Introduction
Breast invasive carcinoma (BIC) refers to a type of
breast cancer where cancer cells invade the surrounding breast
tissue. It encompasses various subtypes such as invasive ductal
carcinoma (IDC) and invasive lobular carcinoma (ILC). IDC, the most
common type of breast cancer, originates in the milk ducts and
invades nearby breast tissue. On the other hand, ILC is
characterized by a single-cell infiltration pattern and loss of
cell cohesion (1,2). Studying BIC holds significant clinical
importance, as breast cancer not only stands as the most commonly
diagnosed malignant neoplasm but also ranks as the second leading
cause of cancer-related mortality among women worldwide (3). Among breast cancers, BIC accounts for
~80% of cases (4). BIC,
particularly IDC, is associated with malignant cells breaching
basement membranes. This infiltration into surrounding breast
tissues often leads to a poorer prognosis (2). Additionally, BIC can present with
unique characteristics such as lymphovascular invasion, nodal
metastasis and lymphatic invasion, which can affect disease
progression and treatment outcomes (1,5).
G protein-coupled estrogen receptor 1 (GPER1) plays
a pivotal role in breast cancers, including BIC. Research conducted
by Yang et al (6)
underscored the involvement of GPER1 activation in estrogen-induced
proliferation and endocrine resistance in breast cancer cells.
Furthermore, studies have identified that GPER1 stabilizes MORC2
via the PRKACA-CMA pathway, leading to amplified proliferation of
breast cancer cells (6-8).
Additionally, GPER1 has been linked to the metastatic behavior of
triple-negative breast carcinoma cells, influencing the response to
selective ERβ agonists and the sensitivity to tamoxifen (9). While some studies have reported a
connection between GPER1 expression and poor postoperative outcomes
in non-small cell lung cancer (10,11),
its prognostic implications in breast cancer patients remain a
topic of debate (12).
GPER1 can signal through Gαs,
Gαi/o and Gβγ subunits, as well as
through β-arrestin, to modulate different signaling cascades
(13). Through the Gαs
subunit, GPER1 can elevate intracellular cyclic AMP (cAMP) levels,
while via Gαi/o, it can activate
the extracellular signal-regulated kinase (ERK) pathway and the
phosphatidylinositol 3-kinase/protein kinase B (PI3K/Akt) pathway
(14,15).
As a G-protein coupled receptor (GPCR), GPER1 is
located on the cell membrane and is considered a promising drug
target for the treatment of various diseases. However, its
multifunctionality has led to controversy within the academic
community on whether to agonize or antagonize this target. Wnuk
et al (16) propose that
blocking or antagonizing GPER1 signaling could have a beneficial
impact on the treatment of estrogen receptor-positive breast
cancer. Conversely, Morelli et al (17) discovered that treating Waldenström
Macroglobulinemia cell lines with the GPER1-selective agonist G-1
could activate the TP53 pathway, halt the G2/M cell
cycle and induce apoptosis. These findings were also confirmed in
mouse models. This research offers valuable insights into using
GPER1 as a therapeutic target for addressing other human
malignancies.
While GPER1 presents itself as a promising drug
target, it is crucial to consider the effect of amino acid
mutations on its ligand binding and signal transduction
capabilities. Researches have shown that mutations in GPCRs can
either weaken or enhance their efficacy as drug targets. For
instance, the F282L mutation in the β2-adrenergic receptor has been
found to weaken information flow compared with the wild type,
leading to alterations in signal transmission pathways (18). In studies on the melanocortin-4
receptor, Tao (19) categorized the
effects of GPCR mutations, particularly missense mutations, into
five types: Defective protein production, intracellular retention,
defective binding, defective signaling and unknown defect. In the
case of GPER1, there are only sporadic reports on the physiological
effects of its mutations. For example, Pupo et al (20) observed that mutations in the
N-linked glycosylation site of GPER1 can result in its localization
to the nucleus, where it functions as a transcription factor.
However, Tutzauer et al (21) found that mutations do not affect the
expression levels of GPER1. Overall, our understanding of GPER1
mutations remains limited and the cellular and physiological
effects of these mutations remain to be elucidated. This limitation
presents a challenge in using GPER1 as a target for BIC treatment,
particularly in the context of precision medicine, where the same
drug may elicit greatly different effects in patients with
different genotypes.
To address these knowledge gaps and explore the
implications of GPER1 mutations in BIC, the present study used the
resources of The Cancer Genome Atlas (TCGA) database to categorize
BIC samples based on GPER1 missense mutations and non-mutation
status. The present study first screened for differentially
expressed genes (DEGs) between the two groups. The identified DEGs
were then subjected to gene ontology (GO), Kyoto Encyclopedia of
Genes and Genomes (KEGG) enrichment analyses and protein-protein
interaction (PPI) network analysis. Based on the PPI network, the
present study further screened for hub genes and evaluated their
prognostic value. Additionally, it analyzed the immune cell
infiltration profiles between the mutation and wild-type (WT)
groups. Finally, using mutation data from the cBioPortal website,
eukaryotic expression vectors were constructed for the mutant and
WT GPER1 proteins, and their pharmacological properties were
examined in cell-based functional assays.
Materials and methods
Chemicals, reagents and plasmids
The highly selective agonist G-1 of GPER1 (purity
≥98%) was obtained from Tocris (cat. no. 3577; Tocris Bioscience).
Dulbecco's modified Eagle's medium (DMEM) and other cell culture
reagents were purchased from Thermo Fisher Scientific, Inc. As G-1
is not soluble in water, it was first dissolved in a 10% DMSO
solution to establish a stock solution with a concentration of 1
mM. Prior to use, the stock solution was diluted to the working
concentration using a serum-free DMEM medium.
The luciferase assay kit was obtained from Beyotime
Institute of Biotechnology. Additionally, the fluorescent
luciferase reporter plasmid pGL4.29 containing the cAMP response
element (CRE) was purchased from Promega Corporation.
The Coding Sequence (CDS) of WT human GPER1 was
obtained from NCBI (accession no. NM_001505.3), while the sequences
of four missense mutations (L129M, E218Q, S235F and A354G) were
sourced from the cBioPortal database (https://www.cbioportal.org; accessed on 22 October
2023). The WT and mutant GPER1 CDS were both synthesized by Beijing
Augct DNA-Syn Biotechnology Co., Ltd. and cloned into the pcDNA3.1
(+) vector (Thermo Fisher Scientific, Inc.).
RNA sequencing data
RNA-seq data for BIC were obtained from TCGA
database (namely TCGA-BRCA dataset; https://portal.gdc.cancer.gov; accessed on 22 October
2023), which was downloaded through UCSC Xena (https://xena.ucsc.edu; accessed on 22 October
2023).
The genetic mutation information and survival status
of these BIC samples were accessed via cBioPortal (https://www.cbioportal.org; accessed on 22 October
2023). Samples from four studies were included in the mutation and
survival analysis: TCGA Cell 2015 (818 samples), TCGA Firehose
Legacy (1,108 samples), TCGA Nature 2012 (825 samples) and TCGA
PanCancer Atlas (1084 samples).
Identification of DEGs
Based on the user's guide, the edgeR package
(version 4.0.16; https://bioconductor.org/packages/edgeR/) in R
software (version 4.3.2; http://www.R-project.org/) (22) was used to identify DEGs between WT
and mutant GPER1 in patients with BIC (23). The threshold for DEGs selection was
set at log2 |fold change (FC)| ≥2 and P<0.05. The heatmap and
volcano plot of DEGs were generated using the pheatmap (version
1.0.12; https://CRAN.R-project.org/package=pheatmap) and
ggplot2 (version 3.5.1; https://CRAN.R-project.org/package=ggplot2) packages
in the R programming platform.
Enrichment analyses of DEGs
GO and KEGG analyses were performed using the
clusterProfiler package in R to identify the categories and
signaling pathways enriched for each gene cluster. To address the
issue of a high false discovery rate (FDR) resulting from multiple
comparisons, q values were calculated. An FDR-adjusted
q<0.25 and P<0.05 were considered statistically
significant.
PPI and hub gene screening
To obtain the PPI network and hub genes associated
with GPER1 in BIC, the DEGs were entered into the search tool for
the retrieval of interacting genes (STRING) database (https://cn.string-db.org/) for analysis with a
confidence score >0.4. Subsequently, the network generated by
STRING was imported into Cytoscape 3.7.2(24) for visualization. The cytoHubba
plugin was used to filter key protein modules and after calculation
with 12 algorithms, the top 10 genes with the highest scores were
selected as hub genes.
Analysis of immune cell
infiltration
CIBERSORT, a deconvolution algorithm, was used to
assess the proportions of 22 immune lymphocyte in various BIC tumor
samples (25). The results obtained
were further used to compare the levels of immune cell infiltration
between WT and mutant GPER1 patients. The number of permutations
was set to 1,000, where P<0.05 serves as the criterion for
successful computation in each sample.
Cell culture
The 293T cells were obtained from Beyotime
Biotechnology (cat. no. C6008, Shanghai, China) and cultured in
DMEM containing 10% fetal bovine serum, in a humidified atmosphere
with 5% CO2 at 37˚C. Upon purchase, all cells underwent
mycoplasma testing and the results were negative. Cell line
authentication was conducted using short tandem repeat (STR)
analysis, which confirmed that the purchased cell line was
293T.
The cells were plated in 6-well plates and cultured
for 24 h prior to assays. The Countess 3 cell counter (Invitrogen;
Thermo Fisher Scientific, Inc.) was used for cell counting.
Detection of intracellular cAMP levels
using a luciferase reporter
The effect of WT and mutant GPER1 on intracellular
cAMP accumulation was assessed using a luciferase reporter assay
that had been previously established (26). Specifically, 500 ng pcDNA3.1 (+)
vectors carrying the WT GPER1 and four mutant variants (designated
as pGPER1-WT, pGPER1-L129M, pGPER1-E218Q, pGPER1-S235F and
pGPER1-A354G) were respectively mixed with 1,000 ng pGL4.29 vector
(Promega Corporation) and 300 ng pEGFP-N1 (serving as an internal
control for transfection efficiency and normalization in luciferase
assays) and then transfected into 293T cells using
polyethyleneimine transfection reagent (Shanghai Fushen
Biotechnology Co., Ltd.).
