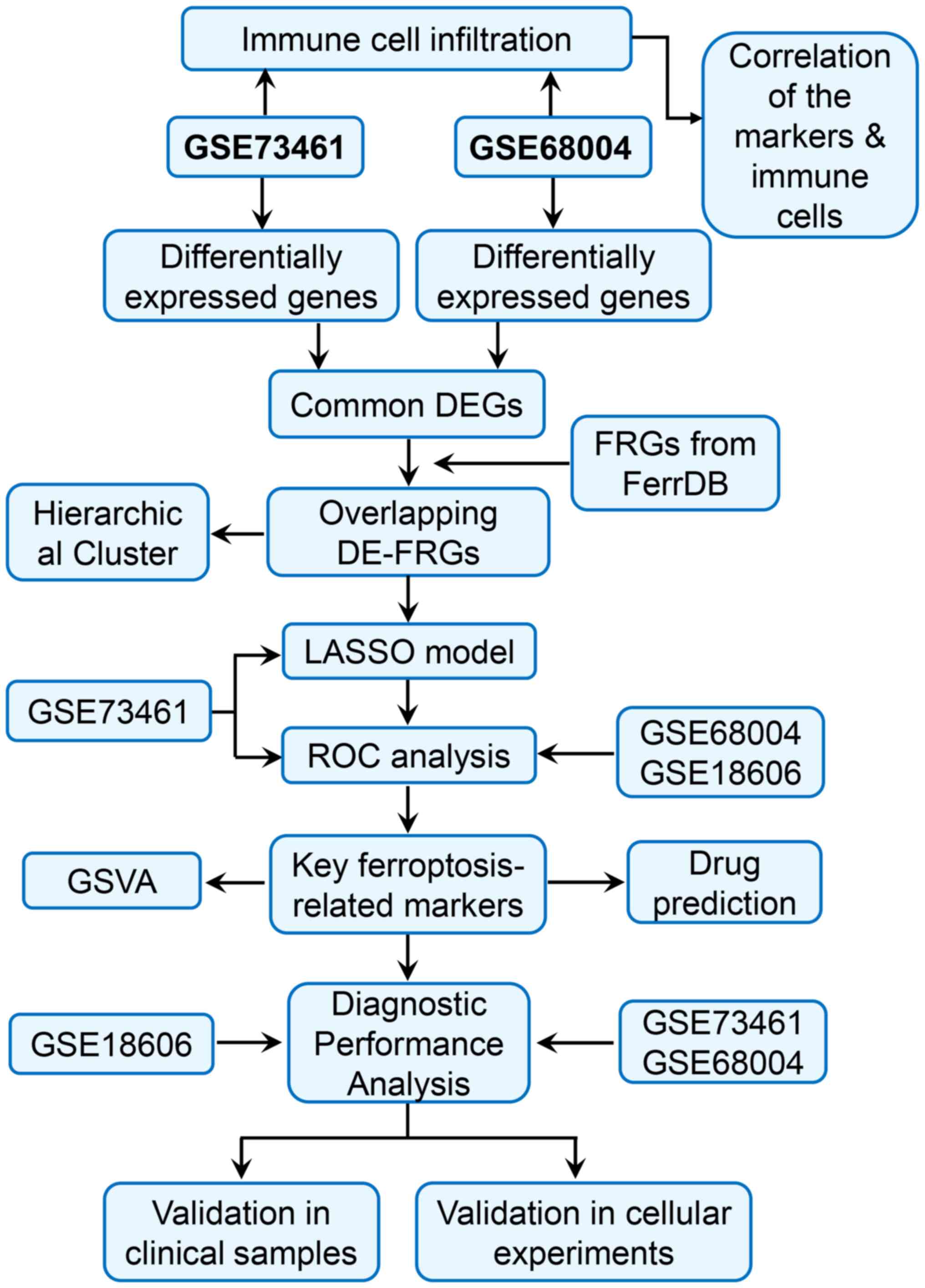
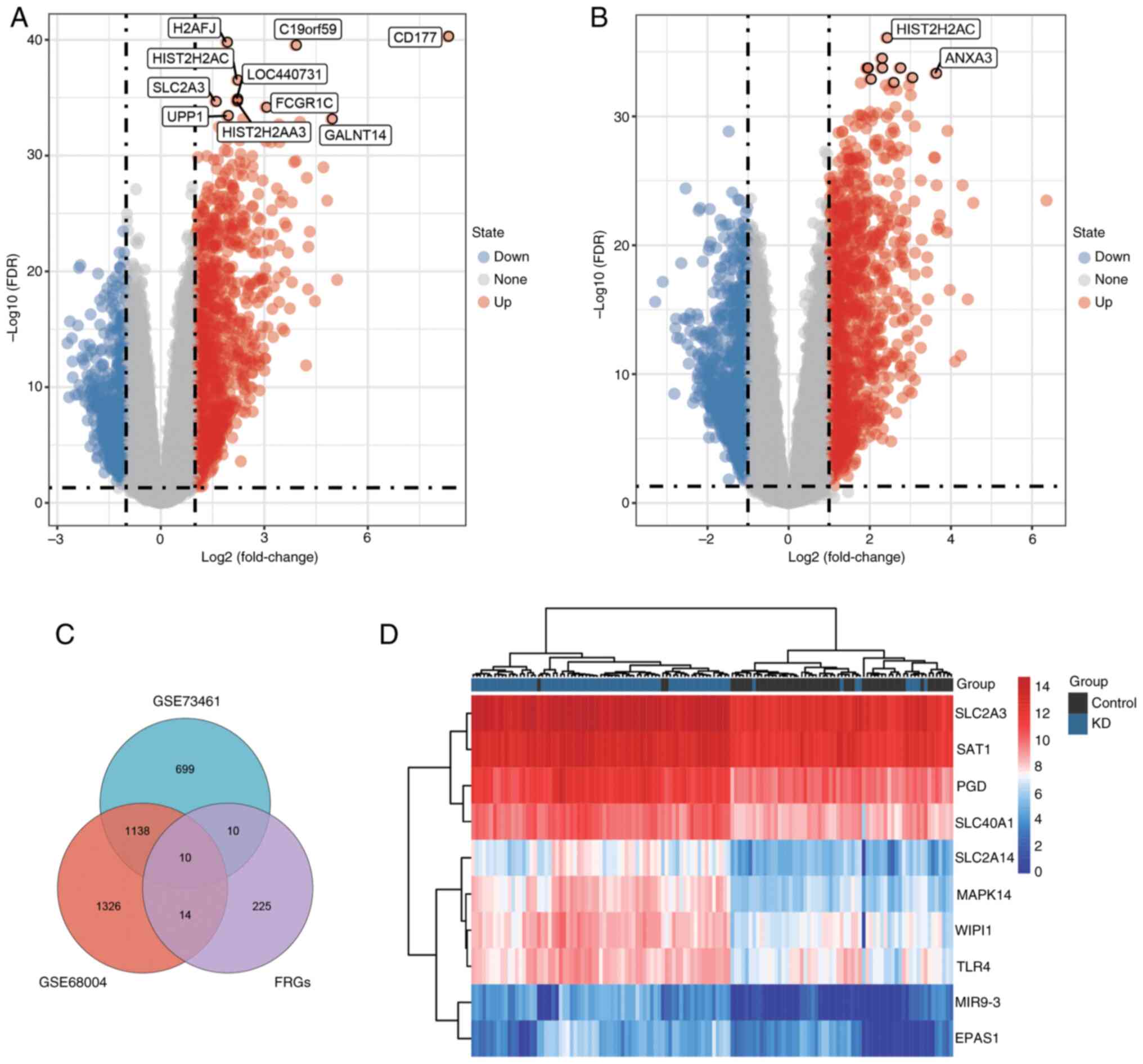
