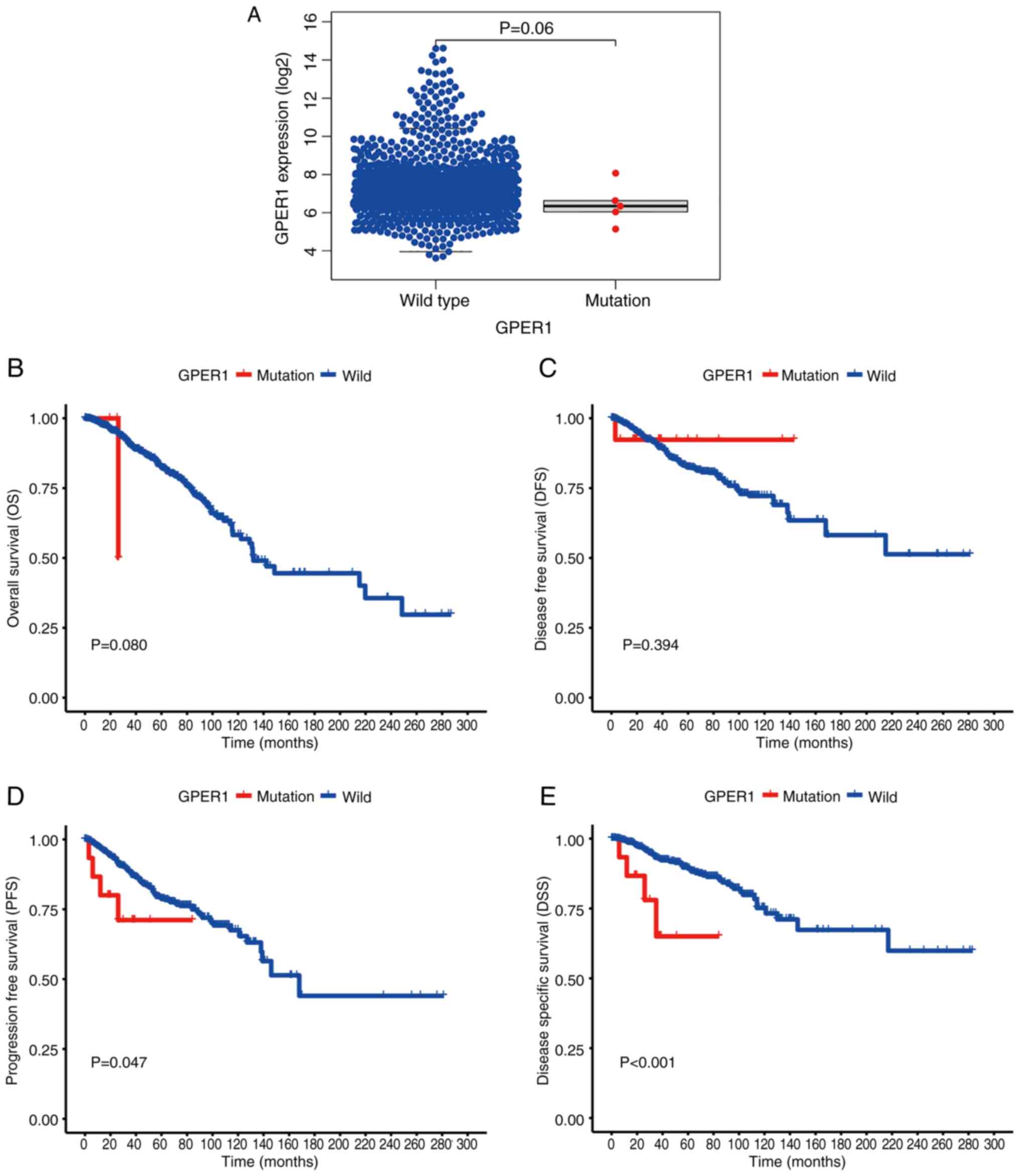
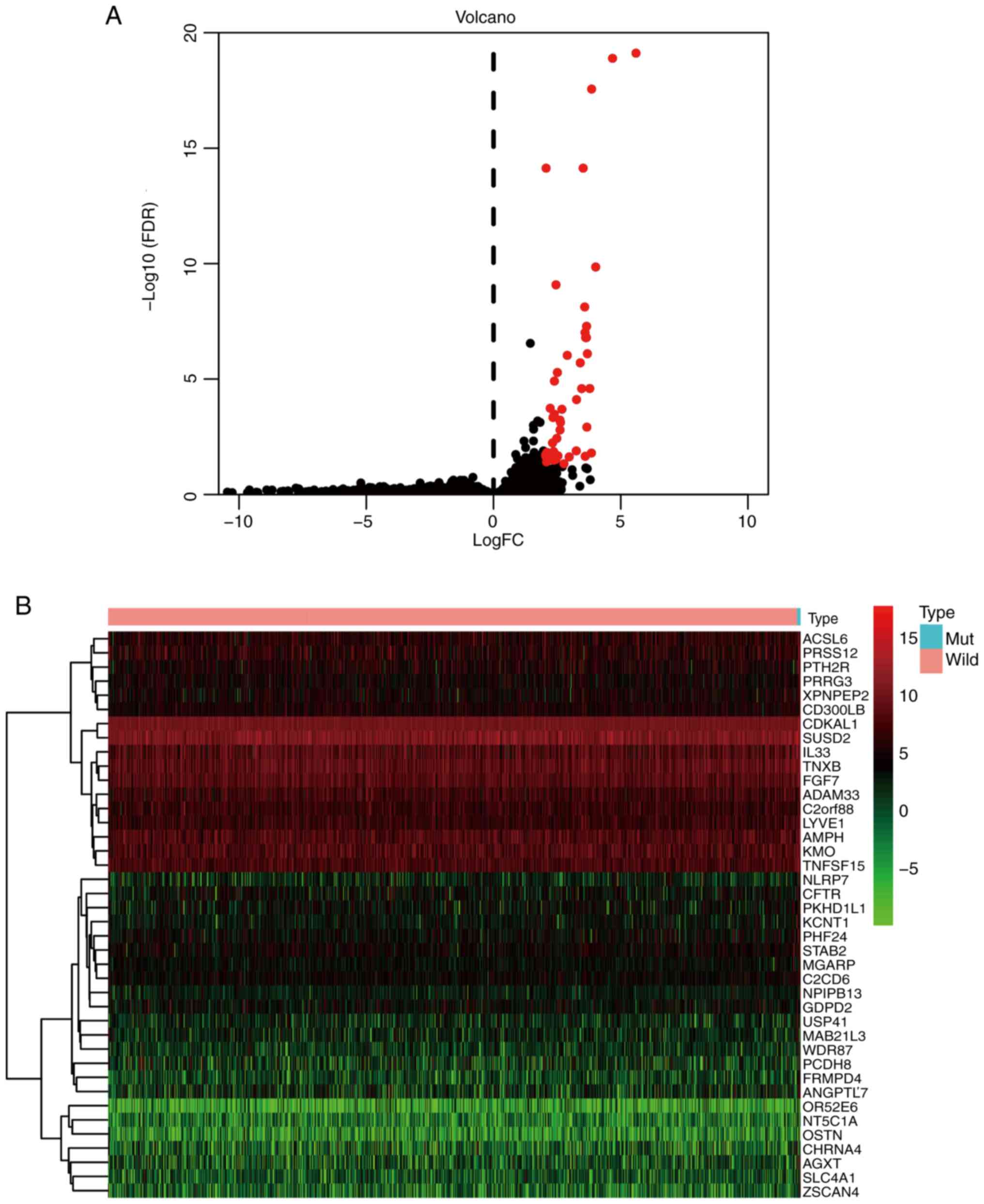
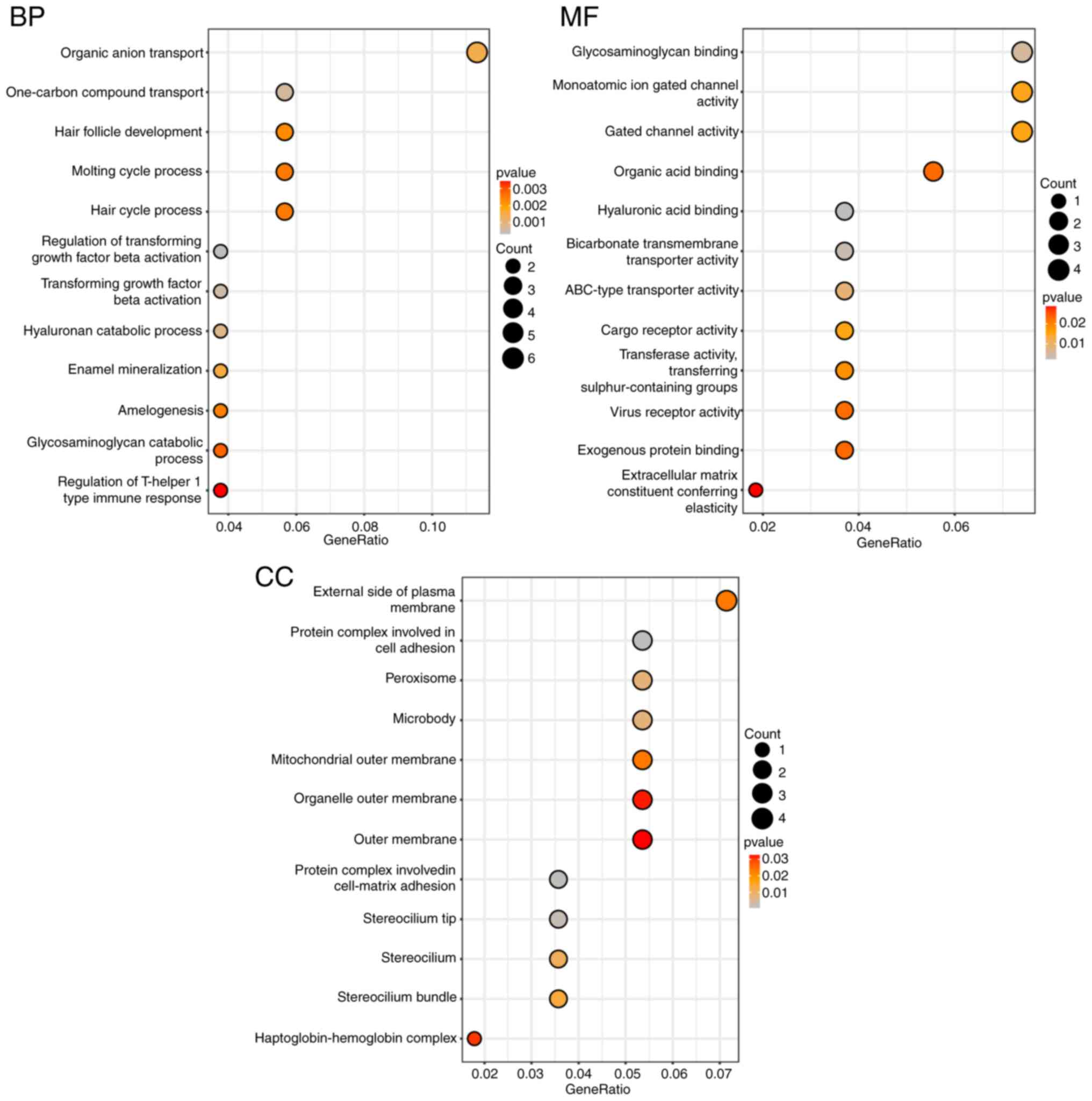
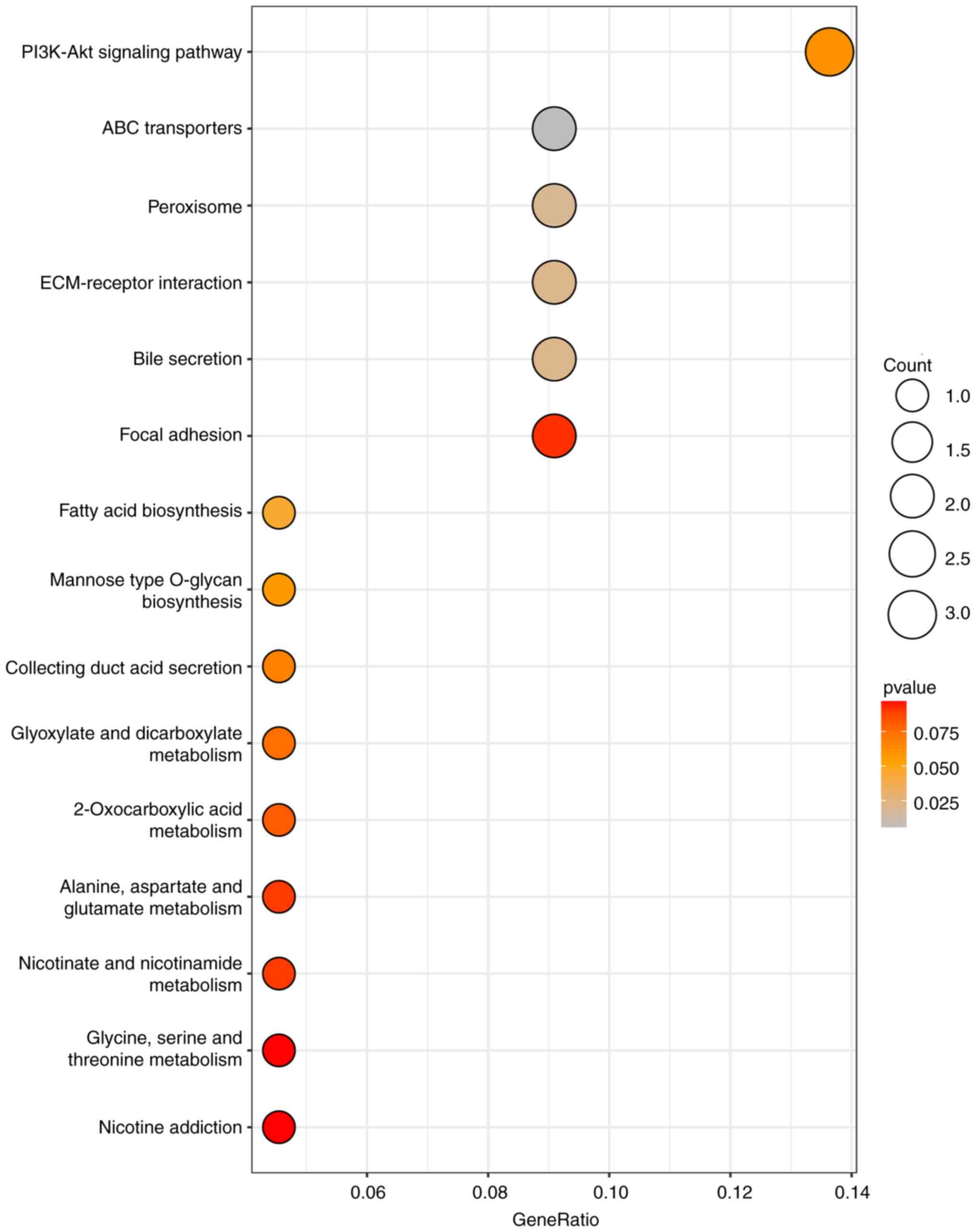
|
1
|
Ide Y, Horii R, Osako T, Ogura K, Yoshida
R, Iwase T and Akiyama F: Clinicopathological significance of
invasive micropapillary carcinoma component in invasive breast
carcinoma. Pathol Int. 61:731–736. 2011.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lwin ZM, Guo C, Salim A, Yip GW, Chew FT,
Nan J, Thike AA, Tan PH and Bay BH: Clinicopathological
significance of calreticulin in breast invasive ductal carcinoma.