Following transfection, the cells were cultured in
the original medium for 24 h at 37˚C, then transferred to 48-well
plates and allowed to grow for an additional 24 h to reach a
density of 2x105 cells per well. Subsequently, the
agonist G-1 (Tocris Bioscience), diluted in serum-free DMEM medium,
was added to the cells in the 48-well plates and incubated for 6
h.
After treatment, the cells were lysed and a
substrate was added to induce luminescence using a Firefly
Luciferase Reporter Gene Assay Kit (cat. no. RG006) obtained from
Beyotime Biotechnology. The luminescence signal was detected using
a Tecan M200 microplate reader (Tecan Group, Ltd.) to determine the
relative luciferase activity in each well. According to a previous
study, enhanced green fluorescent protein (EGFP) was used as an
internal control to normalize the bioluminescent signal of the
luciferase reporter pGL4.29(27).
The fluorescence of EGFP was detected under excitation/emission
wavelengths of 488/507 nm. Firefly luciferase values were divided
by the EGFP fluorescence values to assess normalized reporter gene
activity. Each agonist concentration was tested in triplicate
independent experiments and the data were presented as mean ±
standard error of the mean.
Statistical analyses
The statistical analyses were performed using R
software (version 4.3.2; http://www.R-project.org/) (22) and GraphPad Prism 7.0 (Dotmatics).
The comparison of gene expression between GPER1 wild-type and
mutant patients with BIC was conducted using the Mann-Whitney U
test. The differences in immune cell proportion between the GPER1
wild-type and mutant groups were evaluated using the Mann-Whitney U
test. Kaplan-Meier method with log-rank test was employed for
survival analysis. The luciferase activities after agonist
treatment in cells were converted into fold change relative to the
control group (DMEM serum-free medium) and fitted to a
dose-response curve using a nonlinear regression analysis. For the
half-maximal effective concentration (EC50) and maximum
response concentration (Rmax) of the wild-type and
mutant groups, the Kruskal-Wallis test was performed with Dunn's
post hoc analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
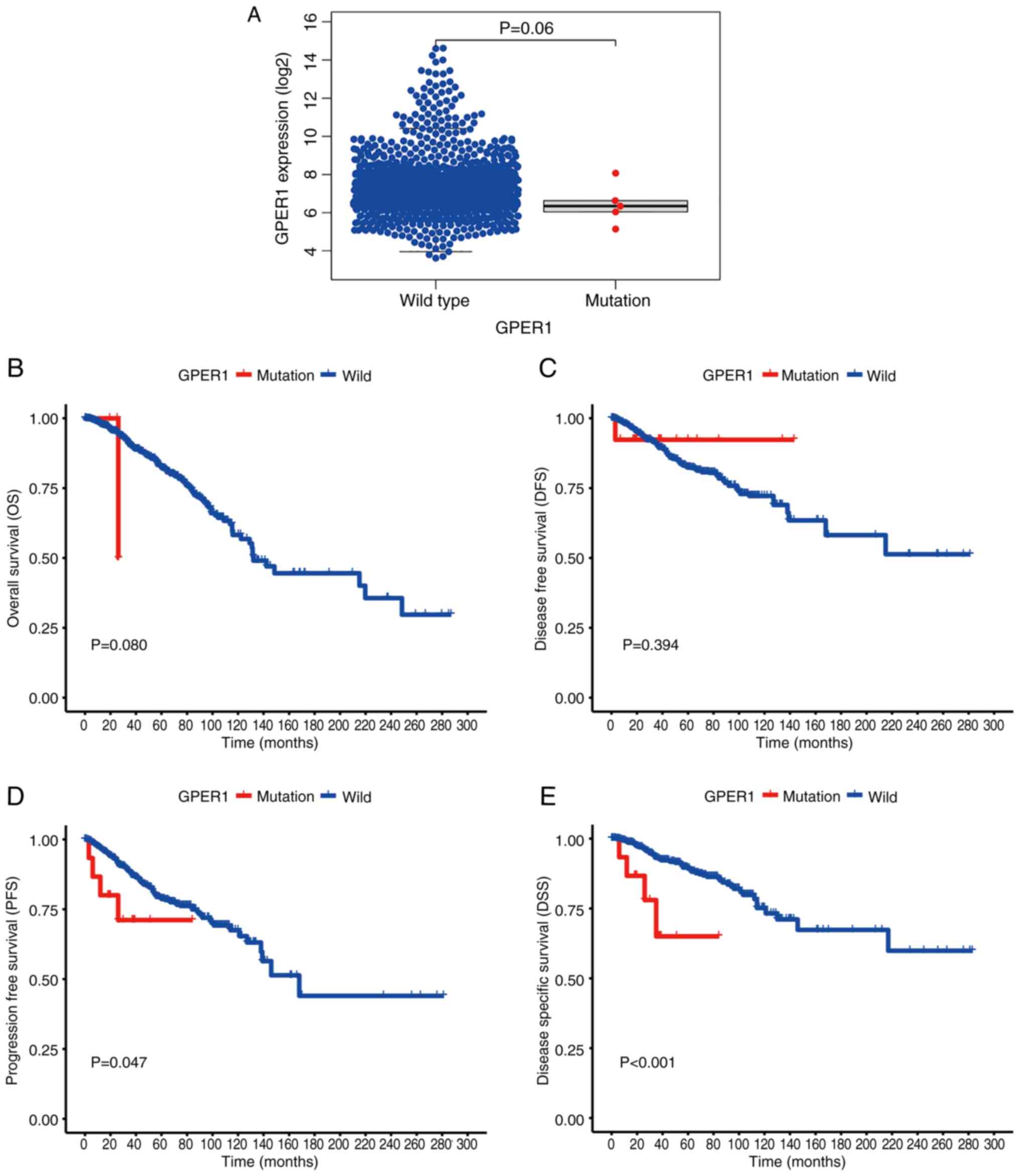
Survival analyses and GPER1
expression
The cBioPortal online survival analysis tool used
data from four studies sourced from TCGA, encompassing a total of
3,835 samples from 3,827 patients. Among these, 29 patients lacked
mutation data, 3,743 patients had no mutations in GPER1 and 55
patients had mutations in GPER1. The findings suggested that,
although no noteworthy disparities were observed in overall
survival (OS) and disease-free survival (DFS) between the GPER1 WT
and mutant cohorts, there were notable variances in
progression-free survival (PFS) and disease-specific survival (DSS)
outcomes across the two groups. The mutant group showed
significantly reduced PFS and DSS compared with the non-mutant
group (P<0.05; Fig. 1B-E).
As for the mRNA expression levels of GPER1 in the WT
and mutant groups, RNAseq samples obtained from the UCSC Xena were
analyzed. Out of the 1,126 samples downloaded, normal adjacent
samples and those with missing clinical data and duplicates were
excluded, resulting in a final dataset of 1,098 samples. Among
these, 5 samples had missense mutations according to cBioPortal.
The results showed no significant difference in GPER1 mRNA
expression between the WT and mutant groups (Fig. 1A).
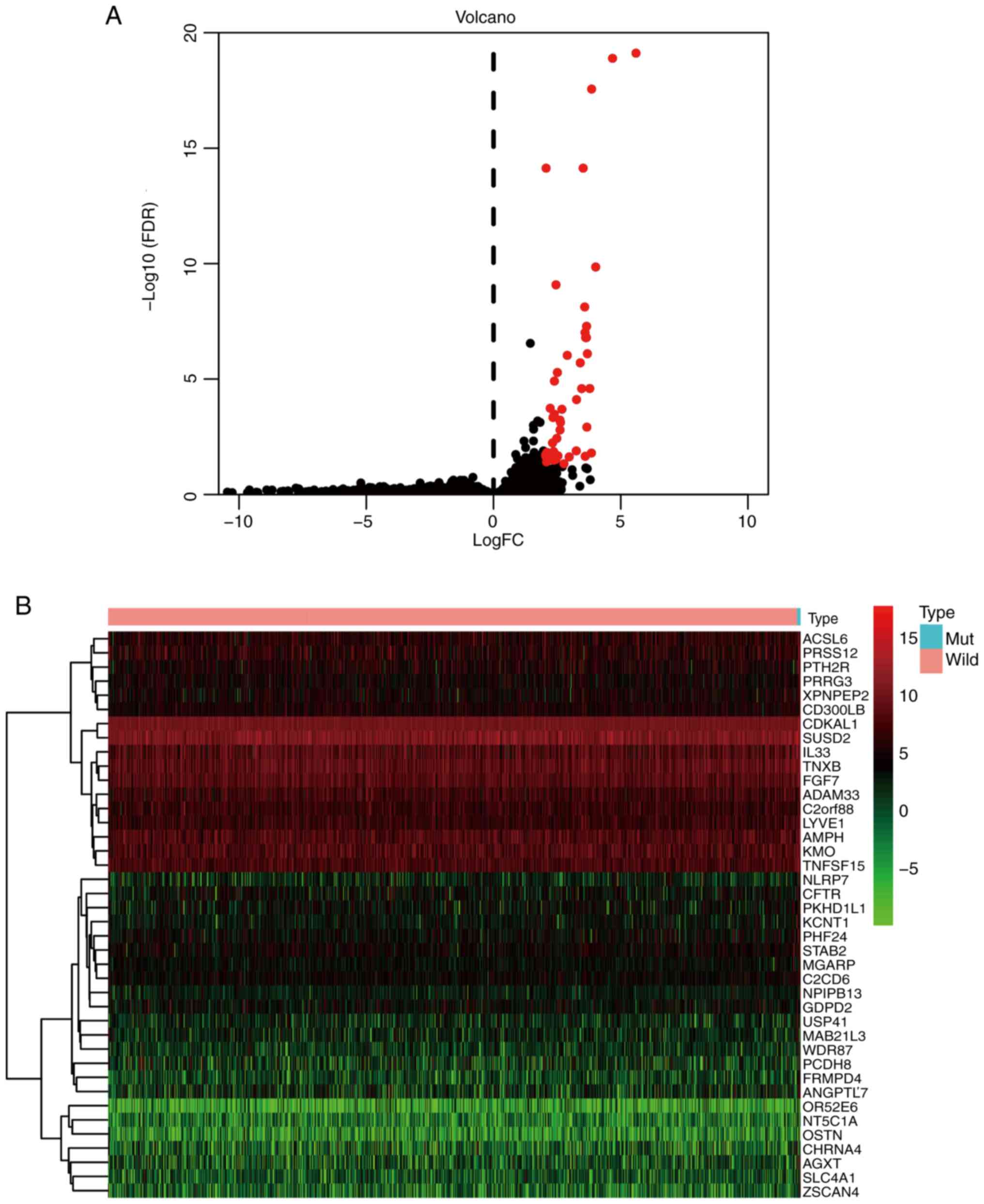
Identification of DEGs
The identification of DEGs was also conducted using
the 1,098 samples obtained from the UCSC Xena. The analysis,
performed with the edgeR package in R, identified 60 DEGs meeting
the criteria of log2 |fold change (FC)| ≥2 and
P<0.05, all of which exhibited upregulation (Table SI). The volcano plot and heatmap
illustrating the DEGs are presented in Fig. 2.