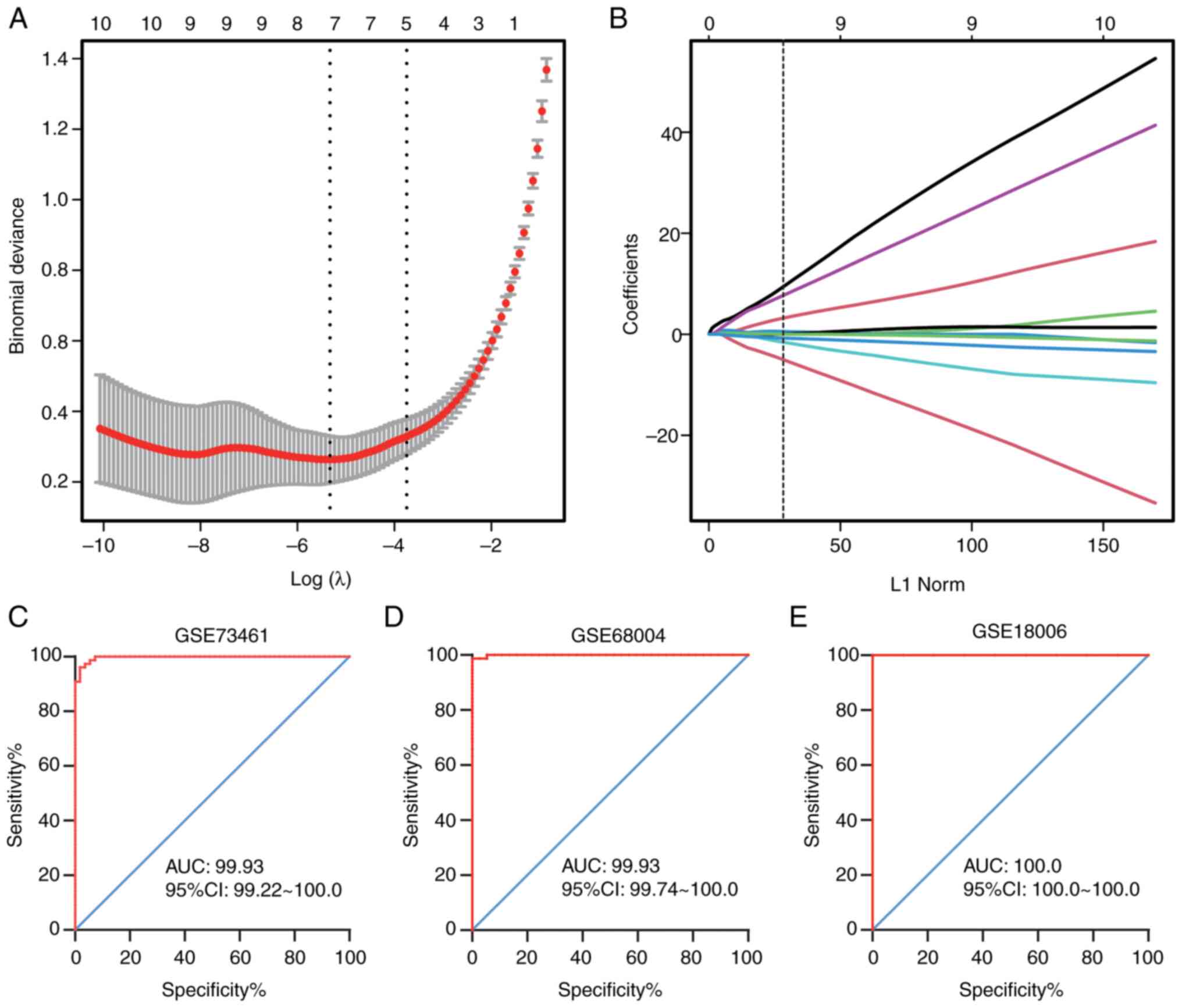
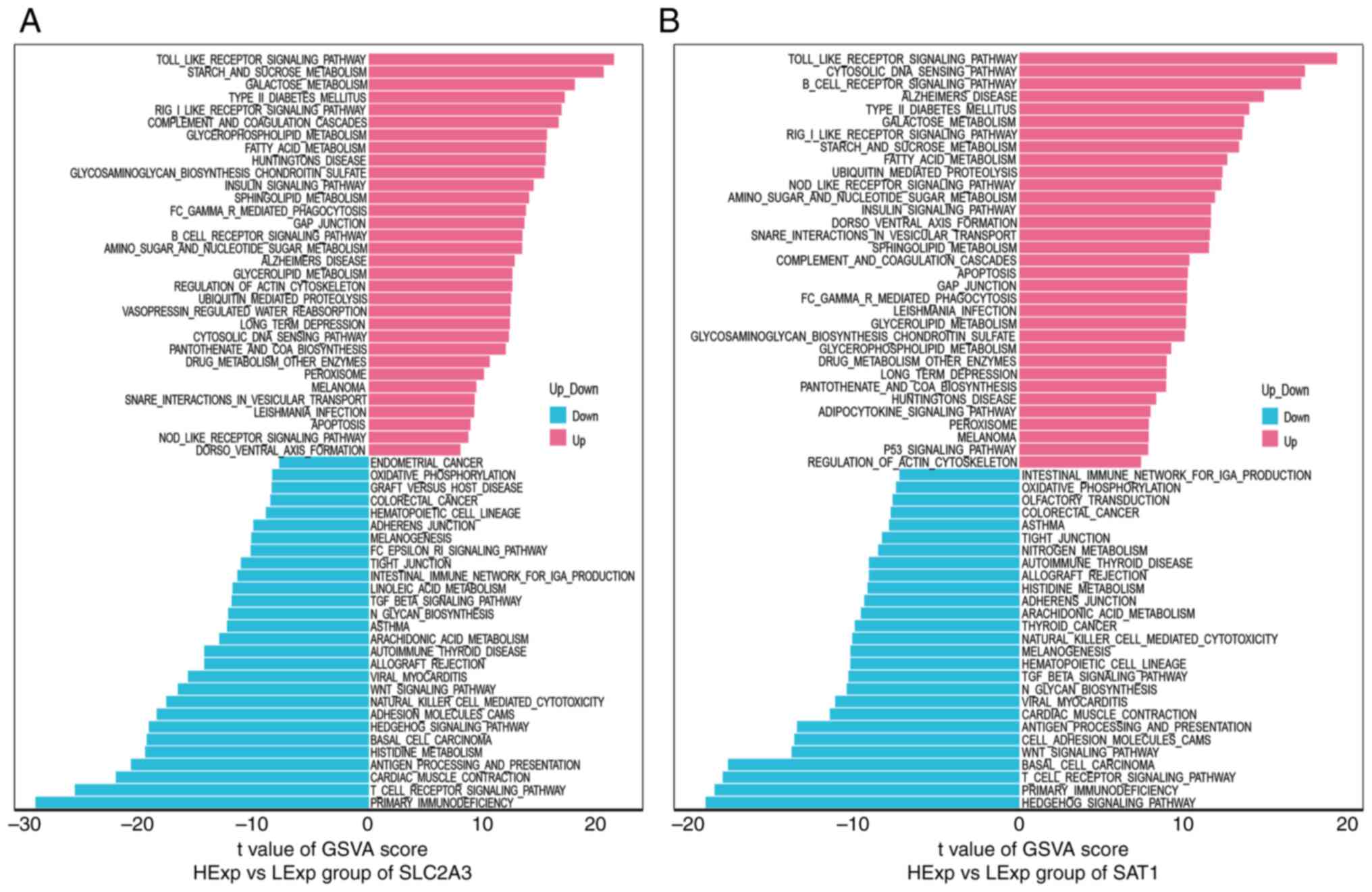