Mod Pathol. 23:1559–1566. 2010.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Siegel RL, Giaquinto AN and Jemal A:
Cancer statistics, 2024. CA Cancer J Clin. 74:12–49.
2024.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Mei L, Wang K and Gu Y: Improved Fuzzy
C-means clustering algorithm-based dynamic contrast-enhanced
magnetic resonance imaging features in the diagnosis of invasive
breast carcinoma before and after menopause. Comput Math Methods
Med. 2022(2917844)2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Rakha EA, Martin S, Lee AHS, Morgan D,
Pharoah PD, Hodi Z, Macmillan D and Ellis IO: The prognostic
significance of lymphovascular invasion in invasive breast
carcinoma. Cancer. 118:3670–3680. 2012.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Yang F, Xie HY, Yang LF, Zhang L, Zhang
FL, Liu HY, Li DQ and Shao ZM: Stabilization of MORC2 by estrogen
and antiestrogens through GPER1-PRKACA-CMA pathway contributes to
estrogen-induced proliferation and endocrine resistance of breast
cancer cells. Autophagy. 16:1061–1076. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Saroha HS, Kumar Guddeti R, Jacob JP,
Kumar Pulukuri K, Karyala P and Pakala SB: MORC2/β-catenin
signaling axis promotes proliferation and migration of breast
cancer cells. Med Oncol. 39(135)2022.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Thomas L, Chutani N, R K, Nair AS, Yellapu
NK, Karyala P and Pakala SB: Microrchidia 2/histone deacetylase 1
complex regulates E-cadherin gene expression and function. Biochem
J. 480:1675–1691. 2023.PubMed/NCBI View Article : Google Scholar
|
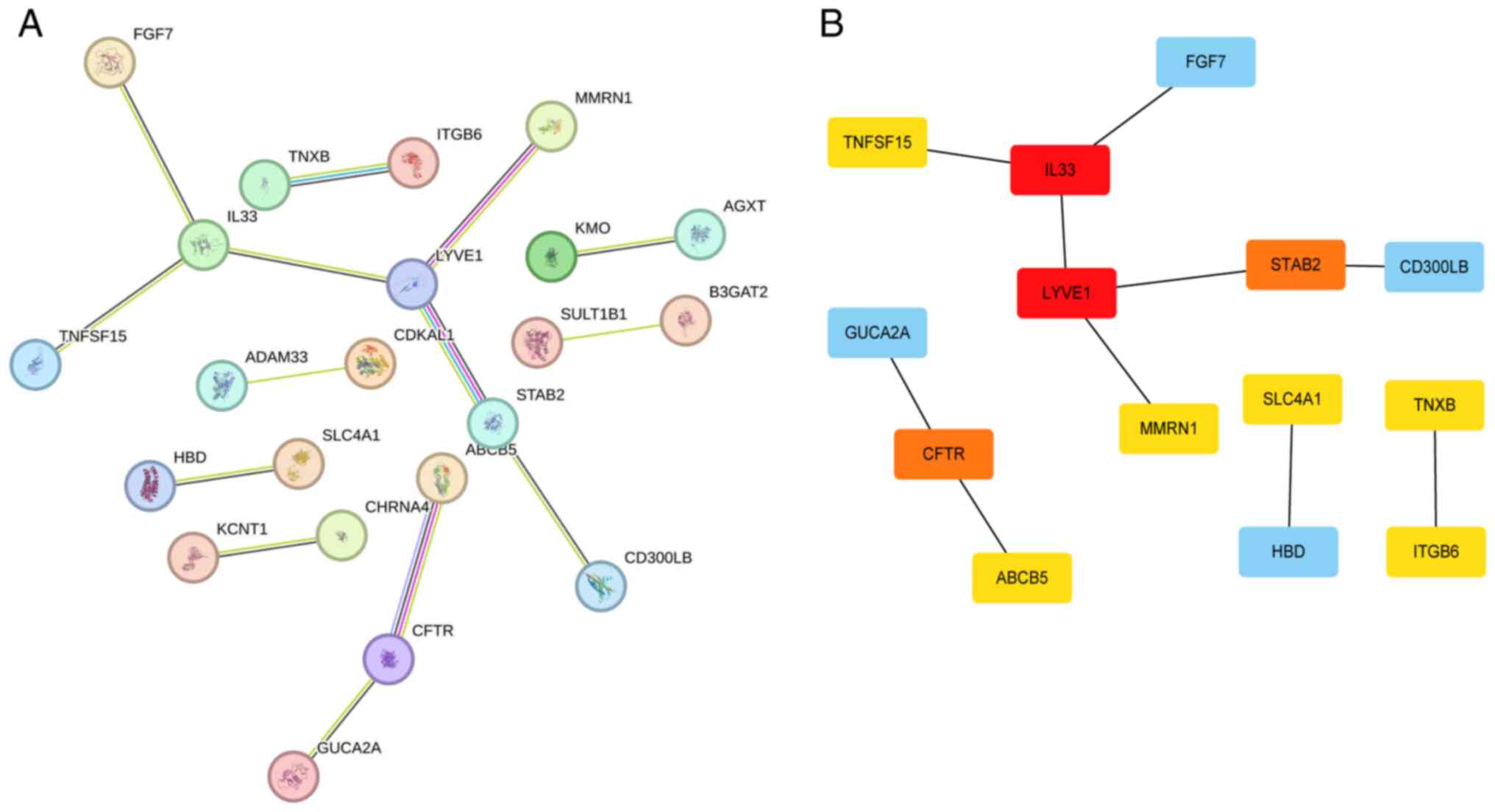
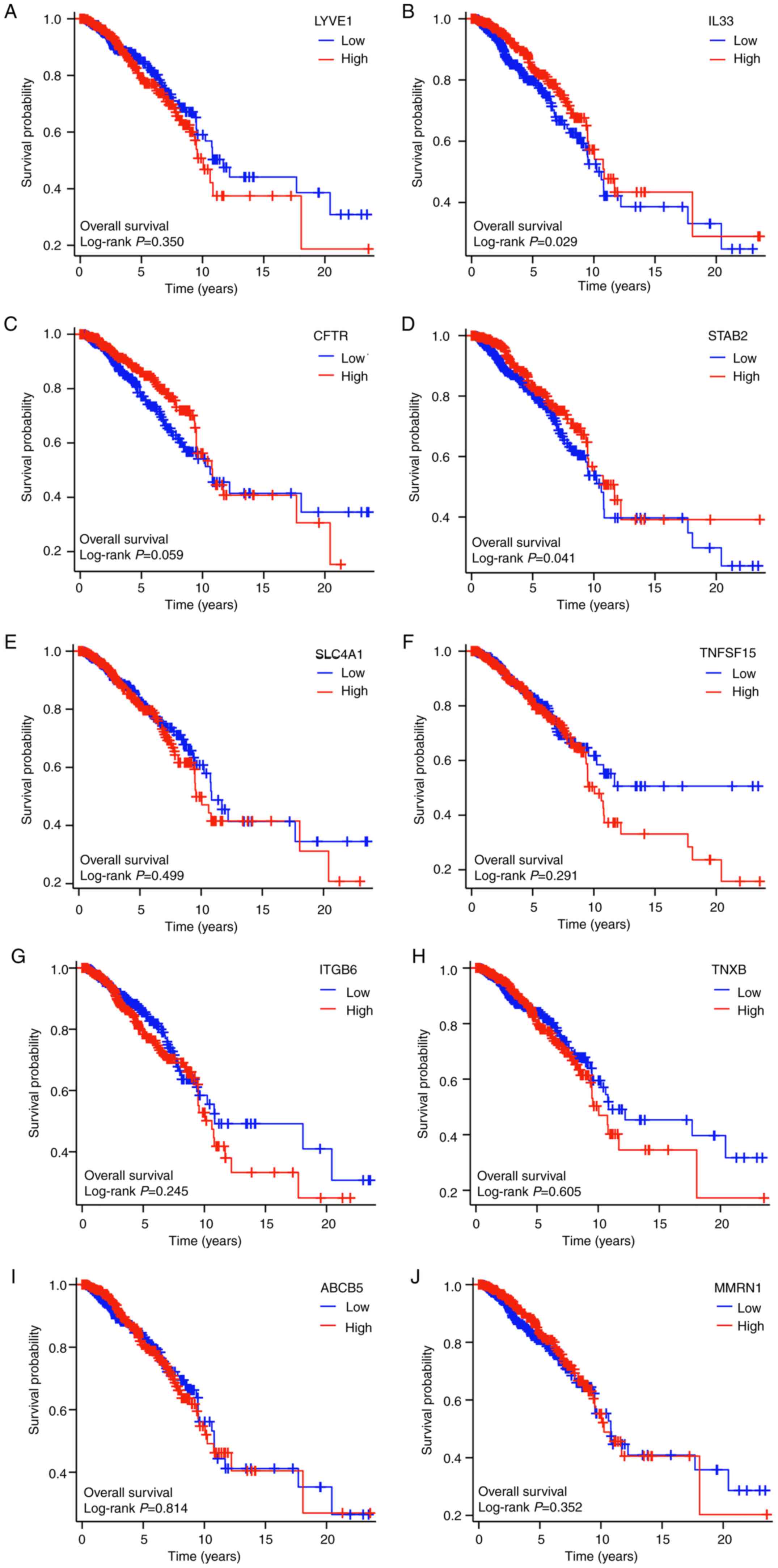
|
9
|
