Introduction
Ultrasound (US) is a sound wave with frequencies
>20 kHz and has been widely used in diagnosis and therapy in
medicine (1-6).
Acoustic intensities for diagnosis are typically <100
mW/cm2 (1-3).
In cancer therapy, high-intensity focused ultrasound is used
(5,6). For example, the acoustic intensity
used for prostate cancer ranges from 100 to 10,000 mW/cm² spatial
average-temporal average intensity (ISATA) (6). Low-intensity pulsed US (LIPUS) is a
well-recognized non-invasive therapy and has garnered attention as
a potential adjunctive therapy for accelerating bone fracture
healing (3,4). LIPUS clinical protocol for fractures
involves a 1.5 MHz sine wave repeated at 1 kHz at 30
mW/cm2 ISATA with a pulsed width of 200 µsec
for 20 min/day (3,4). LIPUS has been shown to augment bone
fracture healing in experimental animal models, using rabbits
(7), rats (8,9) and
mice (10), as well as clinical
settings (11,12). Although molecular mechanisms
underlying the effects of LIPUS remain poorly elucidated, numerous
in vitro studies have demonstrated positive osteogenic
effects of LIPUS on bone cells during fracture healing (13-21).
Mammalian bone is an active mineralized connective
tissue composed of three types of cell: Osteoblasts, osteoclasts
and osteocytes (22). Mechanical
loading has also been widely recognized as an essential factor for
the maintenance of bone. It is well known that both bone matrix and
osteocytes play an important role in sensing mechanical loading of
bone (23-25).
Similarly, the teleost scale is a unique calcified tissue in which
osteoclasts, osteoblasts and two layers of the bone matrix are
present (26-28).
Furthermore, our previous study demonstrated the existence of
osteocyte-like cells in goldfish scales (29). Fish scales are therefore a suitable
model for bone mechanotransduction, the conversion of mechanical
stimulus into a biological response. Osteoblasts and osteoclasts in
goldfish scales respond sensitively to mechanical loading such as
hypergravity (27), microgravity
(30) and ultrasound (16,31,32).
RNA-sequencing (RNA-seq) using next-generation
sequencing (NGS) technology is a powerful tool that applies
genome-wide expression profiles to a wider range of organisms, even
in non-model organisms with no established databases, compared with
DNA microarray (33). Our previous
studies reported that melatonin suppresses osteoclast activation
and cell damage induced by space flight in goldfish scales using
RNA-seq method (30,34). The present study examined the
effects of LIPUS (30 mW/cm2) on the osteoblasts and
osteoclasts of goldfish scales and performed global-scale gene
expression analysis of scales treated with LIPUS using RNA-seq to
determine the underlying mechanism.
Materials and methods
Animals and preparation of fish scales.
Goldfish (Carassius auratus) were obtained from Higashikawa Fish
Farm (Yamatokoriyama, Japan). A total of 12 male fish (weight,
30-40 g; age, ~2 years) were fed a commercial pellet diet (Spectrum
Brands Japan) every morning and were maintained in freshwater (pH,
7.0-7.5) at 26˚C under a 12 h light/12 h dark cycle (32). All experimental procedures were
conducted in accordance with the Guide for Care and Use of
Laboratory Animals and approved by Animal Research Committee of
Kanazawa University (approval no. 242-2023) and were performed
under anesthesia to minimize pain. In addition, all experimental
protocols were in strict accordance with the ARRIVE guidelines
2.0(35).
Regenerating scales that had active osteoclasts and
osteoblasts were used for analysis of LIPUS treatments (27). In brief, goldfish were anesthetized
in freshwater containing ethyl 3-aminobenzoate methanesulfonic acid
salt (MS-222; 330 mg/l; Sigma-Aldrich; Merck KGaA) (34). Adequate anesthesia was indicated
cessation of opercular movements. The normal scales that developed
on the body were removed to allow the regeneration of scales under
anesthesia. The anesthetized fish were returned to freshwater,
allowed to recover and the goldfish were maintained as
aforementioned. Behavior and feeding activity were monitored daily
to check health. At 14 days after the removal of normal scales,
regenerating scales were removed from the goldfish under anesthesia
and were placed in a 6-well cell culture plate (Nippon Genetics,
Co., Ltd.) with 2 ml Leibovitz's L-15 medium (Invitrogen; Thermo
Fisher Scientific, Inc.) containing 1% penicillin-streptomycin
mixture (ICN Biomedicals, Inc.) and incubated for 2 h at 15˚C
before use (27,32). Goldfish were anesthetized using
MS-222. Once opercular movements ceased, anesthesia was continued
for an additional 40 min to euthanize the goldfish. The goldfish,
which were no longer responding to stimuli and not moving, were
then returned to freshwater. Death was confirmed following no
recovery within 20 min.
LIPUS treatment and temperature
measurement of culture medium
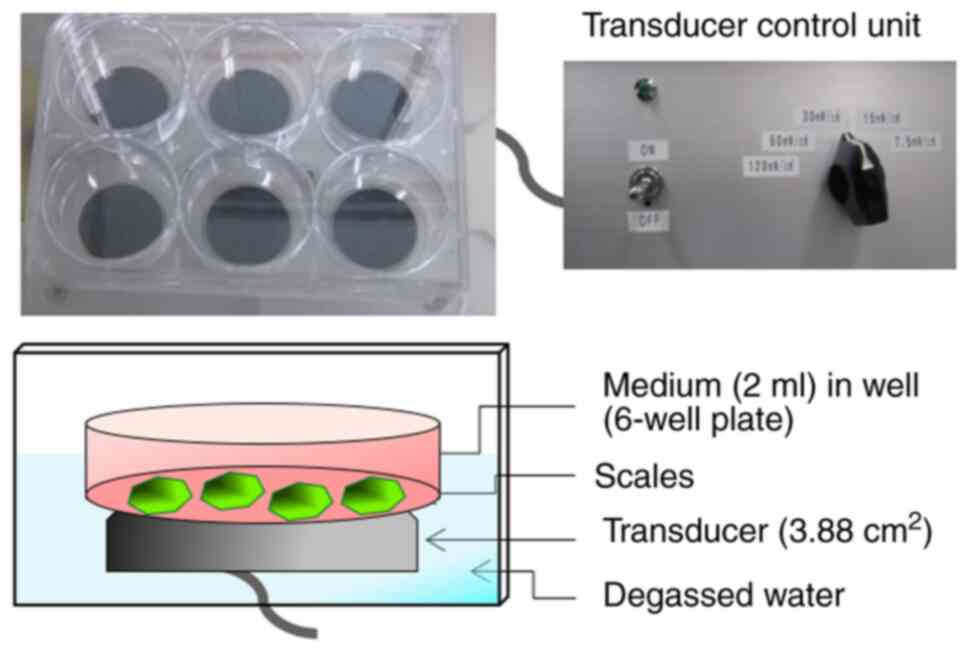
LIPUS treatment was applied using an ultrasound
irradiating system in a 6-well cell culture plate (No.