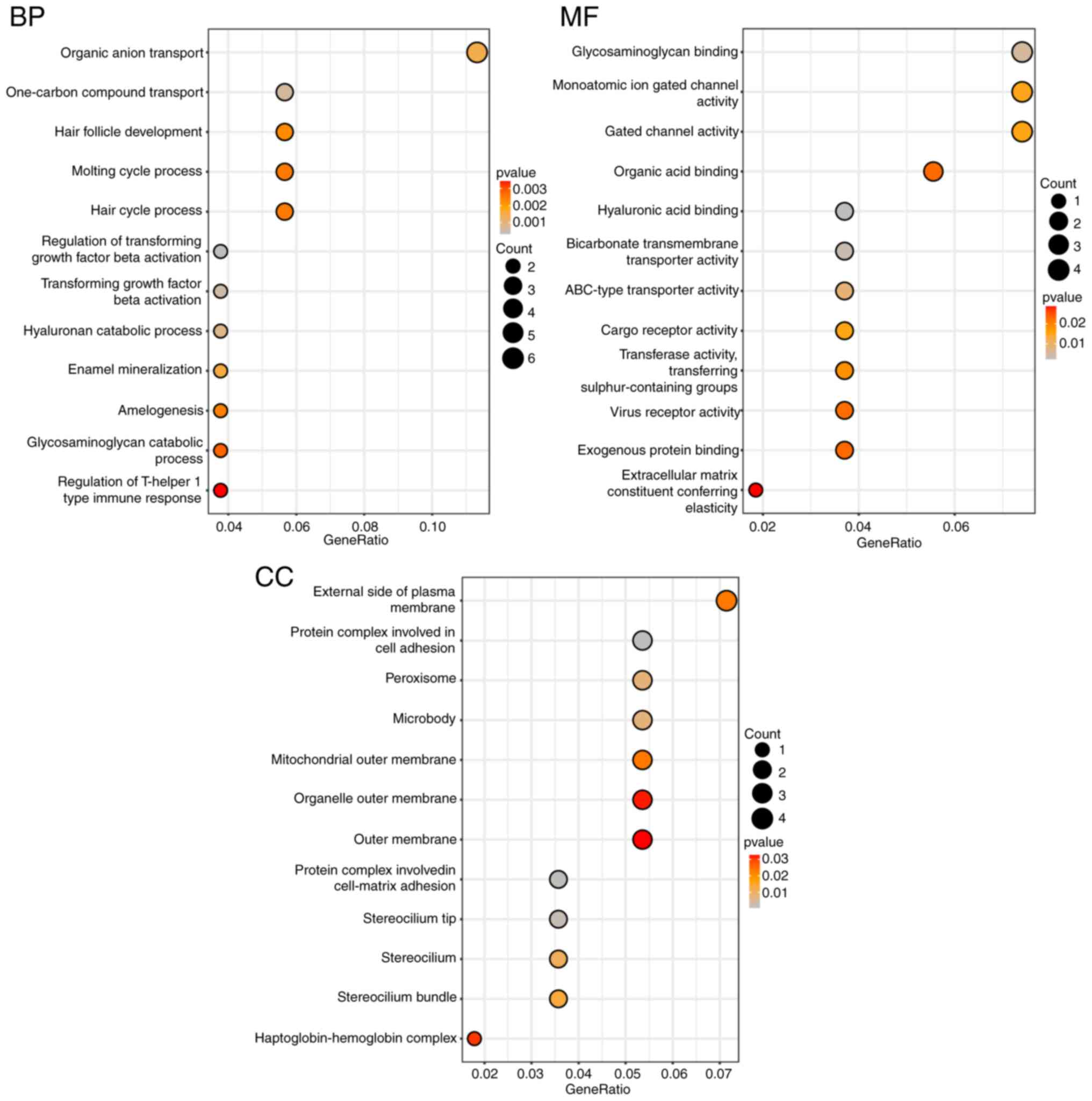
GO and KEGG enrichment analyses
Using the clusterProfiler package in R software, GO
analysis of the 60 DEGs revealed significant enrichment patterns.
Specifically, in terms of Biological Process (BP), the DEGs were
primarily enriched in organic anion transport. For Molecular
Function (MF), the DEGs showed enrichment in glycosaminoglycan
binding, monoatomic ion gated channel activity and gated channel
activity. Regarding Cellular Component (CC), the DEGs were
predominantly enriched in the external side of the plasma membrane
(Fig. 3).
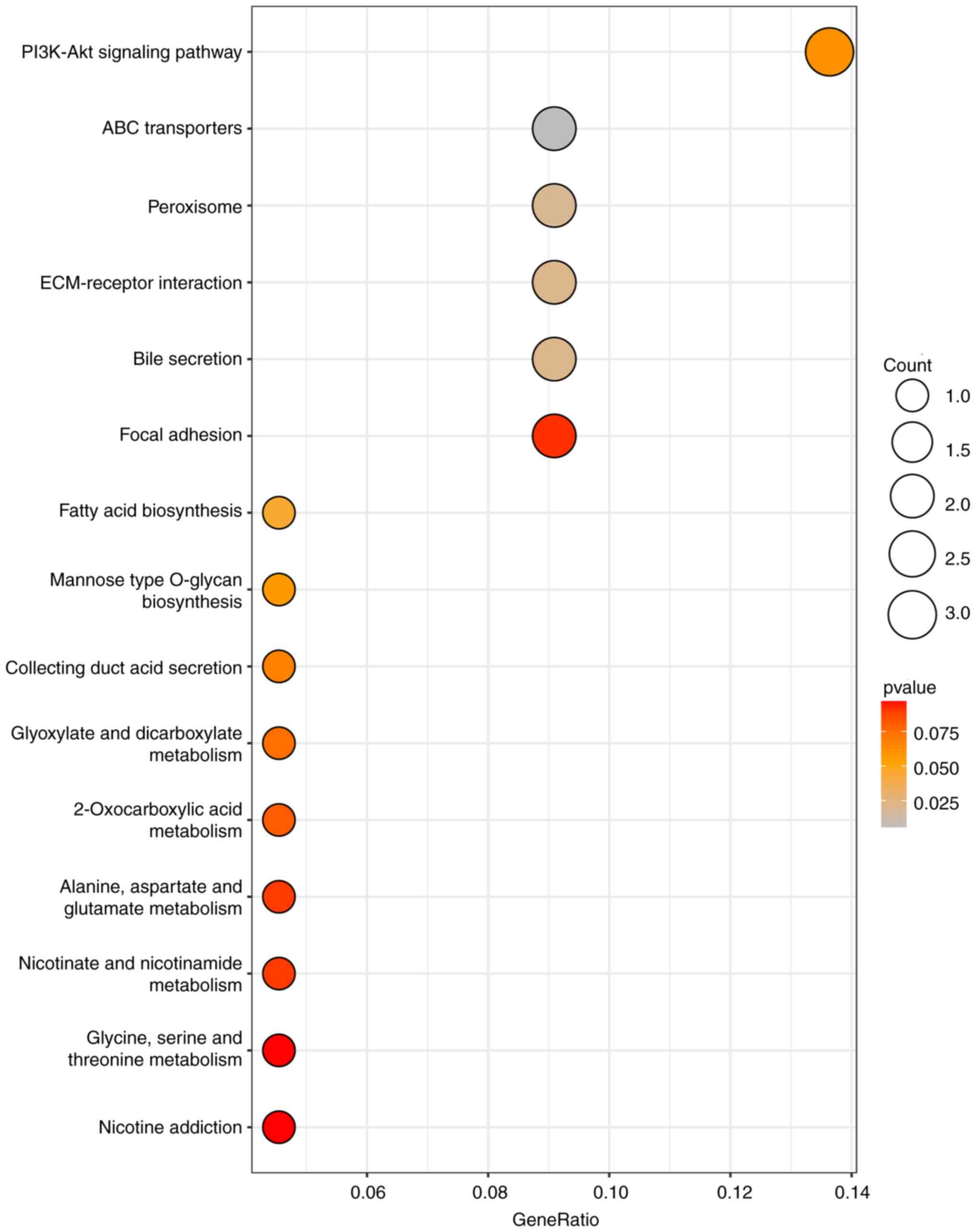
The KEGG analysis of the DEGs using the
clusterProfiler package revealed significant enrichment in the
PI3K-Akt signaling pathway, with subsequent enrichment observed in
ABC transporters, Peroxisome, ECM-receptor interaction and Bile
secretion pathways (Fig. 4).