Schmitz V, Bauerschmitz G, Gallwas J and
Gründker C: Suppression of G Protein-coupled estrogen Receptor 1
(GPER1) Enhances the Anti-invasive efficacy of selective ERβ
Agonists. Anticancer Res. 42:5187–5194. 2022.PubMed/NCBI View Article : Google Scholar
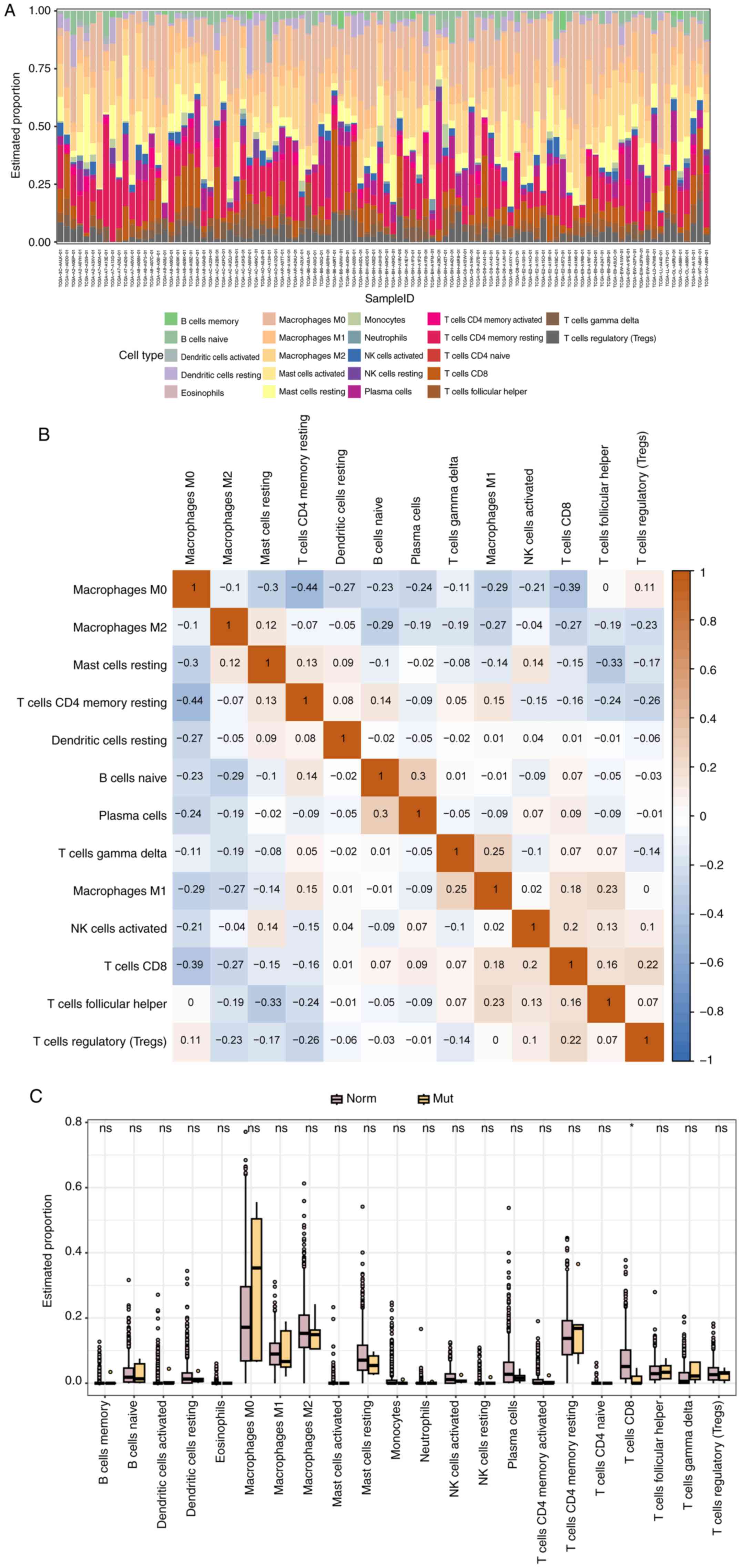
|
|
10
|
Li ZH, Liu C, Liu QH, Wang J, Wang Y, Wang
YF, Deng SJ and Li DB: Cytoplasmic expression of G protein-coupled
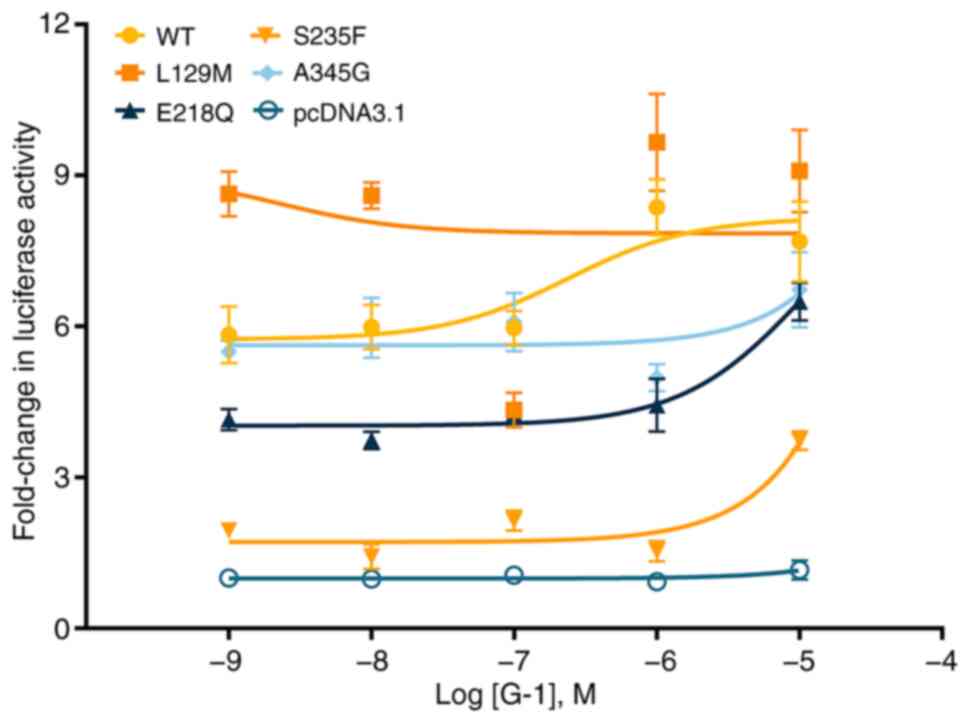
estrogen receptor 1 correlates with poor postoperative prognosis in
non-small cell lung cancer. J Thorac Dis. 14:1466–1477.