US-Vitro-N04-48; Teijin Pharma, Ltd.; Fig. 1) (36). (3,4). The
signal had an ISATA of 30 mW/cm2, with a
frequency of 1.5 MHz in a pulsed-wave mode (0.2-sec burst sine
waves repeated at 1.0 kHz). When the scales were treated with
LIPUS, the 6-well cell culture plate containing scales was placed
on the transducer. LIPUS was transmitted through the bottom of the
cell culture plate. The scales were irradiated with LIPUS for 20
min at room temperature. For control group, the scales were
incubated for 20 min without LIPUS treatment in the aforementioned
LIPUS-exposure setup as described above. The treated scales were
further incubation for 3, 6 or 24 h at 15˚C.
The biophysical effects of ultrasound on living
tissue are divided into thermal and non-thermal effects (2). Temperature of the culture medium was
monitored with a digital thermometer coupled to a type K
thermocouple sensor (Sato Keiryoki MFG Co., Ltd.) at room
temperature (37).
Assay of alkaline phosphatase (ALP)
and tartrate-resistant acid phosphatase (TRAP) activity
ALP or TRAP activity was measured using alkaline (1
mM MgCl2 and 100 mM Tris-HCl, pH 9.5) or acidic buffer
(100 mM sodium acetate and 20 mM tartrate, pH 5.3), respectively.
In short, a 100 µl aliquot of alkaline or acidic buffer was added
to each well at room temperature. Then, each scale was placed into
a well in a 96-well microplate. This microplate was immediately
frozen at -80˚C and then kept at -20˚C until analysis. A 100 µl
aliquot of 20 mM para-nitrophenyl (pNP) phosphate in alkaline or
acidic buffer was added to each well at room temperature. The plate
was incubated at 23˚C for 20 min with shaking. The reaction was
stopped by adding 50 µl 3 M NaOH. A total of 150 µl reacted
solution was transferred to a new plate and the absorbance was
measured at 405 nm. The absorbance was converted into the amount of
pNP produced using a standard curve. After measuring both ALP and
TRAP activity, the scales were measured with Image J software (Ver.
1.53; https://imagej.net/ij/index.html). ALP and TRAP
activities were normalized to the surface area (mm2) of
each scale (27).
RNA isolation
Total RNA was isolated from the regenerating scales
of goldfish using RNeasy Fibrous Tissue Mini kit (cat. no. 74704;
Qiagen GmbH). The concentration of RNA was measured by spectroscopy
with an expected A260/A280 ratio close to 2. RNA quality was
analyzed using a Bioanalyzer 2100 and RNA 6000 Nano kit (Cat. No.
5067-1511; Agilent Technologies, Inc.). Total RNA with RNA
integrity number >9.0 was used for RNA-seq and reverse
transcription-quantitative (RT-q)PCR.
RNA-seq, gene expression and gene
network analyses
RNA-seq was performed by Veritas Genetics Co. The
analysis was conducted with the Novaseq6000 sequencer (Illumina,
Inc.), and data of nucleotide length 150 bp (directional paired-end
reads) was provided.
Quality check of raw read sequences was performed
using FastQC (Ver. 0.11.8; http://www.bioinformatics.babraham.ac.uk/projects/fastqc).
Adaptors and short, low-quality reads (Q<20; <50 bp) were
trimmed using TrimGalore (Ver. 1.18; bioinformatics.babraham.ac.uk/projects/trim_galore/).
For de novo transcriptome assembly and read mapping,
following PCR duplicate removal using seqkit (38), de novo transcriptome assembly
of processed reads was performed using Trinity (r2012-10-05;
cell-innovation.nig.ac.jp/index_en.html) was used with
default parameters to generate a reference sequence of transcripts
(33). To estimate relative RNA
expression levels as transcripts per million reads (TPM), sequence
reads with PCR duplicates were pseudo-mapped to the reference
sequence of transcripts using Kallisto with strict parameters
(k-mer size: 31; bootstrap-samples: 0; min-size: automatically
chosen; ec-max-size: no maximum) (39). To normalize gene expression,
statistical analysis of TPM was performed using Strand NGS v3.3
with the Trimmed Mean of M value (TMM) (40). TMM is a normalized method that is
generally used for analyzing omics data (https://genomebiology.biomedcentral.com/articles/10.1186/gb-2010-11-3-r25).
Furthermore, differentially expressed genes (DEGs) in LIPUS,
compared with LIPUS non-treated scales, were identified using
unpaired t-test and multiple testing Benjamini-Hochberg correction.
Genes were considered differentially expressed when P<0.05 and
fold-change ≥1.2. The obtained data were analyzed using
Ingenuity® Pathway Analysis tools (Qiagen GmbH) to
examine Gene Ontology (geneontology.org/) enrichment, including biological
processes, cellular components, molecular functions, and gene
networks. Upregulated genes at 3, 6 and 24 h after LIPUS treatment
were uploaded to the Ingenuity® Pathway Analysis tools.
Core analysis was performed, followed by analysis of biological
functions, especially osteoblastic differentiation. Candidate genes
were analyzed using Gene Network Analysis to generate gene networks
based on known interactions (37,41).
RT-qPCR
Complementary DNA was produced from total RNA using
a PrimeScript™ RT kit with gDNA Eraser (Takara Bio, Inc.) according
to the manufacturer's protocol. RT-qPCR was performed on an Mx3005P
real-time PCR system (Agilent Technologies, Inc.) using
SYBR® Premix Ex Taq™ II (Tli RNaseH Plus; Takara Bio,
Inc.). The specific primer sequences are listed in Table I. The thermocycling conditions were
as follows: Initial denaturation for 10 min at 95˚C, followed by 40
cycles of 10 sec at 95˚C and 40 sec at 60˚C. For quantification,
the standard curve method was used (42). Moreover, PCR products were
electrophoresed through a 2% agarose gel at 100 V for 30 min using
the Mupid-2plus gel electrophoresis system (Takara Bio Inc.) After
staining with ethidium bromide (0.5 µg/ml) for 15 min at room
temperature, gel bands were visualized using a gel imaging system
(Printgraph TYPE-GX, ATTO Co.). β-actin was used as an internal
control (37,43).
 | Table INucleotide sequences of primers for
target genes. |
Table I
Nucleotide sequences of primers for
target genes.