PPI network and hub genes
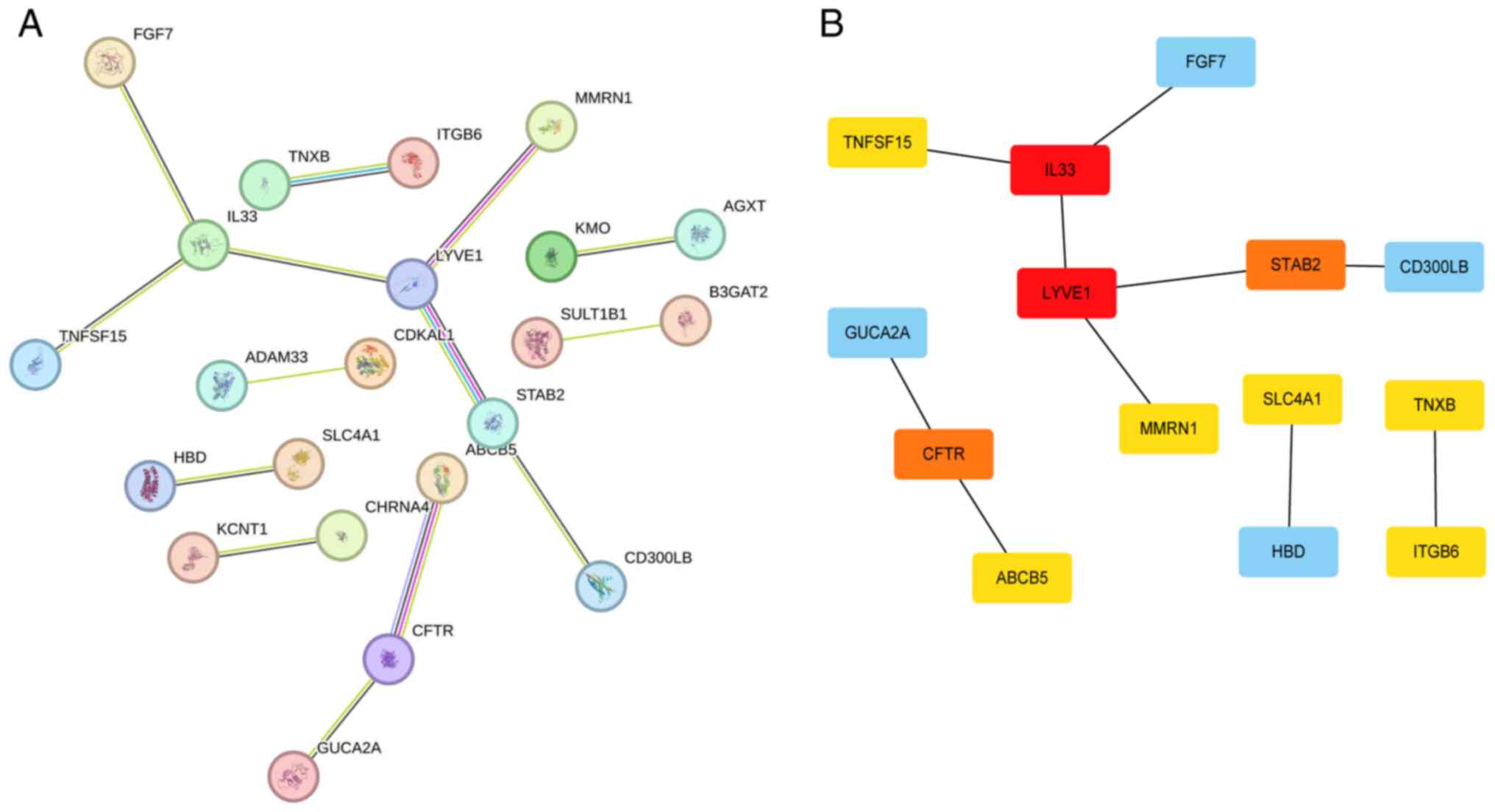
The protein-protein interaction network of DEGs
obtained using the STRING online tool is shown in Fig. 5A. This network was further analyzed
using the cytoHubba plugin in Cytoscape software with 12 algorithms
to calculate the top 10 hub genes. Each algorithm yielded the same
result, identifying IL33 and LYVE1 as the highest-scoring genes,
followed by CFTR and STAB2 and finally FGF7, HBD, SLC4A1, TNFSF15,
ABCB5, ITGB6 (Fig. 5B; Table SII).
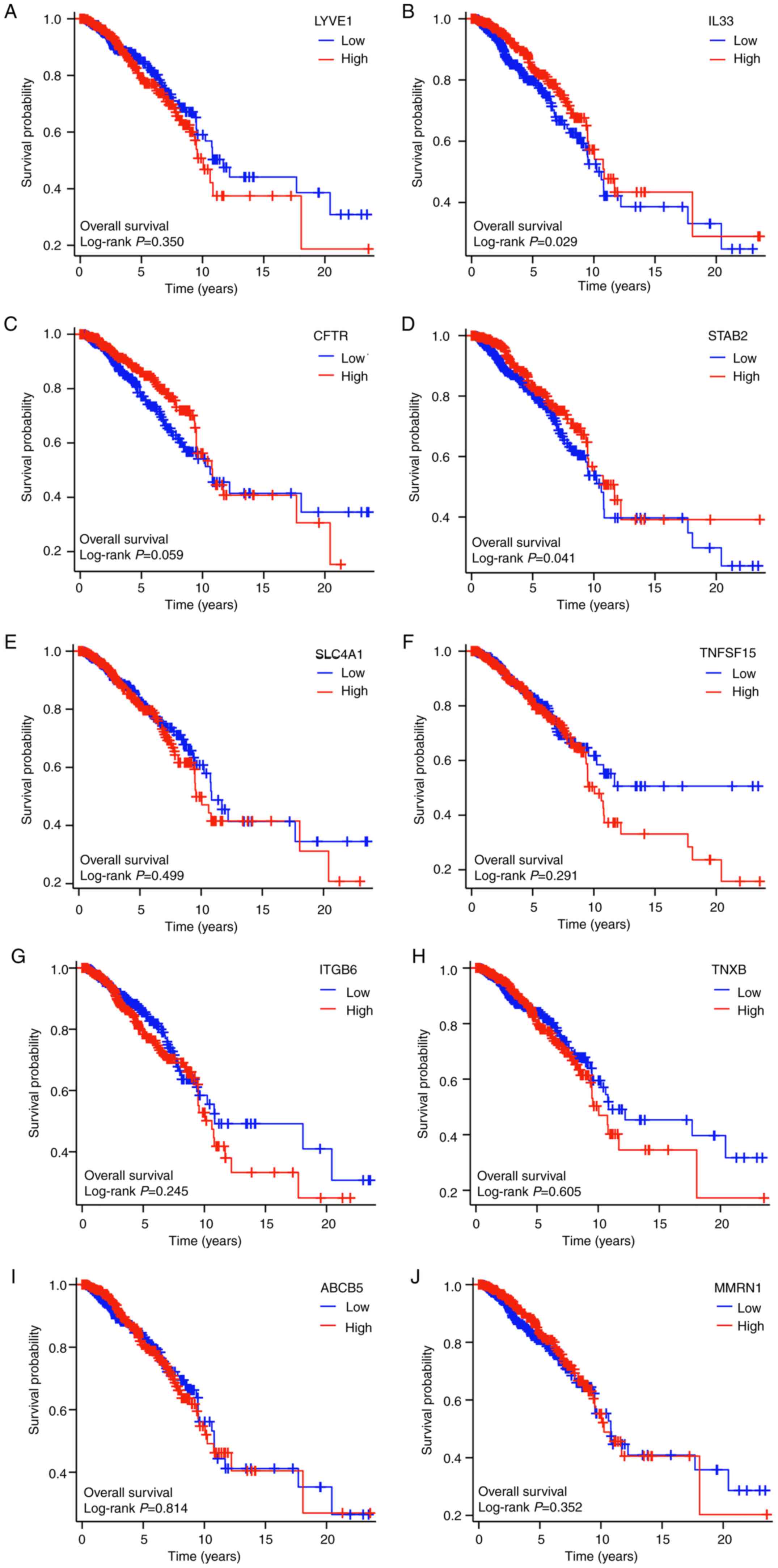
Prognostic values of the hub
genes
Kaplan-Meier survival analysis, employing the
log-rank test, demonstrated that out of the 10 hub genes examined,
only IL33 and STAB2 displayed significant survival disparities
between the high and low expression cohorts (P<0.05).
Additionally, CFTR, another hub gene, exhibited a nearly
significant difference in expression between the two groups
(P=0.059; Fig. 6).