2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Shen Y, Li C, Zhou L and Huang JA: G
protein-coupled oestrogen receptor promotes cell growth of
non-small cell lung cancer cells via YAP1/QKI/circNOTCH1/m6A
methylated NOTCH1 signalling. J Cell Mol Med. 25:284–296.
2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yang F and Shao ZM: Double-edged role of G
protein-coupled estrogen receptor 1 in breast cancer prognosis: An
analysis of 167 breast cancer samples and online data sets. Onco
Targets Ther. 9:6407–6415. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Srivastava DP and Evans PD: G-protein
oestrogen receptor 1: Trials and tribulations of a membrane
oestrogen receptor. J Neuroendocrinol. 25:1219–1230.
2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Evans NJ, Bayliss AL, Reale V and Evans
PD: Characterisation of signalling by the Endogenous GPER1 (GPR30)
Receptor in an Embryonic Mouse Hippocampal Cell Line (mHippoE-18).
PLoS One. 11(e0152138)2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yu M, Xu L, Lei B, Sun S and Yang Y:
Tetrachlorobisphenol A and bisphenol AF induced cell migration by
activating PI3K/Akt signaling pathway via G protein-coupled
estrogen receptor 1 in SK-BR-3 cells. Environ Toxicol. 38:126–135.
2023.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wnuk A, Przepiórska K, Pietrzak BA and
Kajta M: Emerging evidence on membrane estrogen receptors as novel
therapeutic targets for central nervous system pathologies. Int J
Mol Sci. 24(4043)2023.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Morelli E, Hunter ZR, Fulciniti M, Gullà
A, Perrotta ID, Zuccalà V, Federico C, Juli G, Manzoni M, Ronchetti
D, et al: Therapeutic activation of G protein-coupled estrogen
receptor 1 in Waldenström Macroglobulinemia. Exp Hematol Oncol.
11(54)2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Gao N, Liang T, Yuan Y, Xiao X, Zhao Y,
Guo Y, Li M and Pu X: Exploring the mechanism of F282L
mutation-caused constitutive activity of GPCR by a computational
study. Phys Chem Chem Phys. 18:29412–29422. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Tao YX: Molecular mechanisms of the neural
melanocortin receptor dysfunction in severe early onset obesity.
Mol Cell Endocrinol. 239:1–14. 2005.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Pupo M, Bodmer A, Berto M, Maggiolini M,
Dietrich PY and Picard D: A genetic polymorphism repurposes the
G-protein coupled and membrane-associated estrogen receptor GPER to
a transcription factor-like molecule promoting paracrine signaling
between stroma and breast carcinoma cells. Oncotarget.
8:46728–46744. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Tutzauer J, Sjöström M, Bendahl PO, Rydén
L, Fernö M, Fredrik Leeb-Lundberg LM and Alkner S: Plasma membrane
expression of G protein-coupled estrogen receptor (GPER)/G
protein-coupled receptor 30 (GPR30) is associated with worse
outcome in metachronous contralateral breast cancer. PLoS One.
15(e0231786)2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
R Core Team: R Core Team 2023 R: A
language and environment for statistical computing. R foundation
for statistical computing. https://www.R-project.org/. R Foundation for
Statistical Computing, 2023.
|
|
23
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140.
2009.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software Environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wu L, Yu H, Mo H, Lan X, Pan C, Wang L,
Zhao H, Zhou J and Li Y: Functional characterization of
melanocortin-3 receptor in a hibernating cavefish onychostoma
macrolepis. Animals. 12(38)2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Vesuna F, Winnard P and Raman V: Enhanced
green fluorescent protein as an alternative control reporter to
Renilla luciferase. Anal Biochem. 342:345–347. 2005.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Han J, Xiao N, Yang W, Luo S and Zhao J,
Qiang Y, Chaudhary S and Zhao J: MS-ResNet: Disease-specific
survival prediction using longitudinal CT images and clinical data.
Int J Comput Assist Radiol Surg. 17:1049–1057. 2022.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Li Z, Ding B, Xu J, Mao K, Zhang P and Xue
Q: Relevance of STK11 mutations regarding immune cell infiltration,
drug sensitivity, and cellular processes in lung adenocarcinoma.
Front Oncol. 10(580027)2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Sun J, Li S, Wang F, Fan C and Wang J:
Identification of key pathways and genes in pten mutation prostate
cancer by bioinformatics analysis. BMC Med Genet.
20(191)2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Fan C, Zhao C, Shugen Li FW and Wang J:
Significance of PTEN mutation in cellular process, prognosis, and
drug selection in clear cell renal cell carcinoma. Front Oncol.
9(357)2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ono M, Baden A, Okudaira H, Kobayashi M,
Kawai K, Oka S and Yoshimura H: Assessment of amino acid/drug
transporters for renal transport of [18F]fluciclovine
(Anti-[18F]FACBC) in vitro. Int J Mol Sci. 17(1730)2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Becchetti A, Munaron L and Arcangeli A:
The role of ion channels and transporters in cell proliferation and
cancer. Front Physiol. 4(312)2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhou F, Hong M and You G: Regulation of
human organic anion transporter 4 by progesterone and protein
kinase C in human placental BeWo cells. Am J Physiol Endocrinol
Metab. 293:E57–E61. 2007.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Xu W, Tanaka K, Sun AQ and You G:
Functional role of the C terminus of human organic anion
transporter hOAT. J Biol Chem. 281:31178–31183. 2006.PubMed/NCBI View Article : Google Scholar
|
|
36
|
You G: Towards an understanding of organic
anion transporters: Structure-function relationships. Med Res Rev.
24:762–774. 2004.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Baycin-Hizal D, Gottschalk A, Jacobson E,
Mai S, Wolozny D, Zhang H, Krag SS and Betenbaugh MJ: Physiologic
and pathophysiologic consequences of altered sialylation and
glycosylation on Ion channel function. Biochem Biophys Res Commun.