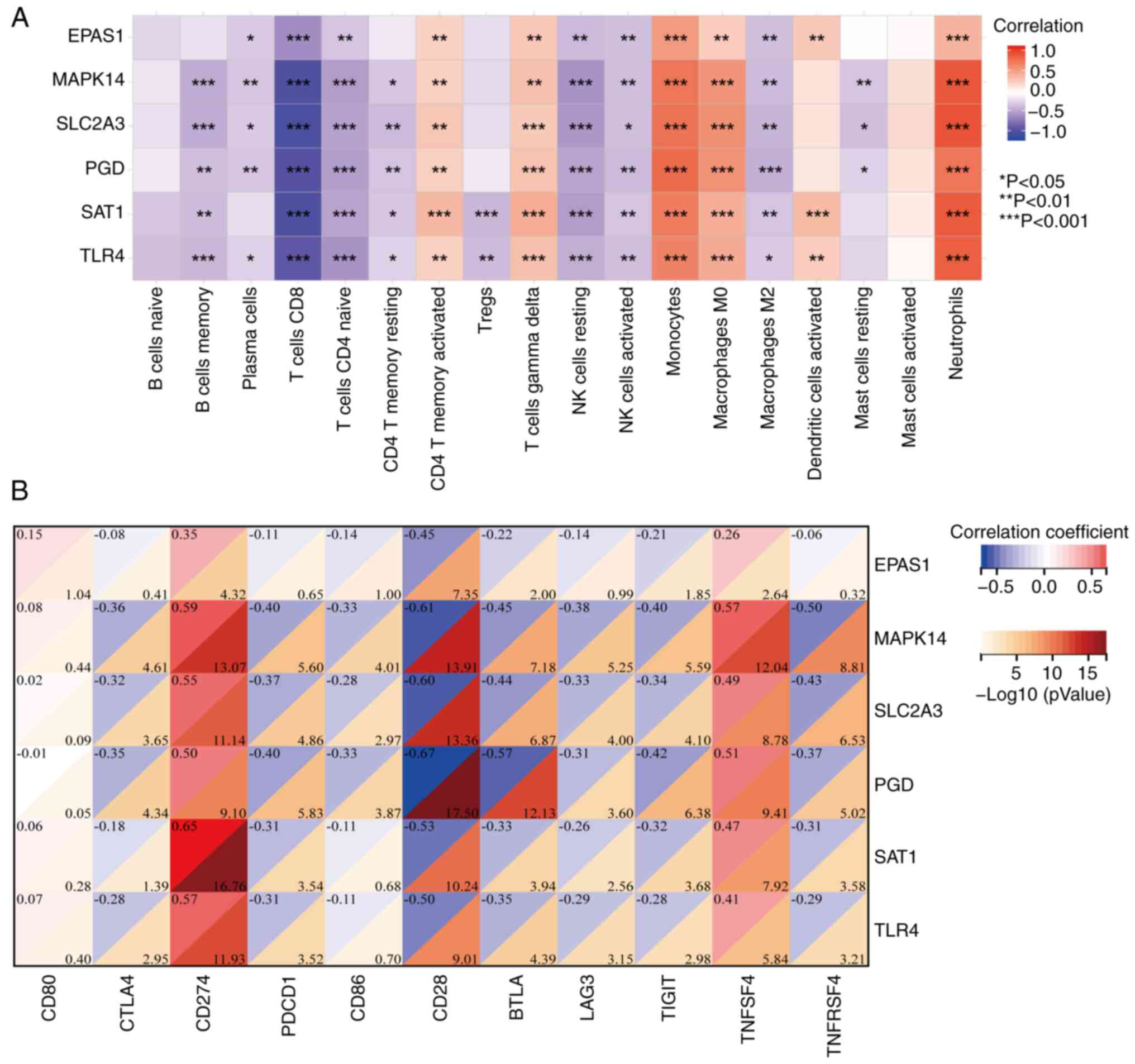