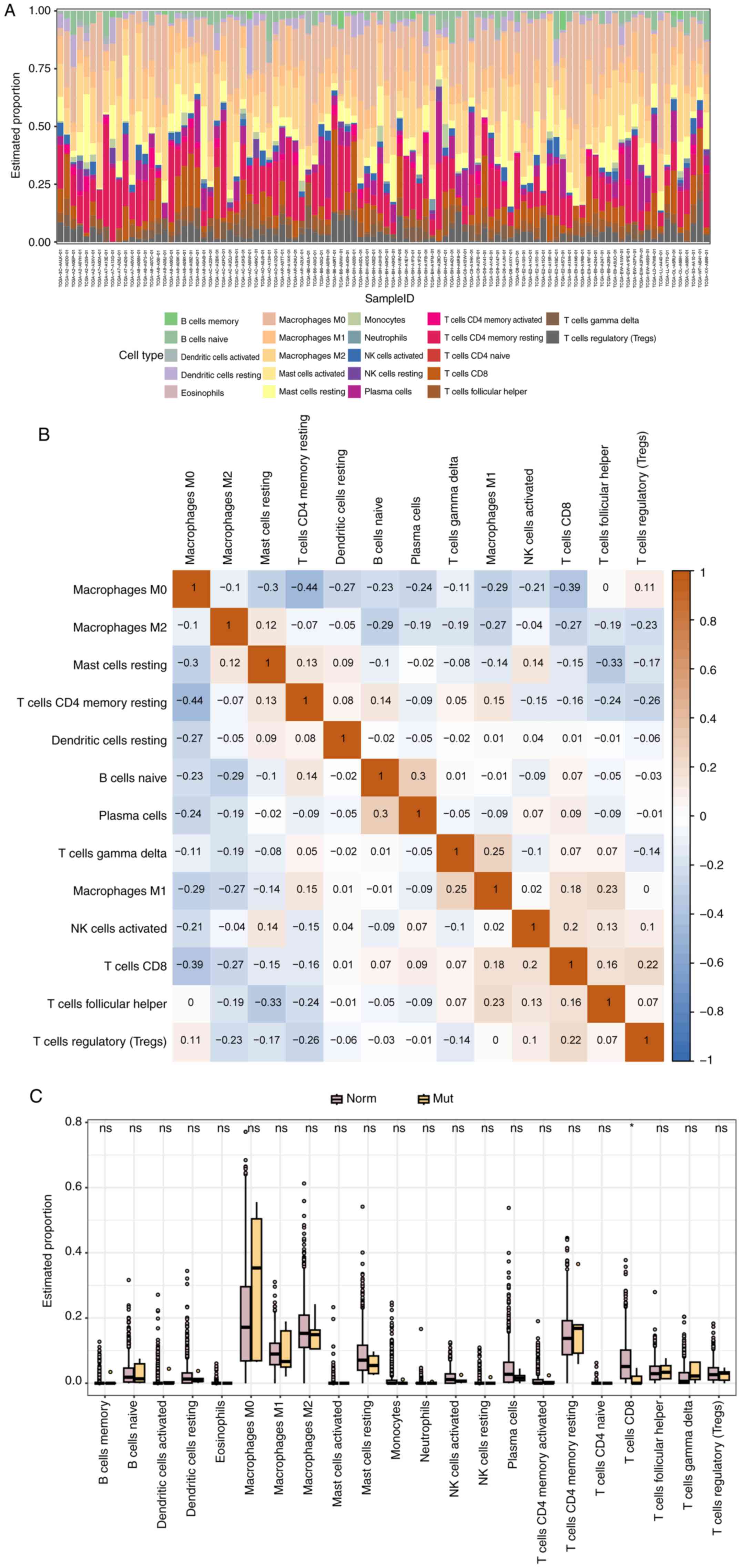
GPER1 mutation-related immune cell
infiltration
The analysis of tumor immune cell infiltration based
on the CIBERSORT algorithm revealed that the proportions of various
immune cells in each sample (Fig.
7A) and found that the content of CD8 T cells in the GPER1
mutation group was significantly lower than that in the WT group
(P<0.05; Fig. 7C). Furthermore,
the correlation matrix of immune cells showed that the content of
memory resting CD4 T cells and CD8 T cells was negatively
correlated with M0 macrophages, while naive B cells and Plasma
cells exhibited a positive correlation. Similarly, gamma and delta
T cells and M1 macrophages also showed a certain degree of positive
correlation (Fig. 7B).
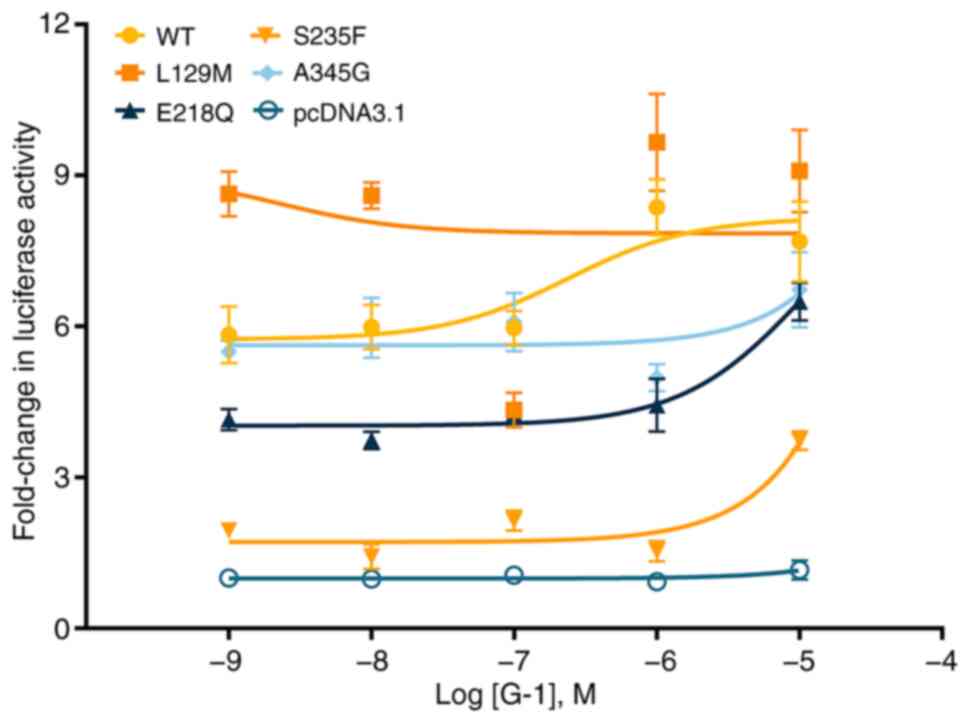
Signaling properties of the GPER1
mutants
As a cell membrane surface receptor, GPER1 is
capable of binding with the agonist G-1, activating intracellular
adenylyl cyclase and facilitating the production of the canonical
second messenger cAMP (14). To
investigate the effect of missense mutations on the signal
transduction of GPER1, a luciferase reporter system was used with
CRE in cells transfected with WT and mutant GPER1 vectors for
analysis. The findings revealed that in comparison with the WT
receptor, the response to the agonist G-1 were significantly
attenuated in all the four missense mutations, as evidenced by a
notable increase in their EC50 values (P<0.05) or undetectable
responses (L129M and A345G) (Fig.
8; Table I). In terms of the
response intensity, three mutants (E218Q, S235F and A345G)
displayed a reduction in maximal response
(Rmax<100%), while one mutant (L129M) exhibited an
enhancement in maximal response (Rmax>100%; Table I).
 | Table IAgonist-stimulated cAMP response of
WT and mutant GPER1s. |
Table I
Agonist-stimulated cAMP response of
WT and mutant GPER1s.
| | G-1 stimulated cAMP
response |
|---|
| WT and mutant
GPER1s | EC50
(µM) | Rmax (%
WT) |
|---|
| WT | 0.20±0.08 | 100 |
| L129M | NDb | 115.40±6.60 |
| E218Q | 157.12a±45.98 | 77.60±2.60 |
| S235F | 10.66±2.70 | 44.60±1.30 |
| A345G | NDb | 80.40±5.10 |
Discussion
Previous studies have demonstrated that GPER1 can
influence the proliferation, metastasis and drug sensitivity of
breast cancer cells (6,9); However, whether mutations in this
receptor, especially missense mutations, will cause a series of
cellular and pharmacological changes similar to those observed in
other GPCRs and thereby affect the regulation of tumor-related
signaling pathways, remains to be elucidated. The present study
analyzed the BIC samples from TCGA database, stratifying them into
GPER1 missense mutation and WT groups. It compared the survival
differences between the two groups, identified DEGs and subjected
the DEGs to GO, KEGG and PPI network analyses. From the PPI
network, 10 hub genes were identified and their prognostic value
assessed. Additionally, the present study compared the immune cell
infiltration profiles between the mutation and WT groups. Using
cell-based pharmacological approaches, it also examined the impact
of the GPER1 mutations on the receptor's regulation of
intracellular cAMP accumulation in response to agonist
stimulation.
Analysis of GPER1 gene expression in both the WT and
mutant groups revealed that the difference between the two groups
nearly reached statistical significance (P=0.06). This
non-significant result, although consistent with the findings of
Tutzauer et al (21), may be
attributed to the limited sample size of mutants, warranting
further sample collection for validation. Regarding survival
analysis, while OS and DFS did not show significant differences
between the two groups, PFS and DSS were significantly lower in the
mutant group compared with the WT group. According to medical
statistics, DSS is generally considered more accurate than OS in
assessing treatment impact on specific diseases (28). Therefore, it can be inferred that
mutations in GPER1 reduce the treatment efficacy for patients with
BIC, resulting in poorer prognosis. Considering the limited number
of mutant individuals included in the study (n=55) and the various
types of mutations, this conclusion requires further support from
additional case reports and larger cohort studies.
Using the criteria of log2 |fold change (FC)| ≥2 and
P<0.05 in the edgeR package of R software, 60 DEGs were
identified between the GPER1 mutant and WT groups. To assess
whether the small number of DEGs identified was due to overly
stringent filtering criteria, the present study further relaxed the
thresholds to log2 |fold change (FC)| ≥1 and P<0.05. Despite
this adjustment, only 80 DEGs were identified. The scarcity of DEGs
hints at the possibility that GPER1 mutations in BIC may primarily
influence a select few cellular functions and signaling pathways.
An intriguing observation is that all identified DEGs were
upregulated. This expression pattern contrasts with findings from
other tumor-related DEG screenings (29-31),
suggesting that GPER1 mutations may elicit a distinct gene
expression profile in BIC.
GO enrichment analysis revealed that the DEGs were
predominantly enriched in organic anion transport,
glycosaminoglycan binding, monoatomic ion-gated channel activity,
gated channel activity and plasma membrane. Organic anion transport
plays a pivotal role in tumor cell proliferation and metastasis
(32,33), influencing the efficacy of
anti-tumor drugs by modulating their intracellular concentrations,
thereby affecting their cytotoxic effects on cancer cells (34-36).
Glycosaminoglycan binding affects crucial cellular functions such
as adhesion, migration and signaling (37). Dysregulation of
glycosaminoglycan-binding proteins has been linked to increased
invasiveness and aggressiveness of tumors (37). Monoatomic ion-gated channel activity
is associated with the regulation of ion flow across cell
membranes, essential for maintaining cellular homeostasis and
involved in processes such as cell signaling, proliferation and
apoptosis (38,39). In tumors, dysregulation of ion
channels can contribute to abnormal cell growth, survival and
migration, influencing the progression and behavior of cancer cells
(38,39). The plasma membrane plays a critical
role in cell-cell interactions, nutrient uptake and signal
transduction, with alterations in its properties affecting cell
adhesion, migration and response to extracellular signals, thereby
impacting cancer development and progression (39). In summary, the terms associated with
the DEGs identified through GO enrichment analysis are all related
to tumor progression and metastasis, potentially influencing the
effectiveness of anti-tumor drugs. This finding is consistent with
the lower PFS and DSS observed in the GPER1 mutant group in the
present study.
