muñoz-Delgado E and Vidal CJ: Identification of hybrid
cholinesterase forms consisting of acetyl- and
butyrylcholinesterase subunits in human glioma. Neuroscience.
107:199–208. 2001.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhao Y, Wang X, Wang T, Hu X, Hui X, Yan
M, Gao Q, Chen T, Li J, Yao M, et al: Acetylcholinesterase, a key
prognostic predictor for hepatocellular carcinoma, suppresses cell
growth and induces chemosensitization. Hepatology. 53:493–503.
2011.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Xu H, Shen Z, Xiao J, Yang Y, Huang W,
Zhou Z, Shen J, Zhu Y, Liu XY and Chu L: Acetylcholinesterase
overexpression mediated by oncolytic adenovirus exhibited potent
anti-tumor effect. BMC Cancer. 14(668)2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Castillo-González AC, Nieto-Cerón S,
Pelegrín-Hernández JP, Montenegro MF, Noguera JA, López-Moreno MF,
Rodríguez-López JN, Vidal CJ, Hellín-Meseguer D and Cabezas-Herrera
J: Dysregulated cholinergic network as a novel biomarker of poor
prognostic in patients with head and neck squamous cell carcinoma.
BMC Cancer. 15(385)2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Castillo-González AC, Pelegrín-Hernández
JP, Nieto-Cerón S, Madrona AP, Noguera JA, López-Moreno MF,
Rodríguez-López JN, Vidal CJ, Hellín-Meseguer D and Cabezas-Herrera
J: Unbalanced acetylcholinesterase activity in larynx squamous cell
carcinoma. Int Immunopharmacol. 29:81–86. 2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Battisti V, Bagatini MD, Maders LDK,
Chiesa J, Santos KF, Gonçalves JF, Abdalla FH, Battisti IE,
Schetinger MR and Morsch VM: Cholinesterase activities and
biochemical determinations in patients with prostate cancer:
Influence of Gleason score, treatment and bone metastasis. Biomed
Pharmacother. 66:249–255. 2012.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yu H, Xia H, Tang Q, Xu H, Wei G, Chen Y,
Dai X, Gong Q and Bi F: Acetylcholine acts through M3 muscarinic
receptor to activate the EGFR signaling and promotes gastric cancer
cell proliferation. Sci Rep. 7(40802)2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Resende RR, Alves AS, Britto LRG and
Ulrich H: Role of acetylcholine receptors in proliferation and
differentiation of P19 embryonal carcinoma cells. Exp Cell Res.
314:1429–1443. 2008.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Cheng K, Samimi R, Xie G, Shant J,
Drachenberg C, Wade M, Davis RJ, Nomikos G and Raufman JP:
Acetylcholine release by human colon cancer cells mediates
autocrine stimulation of cell proliferation. Am J Physiol
Gastrointest Liver Physiol. 295:G591–G597. 2008.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Pettersson A, Nilsson L, Nylund G,
Khorram-Manesh A, Nordgren S and Delbro DS: Is acetylcholine an
autocrine/paracrine growth factor via the nicotinic alpha7-receptor
subtype in the human colon cancer cell line HT-29? Eur J Pharmacol.
609:27–33. 2009.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Song P, Sekhon HS, Jia Y, Keller JA,
Blusztajn JK, Mark GP and Spindel ER: Acetylcholine is synthesized
by and acts as an autocrine growth factor for small cell lung
carcinoma. Cancer Res. 63:214–221. 2003.PubMed/NCBI
|
|
41
|
Lev-Lehman E, Ginzberg D, Hornreich G,
Ehrlich G, Meshorer A, Eckstein F, Soreq H and Zakut H: Antisense
inhibition of acetylcholinesterase gene expression causes transient
hematopoietic alterations in vivo. Gene Ther. 1:127–135.