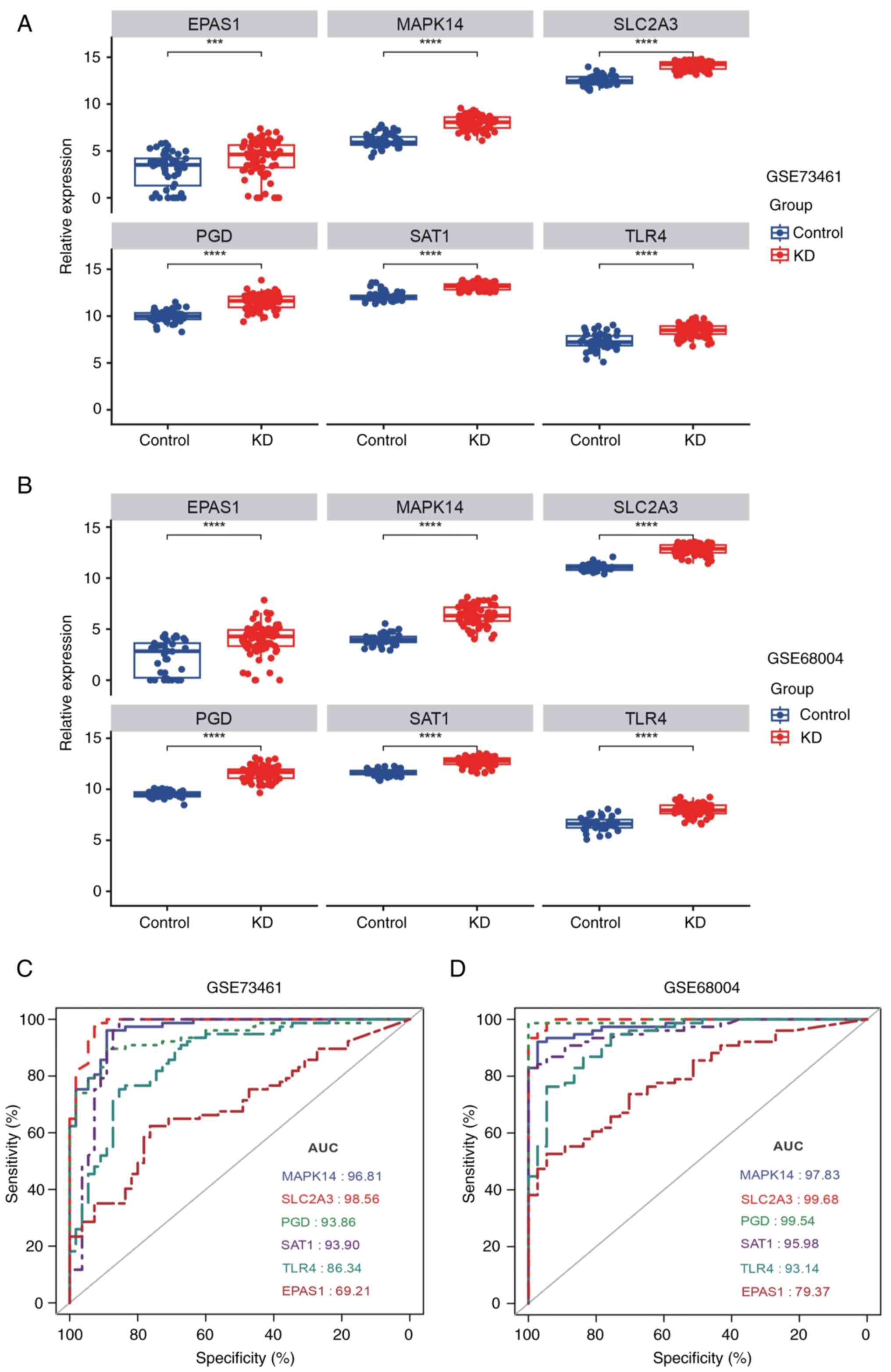
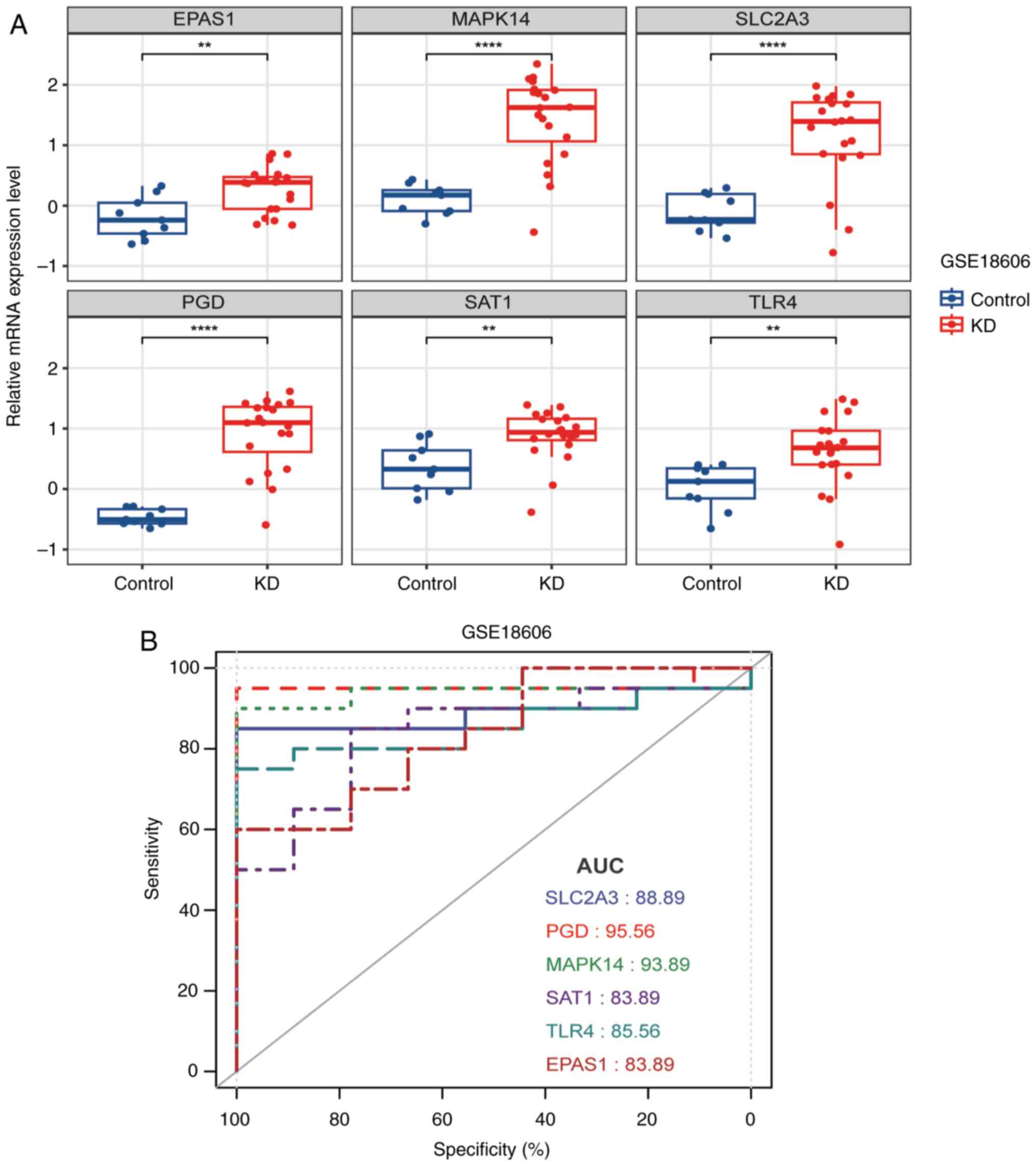