The KEGG analysis of these DEGs revealed enrichment
for the PI3K-Akt, ABC transporters, Peroxisome, ECM-receptor
interaction and bile secretion pathway. This discovery is in line
with numerous studies that have identified this pathway in
differential gene expression analyses comparing tumor and non-tumor
samples (40-42)
Activation of the PI3K-Akt pathway is known to promote oncogenes,
suppress tumor suppressor genes and contribute to the development
of cancer characteristics such as uncontrolled cell growth, evasion
of apoptosis, angiogenesis, tissue invasion and metastasis
(43,44). For instance, in cervical cancer, the
PI3K-Akt pathway plays a role in modulating the expression of HCCR,
a downstream component that influences disease progression
(45). Overall, activation of the
PI3K-Akt signaling pathway is frequently linked to poor prognosis
in cancer (46-48).
Similar to the PI3K-Akt pathway, the ABC transporter pathway is
associated with poor prognosis in cancer patients (49,50). A
study identified transporters such as ABCA1, ABCB1 and ABCG2 as
being involved in the efflux of chemotherapeutic agents, thereby
contributing to the multidrug resistance seen in aggressive breast
cancer subtypes (51). Furthermore,
some other researches indicate that high expression levels of these
transporters are correlated with increased tumor invasiveness
(52,53). The peroxisome signaling pathway,
particularly through peroxisome proliferator-activated receptors
(PPARs), plays a significant role in the development and
progression of breast cancer. PPARs are ligand-dependent nuclear
receptor transcription factors that regulate genes involved in
critical cellular processes such as proliferation, differentiation
and metabolism (54). Dysregulation
of PPARs has been linked to breast cancer. Additionally,
peroxisomes contribute to carcinogenesis by producing reactive
oxygen species (ROS), which can lead to DNA damage and initiate
tumor development (55,56). Components of the extracellular
matrix (ECM), such as fibronectin and collagen, significantly
influence breast cancer cell migration and epithelial-mesenchymal
transition (EMT), a process whereby epithelial cells acquire
mesenchymal traits, enhancing their invasive capabilities (57,58).
ECM-receptor interaction signaling pathways have been reported to
be closely associated with breast cancer metastasis (59,60).
Unlike the previous four signaling pathways, bile acids, although
found to accumulate in human breast tumors, have been shown to
inhibit tumor growth and improve patient survival (61). The enrichment of DEGs in the bile
secretion pathways may be related to the body's mechanisms for
counteracting tumor development. Overall, the KEGG analysis
revealed that DEGs were enriched in signaling pathways, with four
pathways related to tumor progression and drug resistance and one
pathway associated with anti-tumor activity. These findings are
generally consistent with the results obtained from the GO analysis
and can help explain the lower PFS and DSS observed in the GPER1
mutation group.
Through PPI network analysis and hub gene screening,
the present study identified 10 hub genes, among which IL33 and
STAB2 showed significant associations with the survival of BIC
patients, while CFTR demonstrated near-significant relevance
(P=0.059). Of these three genes, only CFTR was found to function as
a hub gene in other cancers and diseases, such as colorectal cancer
(62), chromophobe renal cell
carcinoma (63) and pulmonary
arterial hypertension (64),
whereas IL33 and STAB2 have not been previously identified as hub
genes in other types of cancer. For the other two genes, IL33 has
been shown to promote cancer stemness, tumor growth and metastasis
by recruiting macrophages into the tumor microenvironment (65,66),
while STAB2, a hyaluronic acid receptor, has also been associated
with cancer metastasis (67-69).
Furthermore, the IL33-STAB2 axis has been demonstrated to modulate
immune responses in gastrointestinal cancers (70,71).
In summary, IL33, STAB2 and CFTR are all linked to tumor
progression, but only CFTR has been identified as a hub gene in
some other cancer types, suggesting that IL33 and STAB2 may play
distinct roles in GPER1 mutation-mediated tumor progression.
Based on the analysis of immune cell infiltration
using the CIBERSORT algorithm, it was observed that the proportion
of CD8 T cells in the GPER1 mutation group was notably lower
compared with the WT group. CD8 T cells are essential for
recognizing and eliminating cancer cells and their diminished
presence may lead to immune evasion and tumor progression (72-74).
Furthermore, research has indicated that augmenting the
infiltration of CD8 T cells, either through interventions such as
vitamin D therapy or immune checkpoint blockade, can bolster
anti-tumor immunity and improve treatment outcomes for cancer
patients (75-77).
Consequently, the reduced levels of CD8 T cells in the GPER1
mutation group may contribute to the poorer prognosis observed in
this cohort.
Mutations in GPCRs can lead to diverse
pharmacological effects that significantly affect drug efficacy and
patient responses. These genetic alterations can modify receptor
pharmacology by influencing cell surface expression, receptor
interactions, basal activity and GPCR-G protein coupling. Such
changes can result in varied disease phenotypes or alter drug
response (78). For instance,
mutations in the adhesion G protein-coupled receptor latrophilin 1
can impair receptor trafficking, contributing to conditions such as
obesity (79). Consequently,
understanding the molecular consequences of pathogenic mutations in
GPCRs is crucial for elucidating disease mechanisms and developing
effective therapeutic strategies. To assess the influence of
mutations on the response of GPER1 to agonists, the present study
employed a luciferase reporter system with a CRE element to
quantify cellular cAMP levels downstream of GPER1 activation by the
potent selective agonist G-1. The results revealed that all four
mutations, namely L129M, E218Q, S235F and A345G, attenuated
cellular cAMP levels to varying extents following G-1 stimulation.
While the cAMP signaling pathway demonstrates multifunctionality
and can exert either tumor-suppressive or tumor-promoting effects
across different types of tumor and cellular contexts (80), activation of cAMP signaling by
Gαs in the GPCR system has been implicated as a tumor
suppressor in neoplasms derived from ectodermal cells, including
neural and epidermal stem/progenitor cells (81). Given that GPER1 activates cAMP via
coupling to Gαs, the attenuation of cAMP signaling due
to mutations may contribute to the unfavorable prognosis observed
in patients with BIC with GPER1 mutations. Furthermore, the
diminished ligand activation capacity of mutated GPER1 also
partially accounts for the reduced PFS observed in patients with
BIC with GPER1 mutations.
It is worth noting that, while previous studies have
grouped tumors based on mutations and non-mutations and screened
for DEGs for subsequent bioinformatics analysis (82-84),
the present study has, for the first time, to the best of the
authors' knowledge, integrated cell pharmacological experiments to
detect the potential effect of mutations on key GPCR downstream
signaling pathways in tumors. These intriguing findings provide
important insights for precision medicine targeting GPCRs. However,
the present study has some limitations, such as not grouping
according to the mutation types of GPCRs. Considering the diverse
pharmacological effects of GPCR mutations (19), further subgrouping and studying the
specific effects of different GPCR mutations on tumor cell biology
have become one of the next research priorities.
In summary, the present study explored the molecular
and pharmacological effects of missense mutations in GPER1 using
the BIC sample information from TCGA data. It found that missense
mutations in GPER1 led to adverse prognostic outcomes and reduced
treatment effectiveness. Differential gene expression analysis
revealed that GPER1 mutations caused upregulation of a subset of
genes, which enriched in signaling pathways, such as PI3K-Akt,
implicated in tumor progression. Similarly, hub genes selected
through PPI network screening, including IL33, STAB2 and CFTR, were
all associated with tumor progression. Immune infiltration analysis
also demonstrated a reduction in anti-tumor CD8 T cell content with
GPER1 mutations. Pharmacological analysis revealed that mutations
diminish GPER1's ability to induce cAMP production upon agonist
stimulation. These findings provided insights into the design of
anti-tumor drugs targeting GPER1 and personalized medicine
approaches.
Supplementary Material
List of differentially expressed
genes.
Top 10 in network string interactions
short.tsv ranked by the Maximal Clique Centrality method.
Acknowledgements
The authors appreciate the support of Animal
Genetics and Breeding Laboratory, College of Animal Science and
Technology, Northwest A&F University (Shaanxi, China).
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant nos. 82174164, 81901886 and
31502180), Shaanxi Administration of Traditional Chinese Medicine
(grant no. 2021-ZZ-JC019), the Key Research and Development Program
of Shaanxi (grant nos. 2022SF-411 and 2023-YBSF-565) and the
Fundamental Research Funds for the Central Universities (grant no.
xzy012020040), the Natural Science Foundation of Shaanxi Province
(grant no. 2023-JC-YB-155).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YL conceived the study and conducted the overall
review and revision of the manuscript. CD and SQZ secured the
research funding and supervised the progress of the study. HY and
HLM performed the cellular pharmacology experiments. YZ conducted
the differential analysis of breast cancer data from the TCGA
database and drafted the initial manuscript. QYY carried out the
immune cell infiltration analysis. HY and HLM confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript. HY and HM confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ide Y, Horii R, Osako T, Ogura K, Yoshida
R, Iwase T and Akiyama F: Clinicopathological significance of
invasive micropapillary carcinoma component in invasive breast
carcinoma. Pathol Int. 61:731–736. 2011.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lwin ZM, Guo C, Salim A, Yip GW, Chew FT,
Nan J, Thike AA, Tan PH and Bay BH: Clinicopathological
significance of calreticulin in breast invasive ductal carcinoma.
Mod Pathol. 23:1559–1566. 2010.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Siegel RL, Giaquinto AN and Jemal A:
Cancer statistics, 2024. CA Cancer J Clin. 74:12–49.