453:243–253. 2014.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zhang G, Yang H, Liang H, Yang J, Shi J,
McFarland K, Chen Y and Cui J: A charged residue in S4 regulates
coupling among the activation gate, voltage, and Ca2+ sensors in BK
channels. J Neurosci. 34:12280–12288. 2014.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Thompson AN, Posson DJ, Parsa PV and
Nimigean CM: Molecular mechanism of pH sensing in KcsA potassium
channels. Proc Natl Acad Sci USA. 105:6900–6905. 2008.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Yang R, Zhou Y, Du C and Wu Y:
Bioinformatics analysis of differentially expressed genes in tumor
and paracancerous tissues of patients with lung adenocarcinoma. J
Thorac Dis. 12:7355–7364. 2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Fan Z, Liu Y, Liu X, Nian W, Huang X, Yang
Q, Hou S and Chen F: Phosphorylation of AKT by lysyl oxidase-like 2
activates the PI3K/AKT signaling pathway to promote proliferation,
invasion and metastasis in esophageal squamous carcinoma. Clin
Transl Oncol. 25:2487–2498. 2023.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Chi M, Liu J, Mei C, Shi Y, Liu N, Jiang
X, Liu C, Xue N, Hong H, Xie J, et al: TEAD4 functions as a
prognostic biomarker and triggers EMT via PI3K/AKT pathway in
bladder cancer. J Exp Clin Cancer Res. 41(175)2022.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Fadhal E: A comprehensive analysis of the
PI3K/AKT pathway: Unveiling key proteins and therapeutic targets
for cancer treatment. Cancer Inform.
22(11769351231194273)2023.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Ahmad I, Hoque M, Alam SSM, Zughaibi TA
and Tabrez S: Curcumin and plumbagin synergistically target the
PI3K/Akt/mTOR pathway: A prospective role in cancer treatment. Int
J Mol Sci. 24(6651)2023.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Shi X, Wang J, Lei Y, Cong C, Tan D and
Zhou X: Research progress on the PI3K/AKT signaling pathway in
gynecological cancer (Review). Mol Med Rep. 19:4529–4535.
2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Jin T, Li D, Yang T, Liu F, Kong J and
Zhou Y: PTPN1 promotes the progression of glioma by activating the
MAPK/ERK and PI3K/AKT pathways and is associated with poor patient
survival. Oncol Rep. 42:717–725. 2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Jiang AG, Yu H and Huang JA: Expression
and clinical significance of the phosphatidylinositol
3-kinase/protein kinase B signal transduction pathway in non-small
cell lung carcinoma. Oncol Lett. 8:601–607. 2014.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Li X, Sun H, Hou Y and Jin W:
Comprehensive combined proteomics and genomics analysis identifies
prognostic related transcription factors in breast cancer and
explores the role of dmap1 in breast cancer. J Pers Med.
11(1068)2021.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Andersen V, Vogel LK, Kopp TI, Sæbø M,
Nonboe AW, Hamfjord J, Kure EH and Vogel U: High ABCC2 and low
ABCG2 gene expression are early events in the colorectal
adenoma-carcinoma sequence. PLoS One. 10(e0119255)2015.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Yamada A, Ishikawa T, Ota I, Kimura M,
Shimizu D, Tanabe M, Chishima T, Sasaki T, Ichikawa Y, Morita S, et
al: High expression of ATP-binding cassette transporter ABCC11 in
breast tumors is associated with aggressive subtypes and low
disease-free survival. Breast Cancer Res Treat. 137:773–782.
2013.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Sakil HAM, Stantic M, Wolfsberger J, Brage
SE, Hansson J and Wilhelm MT: ΔNp73 regulates the expression of the
multidrug-resistance genes ABCB1 and ABCB5 in breast cancer and
melanoma cells-a short report. Cell Oncol. 40:631–638.
2017.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Yuan Y, Xiang Z, Xia Y, Xie J, Jiang X and
Lu Z: The role of ATP binding cassette (ABC) transporters in breast
cancer: Evaluating prognosis, predicting immunity, and guiding
treatment. Channels (Austin). 17(2273247)2023.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Modi A, Roy D, Sharma S, Vishnoi JR,
Pareek P, Elhence P, Sharma P and Purohit P: ABC transporters in
breast cancer: Their roles in multidrug resistance and beyond. J
Drug Target. 30:927–947. 2022.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Zhao B, Xin Z, Ren P and Wu H: The role of
PPARs in breast cancer. Cells. 12(130)2023.
|
|
55
|
Nordgren M and Fransen M: Peroxisomal
metabolism and oxidative stress. Biochimie. 98:56–62.
2014.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Zhang J, Tripathi DN, Jing J, Alexander A,
Kim J, Powell RT, Dere R, Tait-Mulder J, Lee JH, Paull TT, et al:
ATM functions at the peroxisome to induce pexophagy in response to
ROS. Nat Cell Biol. 17:1259–1269. 2015.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Brandão-Costa RM, Helal-Neto E, Vieira AM,
Barcellos-De-souza P, Morgado-Diaz J and Barja-Fidalgo C:
Extracellular matrix derived from high metastatic human breast
cancer triggers epithelial-mesenchymal transition in epithelial
breast cancer cells through αvβ3 integrin. Int J Mol Sci.
21(2995)2020.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Yu TY, Zhang G, Chai XX, Ren L, Yin DC and
Zhang CY: Recent progress on the effect of extracellular matrix on
occurrence and progression of breast cancer. Life Sci.
332(122084)2023.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Wu JZ, Yang TJ, Lu P and Ma W: Analysis of
signaling pathways in recurrent breast cancer. Genet Mol Res.
13:10097–10104. 2014.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Huang Z, Xu E, Ma X, Wang Y, Zhu J, Zhu K,
Hu J and Zhang C: Low NT5DC2 expression predicts favorable
prognosis and suppresses soft tissue sarcoma progression via
ECM-receptor interaction pathway. Transl Oncol.
44(101937)2024.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Tang W, Putluri V, Ambati CR, Dorsey TH,
Putluri N and Ambs S: Liver-And Microbiome-derived bile acids
accumulate in human breast tumors and inhibit growth and improve
patient survival. Clin Cancer Res. 25:5972–5983. 2019.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Scott P, Anderson K, Singhania M and
Cormier R: Cystic fibrosis, CFTR, and colorectal cancer. Int J Mol
Sci. 21(2891)2020.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Wang S, Yu ZH and Chai KQ: Identification
of CFTR as a novel key gene in chromophobe renal cell carcinoma
through bioinformatics analysis. Oncol Lett. 18:1767–1774.