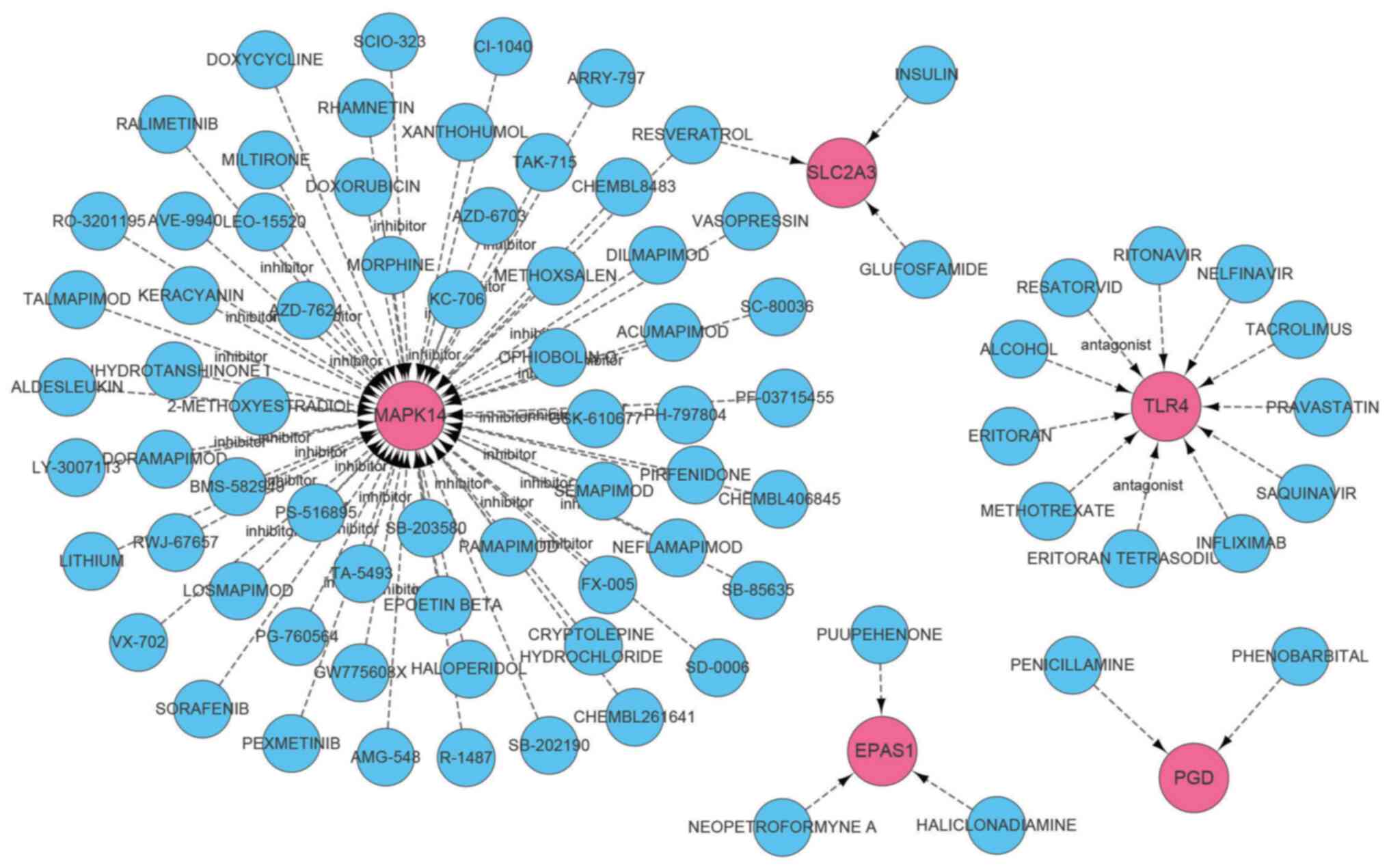
|
1
|
Ramphul K and Mejias SG: Kawasaki disease:
A comprehensive review. Arch Med Sci Atheroscler Dis. 3:e41–e45.