1994.PubMed/NCBI
|
|
42
|
Soreq H, Patinkin D, Lev-Lehman E, Grifman
M, Ginzberg D, Eckstein F and Zakut H: Antisense oligonucleotide
inhibition of acetylcholinesterase gene expression induces
progenitor cell expansion and suppresses hematopoietic apoptosis ex
vivo. Proc Natl Acad Sci USA. 91:7907–7911. 1994.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Brown LM, Blair A, Gibson R, Everett GD,
Cantor KP, Schuman LM, Burmeister LF, Van Lier SF and Dick F:
Pesticide exposures and other agricultural risk factors for
leukemia among men in Iowa and Minnesota. Cancer Res. 50:6585–6591.
1990.PubMed/NCBI
|
|
44
|
Cantor KP, Blair A, Everett G, Gibson R,
Burmeister LF, Brown LM, Schuman L and Dick FR: Pesticides and
other agricultural risk factors for non-Hodgkin's lymphoma among
men in Iowa and Minnesota. Cancer Res. 52:2447–2455.
1992.PubMed/NCBI
|
|
45
|
Cabello G, Valenzuela M, Vilaxa A, Durán
V, Rudolph I, Hrepic N and Calaf G: A rat mammary tumor model
induced by the organophosphorous pesticides parathion and
malathion, possibly through acetylcholinesterase inhibition.
Environ Health Perspect. 109:471–479. 2001.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Nasterlack M: Pesticides and childhood
cancer: An update. Int J Hyg Environ Health. 210:645–657.
2007.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Calaf GM, Parra E and Garrido F: Cell
proliferation and tumor formation induced by eserine, an
acetylcholinesterase inhibitor, in rat mammary gland. Oncol Rep.
17:25–33. 2007.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Kawashima K and Fujii T: Extraneuronal
cholinergic system in lymphocytes. Pharmacol Ther. 86:29–48.
2000.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Kawashima K and Fujii T: The lymphocytic
cholinergic system and its biological function. Life Sci.
72:2101–2109. 2003.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Kawashima K and Fujii T: Expression of
non-neuronal acetylcholine in lymphocytes and its contribution to
the regulation of immune function. Front Biosci. 9:2063–2085.
2004.PubMed/NCBI View
Article : Google Scholar
|
|
51
|
Ellman GL, Courtney KD, Andres V Jr and
Feather-Stone RM: A new and rapid colorimetric determination of
acetylcholinesterase activity. Biochem Pharmacol. 7:88–95.
1961.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Bradford MM: A rapid and sensitive method
for the quantitation of microgram quantities of protein utilizing
the principle of protein-dye binding. Anal Biochem. 72:248–254.
1976.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Martin RG and Ames BN: A method for
determining the sedimentation behavior of enzymes: Application to
protein mixtures. J Biol Chem. 236:1372–1379. 1961.PubMed/NCBI
|
|
54
|
Bartha E, Rakonczay Z, Kása P, Hollán S
and Gyévai A: Molecular form of human lymphocyte membrane-bound
acetylcholinesterase. Life Sci. 41:1853–1860. 1987.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Sine JP and Caye-Vaugien C: Properties and
characterization of soluble forms of lymphocyte
acetylcholinesterase from an ox. Biochimie. 66:203–214.
1984.PubMed/NCBI View Article : Google Scholar : (In French).
|
|
56
|
Toutant JP, Richards MK, Krall JA and
Rosenberry TL: Molecular forms of acetylcholinesterase in two
sublines of human erythroleukemia K562 cells. Sensitivity or
resistance to phosphatidylinositol-specific phospholipase C and
biosynthesis. Eur J Biochem. 187:31–38. 1990.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Massoulié J, Bon S, Perrier N and Falasca
C: The C-terminal peptides of acetylcholinesterase: Cellular
trafficking, oligomerization and functional anchoring. Chem Biol
Interact. 157-158:3–14. 2005.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Liao Z, Jaular LM, Soueidi E, Jouve M,
Muth DC, Schøyen TH, Seale T, Haughey NJ, Ostrowski M, Théry C and
Witwer KW: Acetylcholinesterase is not a generic marker of
extracellular vesicles. J Extracell Vesicles.