| Gene | Sequence,
5'→3' |
|---|
| ACTB | F:
TGTGCTGTCCCTGTATGCCT |
| | R:
ATTTCCCTCTCGGCTGTGGT |
| PRKD1 | F:
GCCATTCTCCAGAACCTC |
| | R:
AGAAACTTGGTGATGCGT |
| PTGS2 | F:
TGTGTTCGGGGAGACTATGG |
| | R:
CCACTTTCCCACCAAACGTG |
| TNFRSF11B | F:
TGACAGGTGTCCTCCAGGAA |
| | R:
TCCAGAACTCCGTGAACAGAC |
| WNT3A | F:
ACCGAAACTGACCTGGTCTAC |
| | R:
CAAGTCGCAGCCATCGATAC |
Statistical analysis
Data are presented as the mean ± SD of three or more
independent experiments. The values of the control scales were
compared with those of LIPUS-treated scales. Differences were
analyzed using a paired t test. Statistical analysis was performed
using R software (Ver. 4.3.3.; r-project.org/). P<0.05 was considered to indicate
a statistically significant difference.
Results
Effect of LIPUS on temperature of
culture medium
Biophysical effects of ultrasound on living tissues
are divided into thermal and nonthermal effects (2). Therefore, the effects of LIPUS on the
temperature of the culture medium were determined. The initial
temperature of the culture medium was 24.9±0.2˚C. The temperature
of the culture medium 20 min after LIPUS treatment was not
significantly increased (25.0±0.2˚C), suggesting thermal effects
were not included in the bioeffects of LIPUS.
Effects of LIPUS on ALP and TRAP
activity of goldfish scales
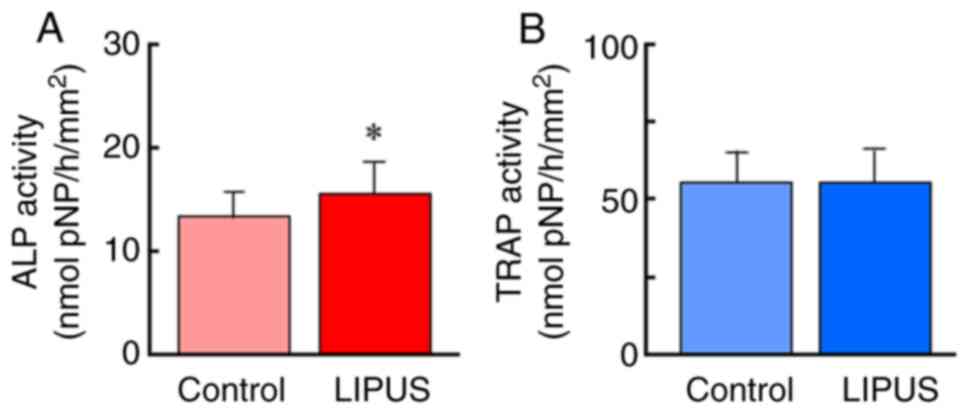
LIPUS treatment significantly increased ALP
activity, a marker of osteoblasts, but did not affect TRAP
activity, a marker of osteoclasts (Fig.
2A). These data indicated that LIPUS induced osteoblastic
activation in goldfish scales.
Gene expression analysis
To identify candidate genes responsive to LIPUS
treatment, the time course of the gene expression profile was
examined using RNA-seq. Gene expression analysis of the scales
exposed to LIPUS revealed 207, 280 and 350 upregulated genes at 3,
6 and 24 h, respectively, and 188, 368 and or 298 downregulated