2024.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Mei L, Wang K and Gu Y: Improved Fuzzy
C-means clustering algorithm-based dynamic contrast-enhanced
magnetic resonance imaging features in the diagnosis of invasive
breast carcinoma before and after menopause. Comput Math Methods
Med. 2022(2917844)2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Rakha EA, Martin S, Lee AHS, Morgan D,
Pharoah PD, Hodi Z, Macmillan D and Ellis IO: The prognostic
significance of lymphovascular invasion in invasive breast
carcinoma. Cancer. 118:3670–3680. 2012.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Yang F, Xie HY, Yang LF, Zhang L, Zhang
FL, Liu HY, Li DQ and Shao ZM: Stabilization of MORC2 by estrogen
and antiestrogens through GPER1-PRKACA-CMA pathway contributes to
estrogen-induced proliferation and endocrine resistance of breast
cancer cells. Autophagy. 16:1061–1076. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Saroha HS, Kumar Guddeti R, Jacob JP,
Kumar Pulukuri K, Karyala P and Pakala SB: MORC2/β-catenin
signaling axis promotes proliferation and migration of breast
cancer cells. Med Oncol. 39(135)2022.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Thomas L, Chutani N, R K, Nair AS, Yellapu
NK, Karyala P and Pakala SB: Microrchidia 2/histone deacetylase 1
complex regulates E-cadherin gene expression and function. Biochem
J. 480:1675–1691. 2023.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Schmitz V, Bauerschmitz G, Gallwas J and
Gründker C: Suppression of G Protein-coupled estrogen Receptor 1
(GPER1) Enhances the Anti-invasive efficacy of selective ERβ
Agonists. Anticancer Res. 42:5187–5194. 2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Li ZH, Liu C, Liu QH, Wang J, Wang Y, Wang
YF, Deng SJ and Li DB: Cytoplasmic expression of G protein-coupled
estrogen receptor 1 correlates with poor postoperative prognosis in
non-small cell lung cancer. J Thorac Dis. 14:1466–1477.
2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Shen Y, Li C, Zhou L and Huang JA: G
protein-coupled oestrogen receptor promotes cell growth of
non-small cell lung cancer cells via YAP1/QKI/circNOTCH1/m6A
methylated NOTCH1 signalling. J Cell Mol Med. 25:284–296.
2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yang F and Shao ZM: Double-edged role of G
protein-coupled estrogen receptor 1 in breast cancer prognosis: An
analysis of 167 breast cancer samples and online data sets. Onco
Targets Ther. 9:6407–6415. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Srivastava DP and Evans PD: G-protein
oestrogen receptor 1: Trials and tribulations of a membrane
oestrogen receptor. J Neuroendocrinol. 25:1219–1230.
2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Evans NJ, Bayliss AL, Reale V and Evans
PD: Characterisation of signalling by the Endogenous GPER1 (GPR30)
Receptor in an Embryonic Mouse Hippocampal Cell Line (mHippoE-18).
PLoS One. 11(e0152138)2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yu M, Xu L, Lei B, Sun S and Yang Y:
Tetrachlorobisphenol A and bisphenol AF induced cell migration by
activating PI3K/Akt signaling pathway via G protein-coupled
estrogen receptor 1 in SK-BR-3 cells. Environ Toxicol. 38:126–135.
2023.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wnuk A, Przepiórska K, Pietrzak BA and
Kajta M: Emerging evidence on membrane estrogen receptors as novel
therapeutic targets for central nervous system pathologies. Int J
Mol Sci. 24(4043)2023.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Morelli E, Hunter ZR, Fulciniti M, Gullà
A, Perrotta ID, Zuccalà V, Federico C, Juli G, Manzoni M, Ronchetti
D, et al: Therapeutic activation of G protein-coupled estrogen
receptor 1 in Waldenström Macroglobulinemia. Exp Hematol Oncol.
11(54)2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Gao N, Liang T, Yuan Y, Xiao X, Zhao Y,
Guo Y, Li M and Pu X: Exploring the mechanism of F282L
mutation-caused constitutive activity of GPCR by a computational
study. Phys Chem Chem Phys. 18:29412–29422. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Tao YX: Molecular mechanisms of the neural
melanocortin receptor dysfunction in severe early onset obesity.
Mol Cell Endocrinol. 239:1–14. 2005.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Pupo M, Bodmer A, Berto M, Maggiolini M,
Dietrich PY and Picard D: A genetic polymorphism repurposes the
G-protein coupled and membrane-associated estrogen receptor GPER to
a transcription factor-like molecule promoting paracrine signaling
between stroma and breast carcinoma cells. Oncotarget.
8:46728–46744. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Tutzauer J, Sjöström M, Bendahl PO, Rydén
L, Fernö M, Fredrik Leeb-Lundberg LM and Alkner S: Plasma membrane
expression of G protein-coupled estrogen receptor (GPER)/G
protein-coupled receptor 30 (GPR30) is associated with worse
outcome in metachronous contralateral breast cancer. PLoS One.
15(e0231786)2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
R Core Team: R Core Team 2023 R: A
language and environment for statistical computing. R foundation
for statistical computing. https://www.R-project.org/. R Foundation for
Statistical Computing, 2023.
|
|
23
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140.
2009.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software Environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wu L, Yu H, Mo H, Lan X, Pan C, Wang L,
Zhao H, Zhou J and Li Y: Functional characterization of
melanocortin-3 receptor in a hibernating cavefish onychostoma
macrolepis. Animals. 12(38)2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Vesuna F, Winnard P and Raman V: Enhanced
green fluorescent protein as an alternative control reporter to
Renilla luciferase. Anal Biochem. 342:345–347. 2005.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Han J, Xiao N, Yang W, Luo S and Zhao J,
Qiang Y, Chaudhary S and Zhao J: MS-ResNet: Disease-specific
survival prediction using longitudinal CT images and clinical data.
Int J Comput Assist Radiol Surg. 17:1049–1057. 2022.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Li Z, Ding B, Xu J, Mao K, Zhang P and Xue
Q: Relevance of STK11 mutations regarding immune cell infiltration,
drug sensitivity, and cellular processes in lung adenocarcinoma.
Front Oncol. 10(580027)2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Sun J, Li S, Wang F, Fan C and Wang J:
Identification of key pathways and genes in pten mutation prostate
cancer by bioinformatics analysis. BMC Med Genet.
20(191)2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Fan C, Zhao C, Shugen Li FW and Wang J:
Significance of PTEN mutation in cellular process, prognosis, and
drug selection in clear cell renal cell carcinoma. Front Oncol.
9(357)2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ono M, Baden A, Okudaira H, Kobayashi M,
Kawai K, Oka S and Yoshimura H: Assessment of amino acid/drug
transporters for renal transport of [18F]fluciclovine
(Anti-[18F]FACBC) in vitro. Int J Mol Sci. 17(1730)2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Becchetti A, Munaron L and Arcangeli A:
The role of ion channels and transporters in cell proliferation and
cancer. Front Physiol. 4(312)2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhou F, Hong M and You G: Regulation of
human organic anion transporter 4 by progesterone and protein
kinase C in human placental BeWo cells. Am J Physiol Endocrinol
Metab. 293:E57–E61. 2007.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Xu W, Tanaka K, Sun AQ and You G:
Functional role of the C terminus of human organic anion
transporter hOAT. J Biol Chem. 281:31178–31183. 2006.PubMed/NCBI View Article : Google Scholar
|
|
36
|
You G: Towards an understanding of organic
anion transporters: Structure-function relationships. Med Res Rev.
24:762–774. 2004.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Baycin-Hizal D, Gottschalk A, Jacobson E,
Mai S, Wolozny D, Zhang H, Krag SS and Betenbaugh MJ: Physiologic
and pathophysiologic consequences of altered sialylation and
glycosylation on Ion channel function. Biochem Biophys Res Commun.
453:243–253. 2014.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zhang G, Yang H, Liang H, Yang J, Shi J,
McFarland K, Chen Y and Cui J: A charged residue in S4 regulates
coupling among the activation gate, voltage, and Ca2+ sensors in BK
channels. J Neurosci. 34:12280–12288. 2014.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Thompson AN, Posson DJ, Parsa PV and
Nimigean CM: Molecular mechanism of pH sensing in KcsA potassium
channels. Proc Natl Acad Sci USA. 105:6900–6905. 2008.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Yang R, Zhou Y, Du C and Wu Y:
Bioinformatics analysis of differentially expressed genes in tumor
and paracancerous tissues of patients with lung adenocarcinoma. J
Thorac Dis. 12:7355–7364. 2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Fan Z, Liu Y, Liu X, Nian W, Huang X, Yang
Q, Hou S and Chen F: Phosphorylation of AKT by lysyl oxidase-like 2
activates the PI3K/AKT signaling pathway to promote proliferation,
invasion and metastasis in esophageal squamous carcinoma. Clin
Transl Oncol. 25:2487–2498. 2023.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Chi M, Liu J, Mei C, Shi Y, Liu N, Jiang
X, Liu C, Xue N, Hong H, Xie J, et al: TEAD4 functions as a
prognostic biomarker and triggers EMT via PI3K/AKT pathway in
bladder cancer. J Exp Clin Cancer Res. 41(175)2022.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Fadhal E: A comprehensive analysis of the
PI3K/AKT pathway: Unveiling key proteins and therapeutic targets
for cancer treatment. Cancer Inform.
22(11769351231194273)2023.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Ahmad I, Hoque M, Alam SSM, Zughaibi TA
and Tabrez S: Curcumin and plumbagin synergistically target the
PI3K/Akt/mTOR pathway: A prospective role in cancer treatment. Int
J Mol Sci. 24(6651)2023.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Shi X, Wang J, Lei Y, Cong C, Tan D and
Zhou X: Research progress on the PI3K/AKT signaling pathway in
gynecological cancer (Review). Mol Med Rep. 19:4529–4535.
2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Jin T, Li D, Yang T, Liu F, Kong J and
Zhou Y: PTPN1 promotes the progression of glioma by activating the
MAPK/ERK and PI3K/AKT pathways and is associated with poor patient
survival. Oncol Rep. 42:717–725. 2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Jiang AG, Yu H and Huang JA: Expression
and clinical significance of the phosphatidylinositol
3-kinase/protein kinase B signal transduction pathway in non-small
cell lung carcinoma. Oncol Lett. 8:601–607. 2014.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Li X, Sun H, Hou Y and Jin W:
Comprehensive combined proteomics and genomics analysis identifies
prognostic related transcription factors in breast cancer and
explores the role of dmap1 in breast cancer. J Pers Med.