2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Senzaki H: Long-term outcome of Kawasaki
disease. Circulation. 118:2763–2772. 2008.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Newburger JW, Takahashi M and Burns JC:
Kawasaki disease. J Am Coll Cardiol. 67:1738–1749. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
McCrindle BW, Rowley AH, Newburger JW,
Burns JC, Bolger AF, Gewitz M, Baker AL, Jackson MA, Takahashi M,
Shah PB, et al: Diagnosis, treatment, and long-term management of
kawasaki disease: A scientific statement for health professionals
from the American heart association. Circulation. 135:e927–e999.
2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Duan C, Du ZD, Wang Y and Jia LQ: Effect
of pravastatin on endothelial dysfunction in children with medium
to giant coronary aneurysms due to Kawasaki disease. World J
Pediatr. 10:232–237. 2014.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Singh S, Vignesh P and Burgner D: The
epidemiology of Kawasaki disease: A global update. Arch Dis Child.
100:1084–1088. 2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Kainth R and Shah P: Kawasaki disease:
Origins and evolution. Arch Dis Child. 106:413–414. 2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Dietz SM, van Stijn D, Burgner D, Levin M,
Kuipers IM, Hutten BA and Kuijpers TW: Dissecting Kawasaki disease:
A state-of-the-art review. Eur J Pediatr. 176:995–1009.
2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Nakamura A, Ikeda K and Hamaoka K:
Aetiological significance of infectious stimuli in Kawasaki
disease. Front Pediatr. 7(244)2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Rypdal M, Rypdal V, Burney JA, Cayan D,
Bainto E, Skochko S, Tremoulet AH, Creamean J, Shimizu C, Kim J and
Burns JC: Clustering and climate associations of Kawasaki disease
in San Diego County suggest environmental triggers. Sci Rep.
8(16140)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Singh S, Jindal AK and Pilania RK:
Diagnosis of Kawasaki disease. Int J Rheum Dis. 21:36–44.
2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ng YM, Sung RYT, So LY, Fong NC, Ho MH,
Cheng YW, Lee SH, Mak WC, Wong DM, Yam MC, et al: Kawasaki disease
in Hong Kong, 1994 to 2000. Hong Kong Med J. 11:331–335.
2005.PubMed/NCBI
|
|
13
|
Jiang X, Stockwell BR and Conrad M:
Ferroptosis: Mechanisms, biology and role in disease. Nat Rev Mol
Cell Biol. 22:266–282. 2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Angeli JPF, Shah R, Pratt DA and Conrad M:
Ferroptosis inhibition: Mechanisms and opportunities. Trends
Pharmacol Sci. 38:489–498. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Stockwell BR, Friedmann Angeli JP, Bayir
H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascón S, Hatzios SK,
Kagan VE, et al: Ferroptosis: A regulated cell death nexus linking
metabolism, redox biology, and disease. Cell. 171:273–285.
2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Matsushita M, Freigang S, Schneider C,
Conrad M, Bornkamm GW and Kopf M: T cell lipid peroxidation induces
ferroptosis and prevents immunity to infection. J Exp Med.
212:555–568. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wu X, Qin K, Iroegbu CD, Xiang K, Peng J,
Guo J, Yang J and Fan C: Genetic analysis of potential biomarkers
and therapeutic targets in ferroptosis from coronary artery
disease. J Cell Mol Med. 26:2177–2190. 2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wen H, Hun M, Zhao M, Han P and He Q:
Serum ferritin as a crucial biomarker in the diagnosis and
prognosis of intravenous immunoglobulin resistance and coronary
artery lesions in Kawasaki disease: A systematic review and
meta-analysis. Front Med (Lausanne). 9(941739)2022.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wright VJ, Herberg JA, Kaforou M, Shimizu
C, Eleftherohorinou H, Shailes H, Barendregt AM, Menikou S, Gormley
S, Berk M, et al: Diagnosis of Kawasaki disease using a minimal
whole-blood gene expression signature. JAMA Pediatr.
172(e182293)2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Jaggi P, Mejias A, Xu Z, Yin H,
Moore-Clingenpeel M, Smith B, Burns JC, Tremoulet AH,
Jordan-Villegas A, Chaussabel D, et al: Whole blood transcriptional
profiles as a prognostic tool in complete and incomplete Kawasaki
disease. PLoS One. 13(e0197858)2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Fury W, Tremoulet AH, Watson VE, Best BM,
Shimizu C, Hamilton J, Kanegaye JT, Wei Y, Kao C, Mellis S, et al:
Transcript abundance patterns in Kawasaki disease patients with
intravenous immunoglobulin resistance. Hum Immunol. 71:865–873.