genes at 3, 6 and 24 h, respectively (Fig. 3). The complete lists of DEGs from
the goldfish scale samples are shown in Table SI, Table SII, Table SIII, Table SIV, Table SV and Table SVI.
Gene network and biological function
analysis
To examine the functional association between
candidate genes, gene network and biological function analyses were
performed using the Ingenuity® Pathways Knowledge Base.
Significant gene networks, GN-3h, GN-6h and GN-24h, were identified
from upregulated genes at 3, 6 and 24 h after LIPUS treatment,
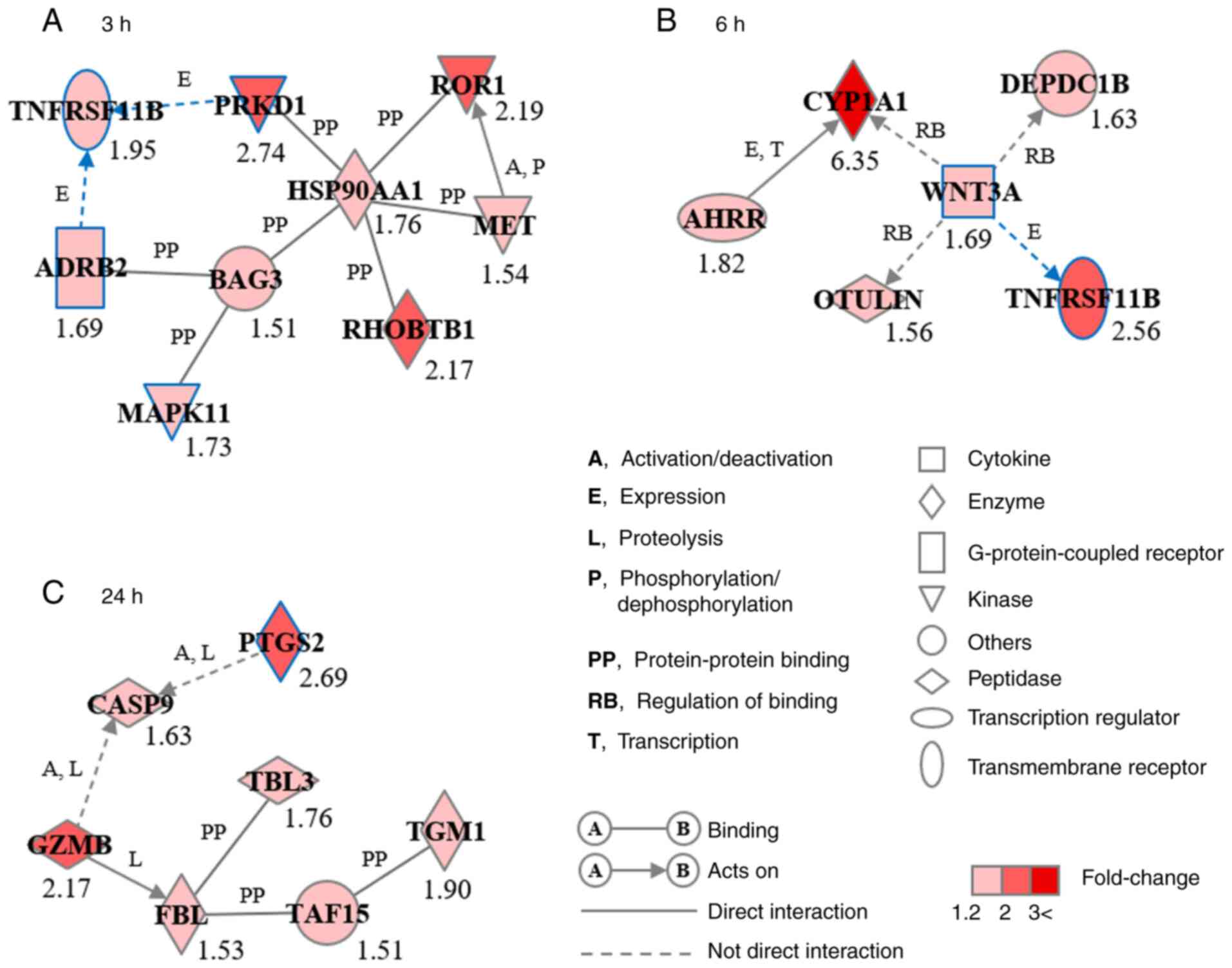
respectively (Fig. 4). GN-3h
consisted of nine genes: Adrenoceptor β2 (ADRB2), BAG cochaperone
3, heat shock protein 90α family class A member 1 (HSP90AA1),
MAPK11, MET proto-oncogene, receptor tyrosine kinase, protein
kinase D1 (PRKD1), Rho-related BTB domain containing 1, receptor
tyrosine kinase-like orphan receptor 1 and tumor necrosis factor
receptor superfamily, member 11b (TNFRSF11B) (Fig. 4A). GN-6h consisted of six genes:
Aryl hydrocarbon receptor repressor, cytochrome P450 family 1
subfamily A member 1, DEP domain-containing 1B, OTU deubiquitinase
with linear linkage specificity, TNFRSF11B and Wnt family member 3A
(WNT3A; Fig. 4B). GN-24h consisted
of seven genes: Caspase 9, fibrillarin, granzyme B,
prostaglandin-endoperoxide synthase 2 (PTGS2), TATA box-binding
protein-associated factor 15, transducin β-like 3 and
transglutaminase 1 (Fig. 4C).
ADRB2, MAPK11, PRKD1, PTGS2, TNFRSF11B and WNT3A were associated
with biological functions of osteogenesis, including osteoblast
differentiation and bone development and formation.
Effects of LIPUS on gene
expression
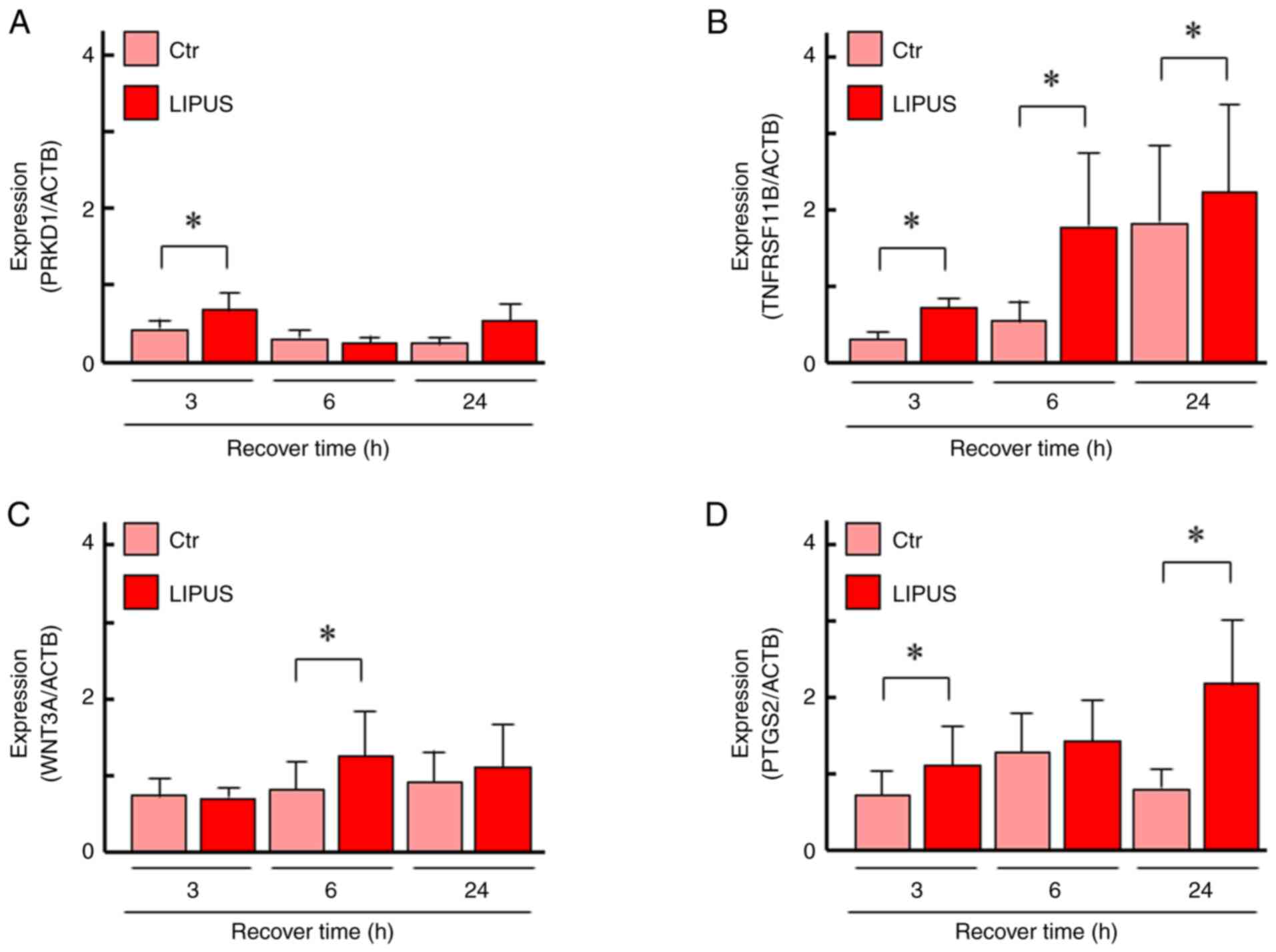
PRKD1, TNFRSF11B, WNT3A and PTGS2–were selected from
the three gene networks. The specificity of the primers was
confirmed by a single band with the correctly amplified fragment
size through agarose gel electrophoresis of the PCR products
(Supplementary material Fig. S1).