2019.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Wei RQ, Zhang WM, Liang Z, Piao C and Zhu
G: Identification of signal pathways and hub genes of pulmonary
arterial hypertension by bioinformatic analysis. Can Respir J.
2022(1394088)2022.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Kudo-Saito C, Miyamoto T, Imazeki H, Shoji
H, Aoki K and Boku N: IL33 is a key driver of treatment resistance
of cancer. Cancer Res. 80:1981–1990. 2021.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Fang M, Li Y, Huang K, Qi S, Zhang J,
Zgodzinski W, Majewski M, Wallner G, Gozdz S, Macek P, et al: IL33
promotes colon cancer cell stemness via JNK activation and
macrophage recruitment. Cancer Res. 77:2735–2745. 2017.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Twarda-Clapa A, Labuzek B, Krzemien D,
Musielak B, Grudnik P, Dubin G and Holak TA: Crystal structure of
the FAS1 domain of the hyaluronic acid receptor stabilin-2. Acta
Crystallogr D Struct Biol. 74:695–701. 2018.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Hirose Y, Saijou E, Sugano Y, Takeshita F,
Nishimura S, Nonaka H, Chen YR, Sekine K, Kido T, Nakamura T, et
al: Inhibition of Stabilin-2 elevates circulating hyaluronic acid
levels and prevents tumor metastasis. Proc Natl Acad Sci USA.
109:4263–4268. 2012.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Han MW, Lee JC, Park SY, Kim YM, Cho KJ,
Kim SW, Lee M, Nam SY, Kim IS and Kim SY: Homotypic interaction of
stabilin-2 plays a critical role in lymph node metastasis of tongue
cancer. Anticancer Res. 36:6611–6618. 2016.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Eissmann MF, Dijkstra C, Jarnicki A,
Phesse T, Brunnberg J, Poh AR, Etemadi N, Tsantikos E, Thiem S,
Huntington ND, et al: IL-33-mediated mast cell activation promotes
gastric cancer through macrophage mobilization. Nat Commun.
10(2735)2019.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Eissmann MF, Buchert M and Ernst M: IL33
and mast cells-the key regulators of immune responses in
gastrointestinal cancers? Front Immunol. 11(1389)2020.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Yoon HH, Orrock JM, Foster NR, Sargent DJ,
Smyrk TC and Sinicrope FA: Prognostic impact of FoxP3+ regulatory T
cells in relation to CD8+ T lymphocyte density in human colon
carcinomas. PLoS One. 7(e42274)2012.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Duhen T, Duhen R, Montler R, Moses J,
Moudgil T, de Miranda NF, Goodall CP, Blair TC, Fox BA, McDermott
JE, et al: Co-expression of CD39 and CD103 identifies
tumor-reactive CD8 T cells in human solid tumors. Nat Commun.
9(2724)2018.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Liu Z, Zhou Q, Wang Z, Zhang H, Zeng H,
Huang Q, Chen Y, Jiang W, Lin Z, Qu Y, et al: Intratumoral
TIGIT+CD8+T-cell infiltration determines poor prognosis and immune
evasion in patients with muscle-invasive bladder cancer. J
Immunother Cancer. 8(e000978)2020.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Ma L, Sun L, Zhao K, Dong Z, Huang Z and
Meng X: The prognostic value of TCF1+CD8+T in primary small cell
carcinoma of the esophagus. Cancer Sci. 112:4968–4976.
2021.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Yang B, Deng B, Jiao XD, Qin BD, Lu Y,
Zhang W, Guo Y, Chen S, Li D, Li B, et al: Low-dose anti-VEGFR2
therapy promotes anti-tumor immunity in lung adenocarcinoma by
down-regulating the expression of layilin on tumor-infiltrating
CD8+T cells. Cell Oncol (Dordr). 45:1297–1309. 2022.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Karkeni E, Morin SO, Bou Tayeh B, Goubard
A, Josselin E, Castellano R, Fauriat C, Guittard G, Olive D and
Nunès JA: Vitamin D controls tumor growth and CD8+ T Cell
infiltration in breast cancer. Front Immunol.
10(1307)2019.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Wang X, Jespers W, de Waal JJ, Wolff KAN,
van Uden L, IJzerman AP, van Westen GJP and Heitman LH:
Cancer-related somatic mutations alter adenosine A1 receptor
pharmacology-A focus on mutations in the loops and C-terminus.
FASEB J. 36(e22358)2022.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Dietzsch AN, Al-Hasani H, Altschmied J,
Bottermann K, Brendler J, Haendeler J, Horn S, Kaczmarek I, Körner
A, Krause K, et al: Dysfunction of the adhesion G protein-coupled
receptor latrophilin 1 (ADGRL1/LPHN1) increases the risk of
obesity. Signal Transduct Target Ther. 9(103)2024.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Zhang H, Kong Q, Wang J, Jiang Y and Hua
H: Complex roles of cAMP-PKA-CREB signaling in cancer. Exp Hematol
Oncol. 9(32)2020.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Rao R, Salloum R, Xin M and Lu QR: The G
protein Gαs acts as a tumor suppressor in sonic hedgehog
signaling-driven tumorigenesis. Cell Cycle. 15:1325–1330.
2016.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Li Y, Zhou X, Liu J, Yin Y, Yuan X, Yang
R, Wang Q, Ji J and He Q: Differentially expressed genes and key
molecules of BRCA1/2-mutant breast cancer: Evidence from
bioinformatics analyses. PeerJ. 8(e8403)2020.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Zhu F, Huang R, Li J, Liao X, Huang Y and
Lai Y: Identification of key genes and pathways associated with
RUNX1 mutations in acute myeloid leukemia using bioinformatics
analysis. Medical Science Monitor. 24:7100–7108. 2018.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Chen S, Chen Y, Lu J, Yuan D, He L, Tan H
and Xu L: Bioinformatics analysis identifies key genes and pathways
in acute myeloid leukemia associated with DNMT3A mutation. Biomed
Res Int. 2020(9321630)2020.PubMed/NCBI View Article : Google Scholar
|