2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Li Y, Li Y, Bai Z, Pan J, Wang J and Fang
F: Identification of potential transcriptomic markers in developing
pediatric sepsis: A weighted gene co-expression network analysis
and a case-control validation study. J Transl Med.
15(254)2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Hara T, Nakashima Y, Sakai Y, Nishio H,
Motomura Y and Yamasaki S: Kawasaki disease: A matter of innate
immunity. Clin Exp Immunol. 186:134–143. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Jackson H, Menikou S, Hamilton S, McArdle
A, Shimizu C, Galassini R, Huang H, Kim J, Tremoulet A, Thorne A,
et al: Kawasaki disease patient stratification and pathway analysis
based on host transcriptomic and proteomic profiles. Int J Mol Sci.
22(5655)2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Shao Y, Saredy J, Yang WY, Sun Y, Lu Y,
Saaoud F, Drummer C IV, Johnson C, Xu K, Jiang X, et al: Vascular
endothelial cells and innate immunity. Arterioscler Thromb Vasc
Biol. 40:e138–e152. 2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Qian G, Xu L, Qin J, Huang H, Zhu L, Tang
Y, Li X, Ma J, Ma Y, Ding Y and Lv H: Leukocyte proteomics coupled
with serum metabolomics identifies novel biomarkers and abnormal
amino acid metabolism in Kawasaki disease. J Proteomics.
239(104183)2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Kuijpers TW, Wiegman A, van Lier RA, Roos
MT, Wertheim-van Dillen PM, Pinedo S and Ottenkamp J: Kawasaki
disease: A maturational defect in immune responsiveness. J Infect
Dis. 180:1869–1877. 1999.PubMed/NCBI View
Article : Google Scholar
|
|
30
|
Ikeda K, Yamaguchi K, Tanaka T, Mizuno Y,
Hijikata A, Ohara O, Takada H, Kusuhara K and Hara T: Unique
activation status of peripheral blood mononuclear cells at acute
phase of Kawasaki disease. Clin Exp Immunol. 160:246–255.
2010.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Hoang LT, Shimizu C, Ling L, Naim AN, Khor
CC, Tremoulet AH, Wright V, Levin M, Hibberd ML and Burns JC:
Global gene expression profiling identifies new therapeutic targets
in acute Kawasaki disease. Genome Med. 6(541)2014.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Hara T, Yamamura K and Sakai Y: The
up-to-date pathophysiology of Kawasaki disease. Clin Transl
Immunology. 10(e1284)2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Sakurai Y: Autoimmune aspects of Kawasaki
disease. J Investig Allergol Clin Immunol. 29:251–261.
2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Popper SJ, Shimizu C, Shike H, Kanegaye
JT, Newburger JW, Sundel RP, Brown PO, Burns JC and Relman DA:
Gene-expression patterns reveal underlying biological processes in
Kawasaki disease. Genome Biol. 8(R261)2007.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Xie Z, Huang Y, Li X, Lun Y, Li X, He Y,
Wu S, Wang S, Sun J and Zhang J: Atlas of circulating immune cells
in Kawasaki disease. Int Immunopharmacol.
102(108396)2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Cargnello M and Roux PP: Activation and
function of the MAPKs and their substrates, the MAPK-activated
protein kinases. Microbiol Mol Biol Rev. 75:50–83. 2011.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zheng T, Zhang B, Chen C, Ma J, Meng D,
Huang J, Hu R, Liu X, Otsu K, Liu AC, et al: Protein kinase p38α
signaling in dendritic cells regulates colon inflammation and
tumorigenesis. Proc Natl Acad Sci USA. 115:E12313–E12322.
2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Yan R and Zhou T: Identification of key
biomarkers in neonatal sepsis by integrated bioinformatics analysis
and clinical validation. Heliyon. 8(e11634)2022.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Reddy ABM, Srivastava SK and Ramana KV:
Aldose reductase inhibition prevents lipopolysaccharide-induced
glucose uptake and glucose transporter 3 expression in RAW264.7
macrophages. Int J Biochem Cell Biol. 42:1039–1045. 2010.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Fu Y, Maianu L, Melbert BR and Garvey WT:
Facilitative glucose transporter gene expression in human
lymphocytes, monocytes, and macrophages: A role for GLUT isoforms
1, 3, and 5 in the immune response and foam cell formation. Blood
Cells Mol Dis. 32:182–190. 2004.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Khan GB, Qasim M, Rasul A, Ashfaq UA and
Alnuqaydan AM: Identification of Lignan compounds as new
6-phosphogluconate dehydrogenase inhibitors for lung cancer.
Metabolites. 13(34)2022.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Liu T, Qi J, Wu H, Wang L, Zhu L, Qin C,
Zhang J and Zhu Q: Phosphogluconate dehydrogenase is a predictive
biomarker for immunotherapy in hepatocellular carcinoma. Front
Oncol. 12(993503)2022.PubMed/NCBI View Article : Google Scholar
|