Expression of PRKD1 was significantly upregulated at 3 h and
TNFRSF11B was significantly upregulated at all time points
(Fig. 5A and B). The expression of WNT3A was
significantly upregulated at 6 h and PTGS2 was significantly
upregulated at 3 and 24 h after LIPUS treatment (Fig. 5C and D). These data were comparable with the
RNA-seq results.
Discussion
Numerous studies have shown that LIPUS induces
osteogenic activity in in vitro experimental models
(13-21).
For example, ALP activity, a marker of osteoblasts, in mouse
preosteoblast MC3T3-E1(14) and
bone marrow mesenchymal stem cells (21) is significantly increased in response
to clinical application of LIPUS (30 mW/cm2 for 20 min).
Kitamura et al (16)
demonstrated that LIPUS at 60 mW/cm2 for 6-20 min
significantly elevates ALP activity in goldfish scales. In
agreement with previous studies (14,16,21),
here, LIPUS (30 mW/cm2 for 20 min) significantly
increased ALP activity but did not affect TRAP activity, a marker
of osteoclasts, suggesting that LIPUS induced osteoblastic
activation in goldfish scales. On the other hand, in our previous
study, the same clinical LIPUS application affected osteoclasts in
fish scales; LIPUS directly caused apoptosis in osteoclasts 3 h
after treatment in both zebrafish and goldfish scales (31) and moderately activated osteoclasts 6
and 12 h after treatment in goldfish scales (32). LIPUS was transmitted through the
bottom of the cell culture plate or directly from the upper side in
the present osteoblast-activating or previous osteoclast-affecting
conditions (31,32), respectively. The discrepancy in the
effects of LIPUS between the present and previous studies may have
been related to this difference in the LIPUS-exposure conditions.
In addition, 2 weeks after daily LIPUS treatment, ALP activity and
regeneration rate are significantly increased in goldfish scales
in vivo (32). The present
results and those of previous studies (16,31,32)
suggest that osteoblasts and osteoclasts in fish scales respond
sensitively to LIPUS mechanical stress.
The present study used RNA-seq and
Ingenuity® pathway analyses to identify DEGs and three
unique gene networks. These networks included ADRB2, MAPK11, PRKD1,
TNFRSF11B, WNT3A and PTGS2, which are known to be involved in
various aspects of osteogenesis, including osteoblast
differentiation and bone development and formation (44-53).
Previous research using knockout mice demonstrated that
ADRB2(44) and PRKD1(45) are key to bone formation, while
TNFRSF11B (46) and MAPK11(47) serve key roles in bone development.
PRKD1(48), TNFRSF11B (49), WNT3A (50) and PTGS2(51) induce osteoblast differentiation in
experimental models such as MC3T3-E1 mouse preosteoblastic cells
(48,49), C2C12(48) and C3H10T1/2(50) mouse mesenchymal stem cells, as well
as PTGS2 knockout mice (51).
Interactions have been reported between TNFRSF11B and
ADRB2(44), PRKD1(52) and WNT3A (53).
The product translated from TNFRSF11B, known as
osteoprotegerin (OPG) and produced by osteoblasts, functions as a
decoy receptor of receptor activator for nuclear factor-κB ligand
(RANKL) and suppresses osteoclastogenesis (54). One of the most effective approaches
for clinically treating osteoporosis involves the use of a specific
antibody to inhibit RANKL, a mechanism akin to that of OPG
(55). Yu et al (49) indicated that ALP activity is
significantly higher in the OPG-overexpressing preosteoblast cell
line MC3T3-E1, indicating that OPG promotes matrix maturation in
preosteoblasts. Moreover, reports indicate that OPG enhances
osteoblastogenesis of human mesenchymal stem cells (18,56)
and LIPUS (400 mW/cm2, 20 min) boosts osteoblastogenesis
of human mesenchymal stem cells by increasing mRNA levels of
TNFRSF11B and ALP (18). Moreover,
Borsje et al (15) reported
that the clinical application of LIPUS significantly increases OPG
mRNA and protein expression in human osteoblast-like Saos-2 cells.
Furthermore, in vivo study using an ovariectomy-induced
osteoporotic fracture rat model found that clinical application of
LIPUS accelerates osteoporotic fracture healing. Moreover, OPG gene
expression is upregulated in LIPUS-treated rats at an earlier stage
of the repair process compared with controls (9). In our previous study using goldfish
scales, OPG was suggested to suppress excessive bone resorption
under osteoclast-activated conditions induced by clinical
application of LIPUS (32). In the
present study, LIPUS induced TNFRSF11B expression and ALP activity
without interfering with TRAP activity. This suggested that in
LIPUS-treated goldfish scales, OPG may be involved in osteoblastic
differentiation rather than suppression of osteoclastogenesis.
Cyclooxygenase 2 (COX-2), which is encoded by PTGS2,
is a key enzyme in prostaglandin E2 (PGE2) biosynthesis. LIPUS
stimulation increases expression of PTGS2 and PGE2 in bone cells
(13,37,57-59).
Tang et al (13)
demonstrated that the clinical application of LIPUS stimulates
mineralization of osteoblasts via the COX-2/PGE2 pathway. COX-2 and
PGE2 are reported to play essential roles in the regulation of
osteoblastic differentiation (13,51,60)
and fracture healing (3,4,10).
Naruse et al (10) used
knockout mice to demonstrate that LIPUS accelerates endochondral
bone healing during senescence in a COX-2-dependent manner. A delay
in fracture healing is also observed in rats and humans
administered COX inhibitors (61,62).
In the present study, expression of PTGS2 was elevated 24 h
after LIPUS treatment in goldfish scales. Omori et al
(63) demonstrated that the
addition of PGE2 to goldfish scales promotes both osteoblastic and
osteoclastic activity. The present data along with the previous
findings suggest that LIPUS stimulation may enhance
osteoblastogenesis via the COX-2/PGE2 pathway. Investigating the
effects of LIPUS on osteoblastgenesis in goldfish scales treated
with either COX-2 inhibitor or small interfering RNA for PTGS2 is
required.
Hsp90α, the product of HSP90AA1, belongs to the HSP
family and exhibits potent chaperone activity (64). This protein is induced by stressors,
including heat stress, and is mediated primarily by heat shock
transcription factor 1(65). In the
present study, expression of HSP90AA1 was significantly upregulated
in the goldfish scales treated with LIPUS, without any temperature
rise in the culture medium. This suggested that temperature had
little involvement in LIPUS-induced HSP90AA1 expression. Similarly,
LIPUS induces HSP90AA1 expression in zebrafish scales (31). Previous studies have also indicated
that LIPUS at 30 mW/cm2 for 15 or 30 min enhances
osteogenic differentiation by elevating HSP90AA1 levels in mouse
calvaria-derived osteoblasts (19)
and human adipose-derived stem cells (20).