8(1628592)2019.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Ben-Ari S, Toiber D, Sas AS, Soreq H and
Ben-Shaul Y: Modulated splicing-associated gene expression in P19
cells expressing distinct acetylcholinesterase splice variants. J
Neurochem. 97 (Suppl 1):S24–S34. 2006.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Fujii T, Takada-Takatori Y and Kawashima
K: Basic and clinical aspects of non-neuronal acetylcholine:
Expression of an independent, non-neuronal cholinergic system in
lymphocytes and its clinical significance in immunotherapy. J
Pharmacol Sci. 106:186–192. 2008.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Jin QH, He HY, Shi YF, Lu H and Zhang XJ:
Overexpression of acetylcholinesterase inhibited cell proliferation
and promoted apoptosis in NRK cells. Acta Pharmacol Sin.
25:1013–1021. 2004.PubMed/NCBI
|
|
62
|
Velan B, Kronman C, Ordentlich A, Flashner
Y, Leitner M, Cohen S and Shafferman A: N-glycosylation of human
acetylcholinesterase: Effects on activity, stability and
biosynthesis. Biochem J. 296:649–656. 1993.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Doctor BP, Chapman TC, Christner CE, Deal
CD, De La Hoz DM, Gentry MK, Ogert RA, Rush RS, Smyth KK and Wolfe
AD: Complete amino acid sequence of fetal bovine serum
acetylcholinesterase and its comparison in various regions with
other cholinesterases. FEBS Lett. 266:123–127. 1990.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Campoy FJ, Cabezas-Herrera J and Vidal CJ:
Interaction of AChE with Lens culinaris agglutinin reveals
differences in glycosylation of molecular forms in sarcoplasmic
reticulum membrane subfractions. J Neurosci Res. 33:568–578.
1992.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Cabezas-Herrera J, Moral-Naranjo MT,
Campoy FJ and Vidal CJ: G4 forms of acetylcholinesterase and
butyrylcholinesterase in normal and dystrophic mouse muscle differ
in their interaction with Ricinus communis agglutinin.
Biochim Biophys Acta. 1225:283–288. 1994.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Nalivaeva NN and Turner AJ:
Post-translational modifications of proteins: Acetylcholinesterase
as a model system. Proteomics. 1:735–747. 2001.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Patinkin D, Seidman S, Eckstein F,
Benseler F, Zakut H and Soreq H: Manipulations of cholinesterase
gene expression modulate murine megakaryocytopoiesis in vitro. Mol
Cell Biol. 10:6046–6050. 1990.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Morell AG, Irvine RA, Sternlieb I,
Scheinberg IH and Ashwell G: Physical and chemical studies on
ceruloplasmin. V. Metabolic studies on sialic acid-free
ceruloplasmin in vivo. J Biol Chem. 243:155–159. 1968.PubMed/NCBI
|
|
69
|
Ashwell G and Harford J:
Carbohydrate-specific receptors of the liver. Annu Rev Biochem.
51:531–554. 1982.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Bon C, Hofer T, Bousquet-Mélou A, Davies
MR and Krippendorff BF: Capacity limits of asialoglycoprotein
receptor-mediated liver targeting. MAbs. 9:1360–1369.
2017.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Chia S, Tay SJ, Song Z, Yang Y, Walsh I
and Pang KT: Enhancing pharmacokinetic and pharmacodynamic
properties of recombinant therapeutic proteins by manipulation of
sialic acid content. Biomed Pharmacother.
163(114757)2023.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Kronman C, Velan B, Marcus D, Ordentlich
A, Reuveny S and Shafferman A: Involvement of oligomerization,
N-glycosylation and sialylation in the clearance of cholinesterases
from the circulation. Biochem J. 311:959–967. 1995.PubMed/NCBI View Article : Google Scholar
|