11(1068)2021.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Andersen V, Vogel LK, Kopp TI, Sæbø M,
Nonboe AW, Hamfjord J, Kure EH and Vogel U: High ABCC2 and low
ABCG2 gene expression are early events in the colorectal
adenoma-carcinoma sequence. PLoS One. 10(e0119255)2015.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Yamada A, Ishikawa T, Ota I, Kimura M,
Shimizu D, Tanabe M, Chishima T, Sasaki T, Ichikawa Y, Morita S, et
al: High expression of ATP-binding cassette transporter ABCC11 in
breast tumors is associated with aggressive subtypes and low
disease-free survival. Breast Cancer Res Treat. 137:773–782.
2013.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Sakil HAM, Stantic M, Wolfsberger J, Brage
SE, Hansson J and Wilhelm MT: ΔNp73 regulates the expression of the
multidrug-resistance genes ABCB1 and ABCB5 in breast cancer and
melanoma cells-a short report. Cell Oncol. 40:631–638.
2017.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Yuan Y, Xiang Z, Xia Y, Xie J, Jiang X and
Lu Z: The role of ATP binding cassette (ABC) transporters in breast
cancer: Evaluating prognosis, predicting immunity, and guiding
treatment. Channels (Austin). 17(2273247)2023.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Modi A, Roy D, Sharma S, Vishnoi JR,
Pareek P, Elhence P, Sharma P and Purohit P: ABC transporters in
breast cancer: Their roles in multidrug resistance and beyond. J
Drug Target. 30:927–947. 2022.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Zhao B, Xin Z, Ren P and Wu H: The role of
PPARs in breast cancer. Cells. 12(130)2023.
|
|
55
|
Nordgren M and Fransen M: Peroxisomal
metabolism and oxidative stress. Biochimie. 98:56–62.
2014.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Zhang J, Tripathi DN, Jing J, Alexander A,
Kim J, Powell RT, Dere R, Tait-Mulder J, Lee JH, Paull TT, et al:
ATM functions at the peroxisome to induce pexophagy in response to
ROS. Nat Cell Biol. 17:1259–1269. 2015.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Brandão-Costa RM, Helal-Neto E, Vieira AM,
Barcellos-De-souza P, Morgado-Diaz J and Barja-Fidalgo C:
Extracellular matrix derived from high metastatic human breast
cancer triggers epithelial-mesenchymal transition in epithelial
breast cancer cells through αvβ3 integrin. Int J Mol Sci.
21(2995)2020.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Yu TY, Zhang G, Chai XX, Ren L, Yin DC and
Zhang CY: Recent progress on the effect of extracellular matrix on
occurrence and progression of breast cancer. Life Sci.
332(122084)2023.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Wu JZ, Yang TJ, Lu P and Ma W: Analysis of
signaling pathways in recurrent breast cancer. Genet Mol Res.
13:10097–10104. 2014.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Huang Z, Xu E, Ma X, Wang Y, Zhu J, Zhu K,
Hu J and Zhang C: Low NT5DC2 expression predicts favorable
prognosis and suppresses soft tissue sarcoma progression via
ECM-receptor interaction pathway. Transl Oncol.
44(101937)2024.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Tang W, Putluri V, Ambati CR, Dorsey TH,
Putluri N and Ambs S: Liver-And Microbiome-derived bile acids
accumulate in human breast tumors and inhibit growth and improve
patient survival. Clin Cancer Res. 25:5972–5983. 2019.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Scott P, Anderson K, Singhania M and
Cormier R: Cystic fibrosis, CFTR, and colorectal cancer. Int J Mol
Sci. 21(2891)2020.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Wang S, Yu ZH and Chai KQ: Identification
of CFTR as a novel key gene in chromophobe renal cell carcinoma
through bioinformatics analysis. Oncol Lett. 18:1767–1774.
2019.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Wei RQ, Zhang WM, Liang Z, Piao C and Zhu
G: Identification of signal pathways and hub genes of pulmonary
arterial hypertension by bioinformatic analysis. Can Respir J.
2022(1394088)2022.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Kudo-Saito C, Miyamoto T, Imazeki H, Shoji
H, Aoki K and Boku N: IL33 is a key driver of treatment resistance
of cancer. Cancer Res. 80:1981–1990. 2021.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Fang M, Li Y, Huang K, Qi S, Zhang J,
Zgodzinski W, Majewski M, Wallner G, Gozdz S, Macek P, et al: IL33
promotes colon cancer cell stemness via JNK activation and
macrophage recruitment. Cancer Res. 77:2735–2745. 2017.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Twarda-Clapa A, Labuzek B, Krzemien D,
Musielak B, Grudnik P, Dubin G and Holak TA: Crystal structure of
the FAS1 domain of the hyaluronic acid receptor stabilin-2. Acta
Crystallogr D Struct Biol. 74:695–701. 2018.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Hirose Y, Saijou E, Sugano Y, Takeshita F,
Nishimura S, Nonaka H, Chen YR, Sekine K, Kido T, Nakamura T, et
al: Inhibition of Stabilin-2 elevates circulating hyaluronic acid
levels and prevents tumor metastasis. Proc Natl Acad Sci USA.
109:4263–4268. 2012.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Han MW, Lee JC, Park SY, Kim YM, Cho KJ,
Kim SW, Lee M, Nam SY, Kim IS and Kim SY: Homotypic interaction of
stabilin-2 plays a critical role in lymph node metastasis of tongue
cancer. Anticancer Res. 36:6611–6618. 2016.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Eissmann MF, Dijkstra C, Jarnicki A,
Phesse T, Brunnberg J, Poh AR, Etemadi N, Tsantikos E, Thiem S,
Huntington ND, et al: IL-33-mediated mast cell activation promotes
gastric cancer through macrophage mobilization. Nat Commun.
10(2735)2019.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Eissmann MF, Buchert M and Ernst M: IL33
and mast cells-the key regulators of immune responses in
gastrointestinal cancers? Front Immunol. 11(1389)2020.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Yoon HH, Orrock JM, Foster NR, Sargent DJ,
Smyrk TC and Sinicrope FA: Prognostic impact of FoxP3+ regulatory T
cells in relation to CD8+ T lymphocyte density in human colon
carcinomas. PLoS One. 7(e42274)2012.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Duhen T, Duhen R, Montler R, Moses J,
Moudgil T, de Miranda NF, Goodall CP, Blair TC, Fox BA, McDermott
JE, et al: Co-expression of CD39 and CD103 identifies
tumor-reactive CD8 T cells in human solid tumors. Nat Commun.
9(2724)2018.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Liu Z, Zhou Q, Wang Z, Zhang H, Zeng H,
Huang Q, Chen Y, Jiang W, Lin Z, Qu Y, et al: Intratumoral
TIGIT+CD8+T-cell infiltration determines poor prognosis and immune
evasion in patients with muscle-invasive bladder cancer. J
Immunother Cancer. 8(e000978)2020.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Ma L, Sun L, Zhao K, Dong Z, Huang Z and
Meng X: The prognostic value of TCF1+CD8+T in primary small cell
carcinoma of the esophagus. Cancer Sci. 112:4968–4976.
2021.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Yang B, Deng B, Jiao XD, Qin BD, Lu Y,
Zhang W, Guo Y, Chen S, Li D, Li B, et al: Low-dose anti-VEGFR2
therapy promotes anti-tumor immunity in lung adenocarcinoma by
down-regulating the expression of layilin on tumor-infiltrating
CD8+T cells. Cell Oncol (Dordr). 45:1297–1309. 2022.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Karkeni E, Morin SO, Bou Tayeh B, Goubard
A, Josselin E, Castellano R, Fauriat C, Guittard G, Olive D and
Nunès JA: Vitamin D controls tumor growth and CD8+ T Cell
infiltration in breast cancer. Front Immunol.
10(1307)2019.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Wang X, Jespers W, de Waal JJ, Wolff KAN,
van Uden L, IJzerman AP, van Westen GJP and Heitman LH:
Cancer-related somatic mutations alter adenosine A1 receptor
pharmacology-A focus on mutations in the loops and C-terminus.
FASEB J. 36(e22358)2022.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Dietzsch AN, Al-Hasani H, Altschmied J,
Bottermann K, Brendler J, Haendeler J, Horn S, Kaczmarek I, Körner
A, Krause K, et al: Dysfunction of the adhesion G protein-coupled
receptor latrophilin 1 (ADGRL1/LPHN1) increases the risk of
obesity. Signal Transduct Target Ther. 9(103)2024.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Zhang H, Kong Q, Wang J, Jiang Y and Hua
H: Complex roles of cAMP-PKA-CREB signaling in cancer. Exp Hematol
Oncol. 9(32)2020.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Rao R, Salloum R, Xin M and Lu QR: The G
protein Gαs acts as a tumor suppressor in sonic hedgehog
signaling-driven tumorigenesis. Cell Cycle. 15:1325–1330.
2016.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Li Y, Zhou X, Liu J, Yin Y, Yuan X, Yang
R, Wang Q, Ji J and He Q: Differentially expressed genes and key
molecules of BRCA1/2-mutant breast cancer: Evidence from
bioinformatics analyses. PeerJ. 8(e8403)2020.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Zhu F, Huang R, Li J, Liao X, Huang Y and
Lai Y: Identification of key genes and pathways associated with
RUNX1 mutations in acute myeloid leukemia using bioinformatics
analysis. Medical Science Monitor. 24:7100–7108. 2018.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Chen S, Chen Y, Lu J, Yuan D, He L, Tan H
and Xu L: Bioinformatics analysis identifies key genes and pathways
in acute myeloid leukemia associated with DNMT3A mutation. Biomed
Res Int. 2020(9321630)2020.PubMed/NCBI View Article : Google Scholar
|