Taken together, the present results provide insight
into the molecular mechanisms underlying LIPUS-induced osteoblast
activation. However, the present study did not conduct
morphological evaluations of the LIPUS-treated goldfish scales. The
biological and morphological roles of genes and their interactions
in LIPUS-treated goldfish scales require further investigation.
Supplementary Material
Confirmation of PCR products with
gene-specific primers. Agarose gel electrophoresis (2%) was
performed and PCR products of the expected size for each gene were
observed. L, DNA size ladder; 1, β-actin (210 bp); 2, tumor
necrosis factor receptor superfamily, member 11b (101 bp); 3, Wnt
family member 3A (129 bp); 4, protein kinase D1 (164 bp); 5,
prostaglandin endoperoxide synthase 2 (116 bp). bp, base pair.
Up-regulated genes 3 h after LIPUS
(low-intensity pulsed ultrasound) exposure.
Up-regulated genes 6 h after LIPUS
(low-intensity pulsed ultrasound) exposure.
Up-regulated genes 24 h after LIPUS
(low-intensity pulsed ultrasound) exposure.
Down-regulated genes 3 h after LIPUS
(low-intensity pulsed ultrasound) exposure.
Down-regulated genes 6 h after LIPUS
(low-intensity pulsed ultrasound) exposure.
Down-regulated genes 24 h after LIPUS
(low-intensity pulsed ultrasound) exposure.
Acknowledgements
The authors would like to thank Dr Hidetada Ohnishi
(Teijin Pharma, Ltd.) for lending ultrasound irradiating
system.
Funding
Funding: The present study was supported in part by Japan
Society for the Promotion of Science KAKENHI (grant nos.
JP17K01353, JP20K12619, JP23K11802 and JP23K10933).
Availability of data and materials
The data generated in the present study may be found
in the DNA Data Bank of Japan under accession number PRJDB17555 or
at the following URL: ddbj.nig.ac.jp/resource/bioproject/PRJDB17555.
Authors' contributions
YT, YF and NS designed the experiments and wrote the
manuscript. YT, KK, YF, TH, RN, MO, HH, JH and NS performed
experiments. YT, KK, YF and NS analyzed the data. YT and NS confirm
the authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
All animal experiments were approved by the Animal
Research Committee of Kanazawa University (Kanazawa, Japan
(approval no. 242-2023).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Barnett SB, Ter Haar GR, Ziskin MC, Rott
HD, Duck FA and Maeda K: International recommendations and
guidelines for the safe use of diagnostic ultrasound in medicine.
Ultrasound Med Biol. 26:355–366. 2000.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Snehota M, Vachutka J, Ter Haar G, Dolezal
L and Kolarova H: Therapeutic ultrasound experiments in vitro:
Review of factors influencing outcomes and reproducibility.
Ultrasonics. 107(106167)2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Padilla F, Puts R, Vico L and Raum K:
Stimulation of bone repair with ultrasound: A review of the
possible mechanic effects. Ultrasonics. 54:1125–1145.
2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Harrison A, Lin S, Pounder N and
Mikuni-Takagaki Y: Mode & mechanism of low intensity pulsed
ultrasound (LIPUS) in fracture repair. Ultrasonics. 70:45–52.
2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kennedy JE: High-intensity focused
ultrasound in the treatment of solid tumours. Nat Rev Cancer.
5:321–327. 2005.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Uchida T, Nakano M, Hongo S, Shoji S,
Nagata Y, Satoh T, Baba S, Usui Y and Terachi T: High-intensity
focused ultrasound therapy for prostate cancer. Int J Urol.
19:187–201. 2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Duarte LR: The stimulation of bone growth
by ultrasound. Arch Orthop Trauma Surg (1978). 101:153–159.
1983.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Azuma Y, Ito M, Harada Y, Takagi H, Ohta T
and Jingushi S: Low-intensity pulsed ultrasound accelerates rat
femoral fracture healing by acting on the various cellular
reactions in the fracture callus. J Bone Miner Res. 16:671–680.
2001.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Cheung WH, Chow SK, Sun MH, Qin L and
Leung KS: Low-intensity pulsed ultrasound accelerated callus
formation, angiogenesis and callus remodeling in osteoporotic
fracture healing. Ultrasound Med Biol. 37:231–238. 2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Naruse K, Sekiya H, Harada Y, Iwabuchi S,
Kozai Y, Kawamata R, Kashima I, Uchida K, Urabe K, Seto K, et al:
Prolonged endochondral bone healing in senescence is shortened by
low-intensity pulsed ultrasound in a manner dependent on COX-2.
Ultrasound Med Biol. 36:1098–1108. 2010.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Heckman JD, Ryaby JP, McCabe J, Frey JJ
and Kilcoyne RF: Acceleration of tibial fracture-healing by
non-invasive, low-intensity pulsed ultrasound. J Bone Joint Surg
Am. 76:26–34. 1994.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Kristiansen TK, Ryaby JP, McCabe J, Frey
JJ and Roe LR: Accelerated healing of distal radial fractures with
the use of specific, low-intensity ultrasound. A multicenter,
prospective, randomized, double-blind, placebo-controlled study. J
Bone Joint Surg Am. 79:961–973. 1997.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Tang CH, Yang RS, Huang TH, Lu DY, Chuang
WJ, Huang TF and Fu WM: Ultrasound stimulates cyclooxygenase-2
expression and increases bone formation through integrin, focal
adhesion kinase, phosphatidylinositol 3-kinase, and Akt pathway in
osteoblasts. Mol Pharmacol. 69:2047–2057. 2006.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Unsworth J, Kaneez S, Harris S, Ridgway J,
Fenwick S, Chenery D and Harrison A: Pulsed low intensity
ultrasound enhances mineralisation in preosteoblast cells.
Ultrasound Med Biol. 9:1468–1474. 2007.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Borsje MA, Ren Y, de Haan-Visser HW and
Kuijer R: Comparison of low-intensity pulsed ultrasound and pulsed
electromagnetic field treatments on OPG and RANKL expression in
human osteoblast-like cells. Angle Orthod. 80:498–503.
2010.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Kitamura K, Suzuki N, Sato Y, Nemoto T,
Ikegame M, Shimizu N, Kondo T, Furusawa Y, Wada S and Hattori A:
Osteoblast activity in the goldfish scale responds sensitively to
mechanical stress. Comp Biochem Physiol A Mol Integr Physiol.
156:357–363. 2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Costa V, Carina V, Fontana S, De Luca A,
Monteleone F, Pagani S, Sartori M, Setti S, Faldini C, Alessandro
R, et al: Osteogenic commitment and differentiation of human
mesenchymal stem cells by low-intensity pulsed ultrasound
stimulation. J Cell Physiol. 233:1558–1573. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chiu CY, Tsai TL, Vanderby R Jr, Bradica
G, Lou SL and Li WJ: Osteoblastogenesis of mesenchymal stem cells
in 3-D culture enhanced by low-intensity pulsed ultrasound through
soluble receptor activator of nuclear factor kappa B ligand.
Ultrasound Med Biol. 41:1842–1852. 2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Miyasaka M, Nakata H, Hao J, Kim YK,
Kasugai S and Kuroda S: Low-intensity pulsed ultrasound stimulation
enhances heat-shock protein 90 and mineralized nodule formation in
mouse calvaria-derived osteoblasts. Tissue Eng Part A.
21:2829–2839. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhang Z, Ma Y, Guo S, He Y, Bai G and
Zhang W: Low-intensity pulsed ultrasound stimulation facilitates in
vitro osteogenic differentiation of human adipose-derived stem
cells via up-regulation of heat shock protein (HSP)70, HSP90, and
bone morphogenetic protein (BMP) signaling pathway. Biosci Rep.
38(BSR20180087)2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhou J, Zhu Y, Ai D, Zhou M, Li H, Fu Y
and Song J: Low-intensity pulsed ultrasound regulates
osteoblast-osteoclast crosstalk via EphrinB2/EphB4 signaling for
orthodontic alveolar bone remodeling. Front Bioeng Biotechnol.
11(1192720)2023.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Florencio-Silva R, Sasso GR, Sasso-Cerri
E, Simões MJ and Cerri PS: Biology of bone tissue: Structure,
function, and factors that influence bone cells. Biomed Res Int.
2015(421746)2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Mikuni-Takagaki Y: Mechanical responses
and signal transduction pathways in stretched osteocytes. J Bone
Miner Metab. 17:57–60. 1999.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Klein-Nulend J, Bakker AD, Bacabac RG,
Vatsa A and Weinbaum S: Mechanosensation and transduction in
osteocytes. Bone. 54:182–190. 2013.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ma Q, Miri Z, Haugen HJ, Moghanian A and
Loca D: Significance of mechanical loading in bone fracture
healing, bone regeneration, and vascularization. J Tissue Eng.
14(20417314231172573)2023.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Bereiter-Hahn J and Zylberberg L:
Regeneration of teleost fish scale. Comp Biochem Physiol.
105A:625–641. 1993.
|
|
27
|
Suzuki N, Kitamura K, Omori K, Nemoto T,
Satoh Y, Tabata MJ, Ikegame M, Yamamoto T, Ijiri K, Furusawa Y, et
al: Response of osteoblasts and osteoclasts in regenerating scales
to gravity loading. Biol Sci Space. 23:211–217. 2009.
|
|
28
|
Hirayama J, Hattori A, Takahashi A,
Furusawa Y, Tabuchi Y, Shibata M, Nagamatsu A, Yano S, Maruyama Y,
Matsubara H, et al: Physiological consequences of space flight,
including abnormal bone metabolism, space radiation injury, and
circadian clock dysregulation: Implications of melatonin use and
regulation as a countermeasure. J Pineal Res.
74(e12834)2023.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Yamamoto T, Ikegame M, Hirayama J,
Kitamura KI, Tabuchi Y, Furusawa Y, Sekiguchi T, Endo M, Mishima H,
Seki A, et al: Expression of sclerostin in the regenerating scales
of goldfish and its increase under microgravity during space
flight. Biomed Res. 41:279–288. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ikegame M, Hattori A, Tabata MJ, Kitamura
KI, Tabuchi Y, Furusawa Y, Maruyama Y, Yamamoto T, Sekiguchi T,
Matsuoka R, et al: Melatonin is a potential drug for the prevention
of bone loss during space flight. J Pineal Res.
67(e12594)2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Suzuki N, Hanmoto T, Yano S, Furusawa Y,
Ikegame M, Tabuchi Y, Kondo T, Kitamura K, Endo M, Yamamoto T, et
al: Low-intensity pulsed ultrasound induces apoptosis in
osteoclasts: Fish scales are a suitable model for the analysis of
bone metabolism by ultrasound. Comp Biochem Physiol A Mol Integr
Physiol. 195:26–31. 2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Hanmoto T, Tabuchi Y, Ikegame M, Kondo T,
Kitamura KI, Endo M, Kobayashi I, Mishima H, Sekiguchi T, Urata M,
et al: Effects of low-intensity pulsed ultrasound on osteoclasts:
Analysis with goldfish scales as a model of bone. Biomed Res.
38:71–77. 2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Haas BJ, Papanicolaou A, Yassour M,
Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber
M, et al: De novo transcript sequence reconstruction from RNA-seq
using the Trinity platform for reference generation and analysis.
Nat Protoc. 8:1494–1512. 2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Furusawa Y, Yamamoto T, Hattori A, Suzuki
N, Hirayama J, Sekiguchi T and Tabuchi Y: De novo transcriptome
analysis and gene expression profiling of fish scales isolated from
Carassius auratus during space flight: Impact of melatonin on gene
expression in response to space radiation. Mol Med Rep.
22:2627–2636. 2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Percie du Sert N, Ahluwalia A, Alam S,
Avey MT, Baker M, Browne WJ, Clark A, Cuthill IC, Dirnagl U,
Emerson M, et al: Reporting animal research: Explanation and
elaboration for the ARRIVE guidelines 2.0. PLoS Biol.
18(e3000411)2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Iwabuchi S, Ito M, Hata J, Chikanishi T,
Azuma Y and Haro H: In vitro evaluation of low-intensity pulsed
ultrasound in herniated disc resorption. Biomaterials.
26:7104–7114. 2005.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Tabuchi Y, Hasegawa H, Suzuki N, Furusawa
Y, Hirano T, Nagaoka R, Hirayama J, Hoshi N and Mochizuki T:
Genetic response to low-intensity ultrasound on mouse ST2 bone
marrow stromal cells. Mol Med Rep. 23(173)2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Shen W, Le S, Li Y and Hu F: SeqKit: A
cross-platform and ultrafast toolkit for FASTA/Q file manipulation.
PLoS One. 11(e0163962)2016.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Bray NL, Pimentel H, Melsted P and Pachter
L: Near-optimal probabilistic RNA-seq quantification. Nat
Biotechnol. 34:525–527. 2016.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Robinson MD and Oshlack A: A scaling
normalization method for differential expression analysis of
RNA-seq data. Genome Biol. 11(R25)2010.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Tabuchi Y, Takasaki I, Doi T, Ishii Y,
Sakai H and Kondo T: Genetic networks responsive to sodium butyrate
in colonic epithelial cells. FEBS Lett. 580:3035–3041.
2006.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Larionov A, Krause A and Miller W: A
standard curve based method for relative real time PCR data
processing. BMC Bioinformatics. 6(62)2005.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Tabuchi Y, Ohta S, Arai Y, Kawahara M,
Ishibashi K, Sugiyama N, Horiuchi T, Furusawa M, Obinata M, Fuse H,
et al: Establishment and characterization of a colonic epithelial
cell line MCE301 from transgenic mice harboring
temperature-sensitive simian virus 40 large T-antigen gene. Cell
Struct Funct. 25:297–307. 2000.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Hanyu R, Wehbi VL, Hayata T, Moriya S,
Feinstein TN, Ezura Y, Nagao M, Saita Y, Hemmi H, Notomi T, et al:
Anabolic action of parathyroid hormone regulated by the
β2-adrenergic receptor. Proc Natl Acad Sci USA. 109:7433–7438.
2012.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Bollag WB, Choudhary V, Zhong Q, Ding KH,
Xu J, Elsayed R, Yu K, Su Y, Bailey LJ, Shi XM, et al: Deletion of
protein kinase D1 in osteoprogenitor cells results in decreased
osteogenesis in vitro and reduced bone mineral density in vivo. Mol
Cell Endocrinol. 461:22–31. 2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Bucay N, Sarosi I, Dunstan CR, Morony S,
Tarpley J, Capparelli C, Scully S, Tan HL, Xu W, Lacey DL, et al:
Osteoprotegerin-deficient mice develop early onset osteoporosis and
arterial calcification. Genes Dev. 12:1260–1268. 1998.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Greenblatt MB, Shim JH, Zou W, Sitara D,
Schweitzer M, Hu D, Lotinun S, Sano Y, Baron R, Park JM, et al: The
p38 MAPK pathway is essential for skeletogenesis and bone
homeostasis in mice. J Clin Invest. 120:2457–2473. 2010.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Jensen ED, Gopalakrishnan R and Westendorf
JJ: Bone morphogenic protein 2 activates protein kinase D to
regulate histone deacetylase 7 localization and repression of
Runx2. J Biol Chem. 284:2225–2234. 2009.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Yu H, de Vos P and Ren Y: Overexpression
of osteoprotegerin promotes preosteoblast differentiation to mature
osteoblasts. Angle Orthod. 81:100–106. 2011.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Si W, Kang Q, Luu HH, Park JK, Luo Q, Song
WX, Jiang W, Luo X, Li X, Yin H, et al: CCN1/Cyr61 is regulated by
the canonical Wnt signal and plays an important role in
Wnt3A-induced osteoblast differentiation of mesenchymal stem cells.
Mol Cell Biol. 26:2955–2964. 2006.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Zhang X, Schwarz EM, Young DA, Puzas JE,
Rosier RN and O'Keefe RJ: Cyclooxygenase-2 regulates mesenchymal
cell differentiation into the osteoblast lineage and is critically
involved in bone repair. J Clin Invest. 109:1405–1415.
2002.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Ford JJ, Yeh LC, Schmidgal EC, Thompson
JF, Adamo ML and Lee JC: Protein kinase D1 is essential for bone
acquisition during pubertal growth. Endocrinology. 154:4182–4191.
2013.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Yang B, Li S, Chen Z, Feng F, He L, Liu B,
He T, Wang X, Chen R, Chen Z, et al: Amyloid β peptide promotes
bone formation by regulating Wnt/β-catenin signaling and the
OPG/RANKL/RANK system. FASEB J. 34:3583–3593. 2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Lacey DL, Boyle WJ, Simonet WS, Kostenuik
PJ, Dougall WC, Sullivan JK, San Martin J and Dansey R: Bench to
bedside: elucidation of the OPG-RANK-RANKL pathway and the
development of denosumab. Nat Rev Drug Discov. 11:401–419.
2012.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Hoter A, El-Sabban ME and Naim HY: The
HSP90 family: Structure, regulation, function, and implications in
health and disease. Int J Mol Sci. 19(2560)2018.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Palumbo S and Li WJ: Osteoprotegerin
enhances osteogenesis of human mesenchymal stem cells. Tissue Eng
Part A. 19:2176–2187. 2013.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Sena K, Leven RM, Mazhar K, Sumner DR and
Virdi AS: Early gene response to low-intensity pulsed ultrasound in
rat osteoblastic cells. Ultrasound Med Biol. 31:703–708.
2005.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Tabuchi Y, Hasegawa H, Suzuki N, Furusawa
Y, Hirano T, Nagaoka R, Takeuchi SI, Shiiba M and Mochizuki T:
Low-intensity pulsed ultrasound promotes the expression of
immediate-early genes in mouse ST2 bone marrow stromal cells. J Med
Ultrason (2001). 47:193–201. 2020.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Veronick JA, Assanah F, Piscopo N, Kutes
Y, Vyas V, Nair LS, Huey BD and Khan Y: Mechanically loading
cell/hydrogel constructs with low-intensity pulsed ultrasound for
bone repair. Tissue Eng Part A. 24:254–263. 2018.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Choudhary S, Halbout P, Alander C, Raisz L
and Pilbeam C: Strontium ranelate promotes osteoblastic
differentiation and mineralization of murine bone marrow stromal
cells: Involvement of prostaglandins. J Bone Miner Res.
22:1002–1010. 2007.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Allen HL, Wase A and Bear WT: Indomethacin
and aspirin: Effect of nonsteroidal anti-inflammatory agents on the
rate of fracture repair in the rat. Acta Orthop Scand. 51:595–600.
1980.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Elmstedt E, Lindholm TS, Nilsson OS and
Törnkvist H: Effect of ibuprofen on heterotopic ossification after
hip replacement. Acta Orthop Scand. 56:25–27. 1985.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Omori K, Wada S, Maruyama Y, Hattori A,
Kitamura K, Sato Y, Nara M, Funahashi H, Yachiguchi K, Hayakawa K,
et al: Prostaglandin E2 increases both osteoblastic and
osteoclastic activities in the scales of goldfish and participates
in the calcium metabolism in goldfish. Zoolog Sci. 29:499–504.
2012.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Zuehlke AD, Beebe K, Neckers L and Prince
T: Regulation and function of the human HSP90AA1 gene. Gene.
570:8–16. 2015.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Akerfelt M, Morimoto RI and Sistonen L:
Heat shock factors: Integrators of cell stress, development and
lifespan. Nat Rev Mol Cell Biol. 11:545–555. 2010.PubMed/NCBI View Article : Google Scholar
|