Introduction
Osteosarcoma is the eighth most common type of
cancer found in children and adolescents, accounting for 2.4% of
all malignancies in pediatric patients and ~20% of all primary bone
cancers (1). Chemotherapy is the
first choice treatment for osteosarcoma, with multiple anticancer
drugs, including doxorubicin, cisplatin and high-dose methotrexate
(2,3). In the last three decades, neoadjuvant
chemotherapy has increased the long-term survival rate of
osteosarcoma patients from <20 to ~80% (4–6).
However, patients that are less responsive to these drugs have a
poor prognosis. In addition, the frequent acquisition of drug
resistance and the occurrence of ‘secondary malignancies’ are often
associated with chemotherapy and are significant obstacles to
achieving favorable outcomes. Thus, it is important to identify the
molecular mechanisms underlying the drug resistance of osteosarcoma
cancer cells.
Drug resistance of osteosarcoma cancer cells has
been attributed to various mechanisms, including dysfunctional
membrane transport (7), resistance
to apoptosis (8), persistence of
stem cell-like tumor cells (9) or
autophagy (7). In addition,
osteosarcoma tumors have been reported to exhibit a multidrug
resistance phenotype (10).
Autophagy is a lysosomal degradation pathway that is essential for
cell growth, survival, differentiation, development and homeostasis
(11). It is a tightly regulated
process that helps to maintain a balance among the synthesis,
degradation and subsequent recycling of cellular products. A number
of studies have demonstrated a critical role for autophagy in
cancer development and therapy (9,12,13).
Previous studies hypothesized that autophagy is facilitated
following treatment with cytotoxic agents (14–17).
However, the mechanisms underlying autophagic drug resistance in
osteosarcoma therapy remain largely unknown.
MicroRNAs (miRNAs) are family of endogenous
non-coding RNA molecules that are comprised of 22 nucleotides,
which regulate gene expression (18) in organisms ranging between
nematodes and humans and in a broad array of mammalian cell
processes (19). Recently, miRNAs
have been associated with cell chemosensitivity or chemotherapy
resistance in a variety of cancer cell types (20–24),
including osteosarcoma (25).
miR-140 was reported to be involved in the chemoresistance of
osteosarcoma cells via the suppression of histone deacetylase 4,
which in turn reduced cell proliferation (25). Furthermore, an increasing number of
studies have demonstrated that miRNA molecules regulate cellular
autophagy processes (26–28). Zhu et al (27) reported that miR-30a targets beclin
1, resulting in decreased autophagic activity. In addition, Brest
et al (28) showed that a
miR-196-based alteration in the expression of immunity-related
GTPase family M protein can affect the efficacy of autophagy.
However, the role of miRNAs in autophagy-mediated chemotherapy
resistance in osteosarcoma remains unknown.
The aim of the present study was to investigate the
expression levels of miR-155 in osteosarcoma cells following
chemotherapy and analyze the association with chemotherapy
resistance in vitro. The effect that miR-155 overexpression
exhibited on autophagy was also investigated with the aim of
demonstrating a novel role for miR-155 in chemotherapy resistance
during the treatment of osteosarcoma.
Materials and methods
Cell culture and reagents
Human osteosarcoma cell lines (Saos-2 and MG-63)
were obtained from the Cell Resource Center of the Chinese Academy
of Medical Sciences (Beijing, China). The cells were cultured in
Eagle’s Minimum Essential Medium (Invitrogen, Carlsbad, CA, USA) or
McCoy’s 5A Modified Medium (Invitrogen) supplemented with 10% fetal
bovine serum (GIBCO, Rockville, MD, USA), and were incubated at
37°C with 5% CO2. Antibodies against GAPDH, LC3-II and
autophagy protein 5 (Atg5) were obtained from Santa Cruz
Biotechnology, Inc. (Santa Cruz, CA, USA) and rapamycin was
purchased from Sigma-Aldrich (St. Louis, MO, USA). The coding
sequence of microtubule-associated protein 1-LC3 fusion with green
fluorescent protein (GFP) was synthesized and cloned into
pcDNA3.1(+) to construct the LC3-GFP-expressing plasmid.
Autophagic vesicles quantified by
GFP-LC3
Quantitative GFP-LC3 light microscopy autophagy
assays were performed in Saos-2 cells with various treatments.
Cells were grown to 80% confluency and were transfected with a
GFP-LC3-expressing plasmid using Lipofectamine 2000 (Invitrogen
Life Technologies, Carlsbad, CA, USA). At 24 h following
transfection, the cells were subjected to 50 nM rapamycin
(Sigma-Aldrich), 0.2 μg/ml doxorubicin (Dox; Sigma-Aldrich) or 20
μM cisplatin (Cis; Sigma-Aldrich) for an additional 24 h. In a
separate experiment, cells were simultaneously and additionally
transfected with 20 nM miR-155 and analyzed with fluorescence
microscopy. The number of punctate GFP-LC3 dots in each cell was
counted and at least 100 cells were included for each group.
miRNA extraction and quantitative
polymerase chain reaction (qPCR)
Total miRNA extraction was performed using a mirVana
miRNA Isolation kit (Ambion, Inc., Austin, TX, USA). Quantification
of miR-155 expression was conducted using the mirVana qRT-PCR miRNA
Detection kit (Ambion, Inc.), where U6 small nuclear RNA was used
as an internal control. The ΔΔCt method was used for
relative quantification (29). A
non-radioactive northern blot method (LED) for small RNA (15–40
bases) detection, using digoxigenin-labeled oligonucleotide probes
containing locked nucleic acids (Roche Diagnostics, GmbH, Mannheim,
Germany) and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide,
(Sigma-Aldrich was utilized to confirm the miR-155 and U6
expression levels, according to the protocol previously described
(30).
Western blot analysis
Cell extracts were prepared according to the
standard protocol, and protein expression levels were detected by
western blot analysis using polyclonal (rabbit) anti-LC3-II,
anti-Atg5 or anti-GAPDH antibodies. Goat anti-mouse IgG or goat
anti-rabbit IgG (Pierce Biotechnology, Inc., Rockford, IL, USA)
secondary antibodies, that were conjugated to horseradish
peroxidase, were used for detection via an enhanced
chemiluminescence detection system (Super Signal West Femto, Pierce
Biotechnology, Inc.).
Cell proliferation assay
Cell viability was expressed as the relative
percentage of viable cells to control human umbilical vein
endothelial cells. For the proliferation assay, following
transfection with miR-155 mimics or miR-155 control, cells were
incubated with Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc., Kumamoto, Japan). The absorbance of each well
at 450 nm was detected following visual color occurrence at 24, 48
or 72 h. Independent experiments were performed in triplicate.
Statistical analysis
For GFP-LC3 dot number analysis, relative miR-155
expression, conversion of LC3-I to LC3-II, relative expression of
Atg5 against GAPDH and CCK-8 measurements, the statistical
evaluations are presented as the mean ± SE. Data were analyzed
using the Student’s t test. All data were analyzed by the SPSS
v16.0 (SPSS Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant result.
Results
Autophagy is induced during chemotherapy
in osteosarcoma cells
An increasing number of studies have indicated that
autophagy is induced in cancer cells following treatment with
cytotoxic agents (14–17). To confirm the autophagy level in
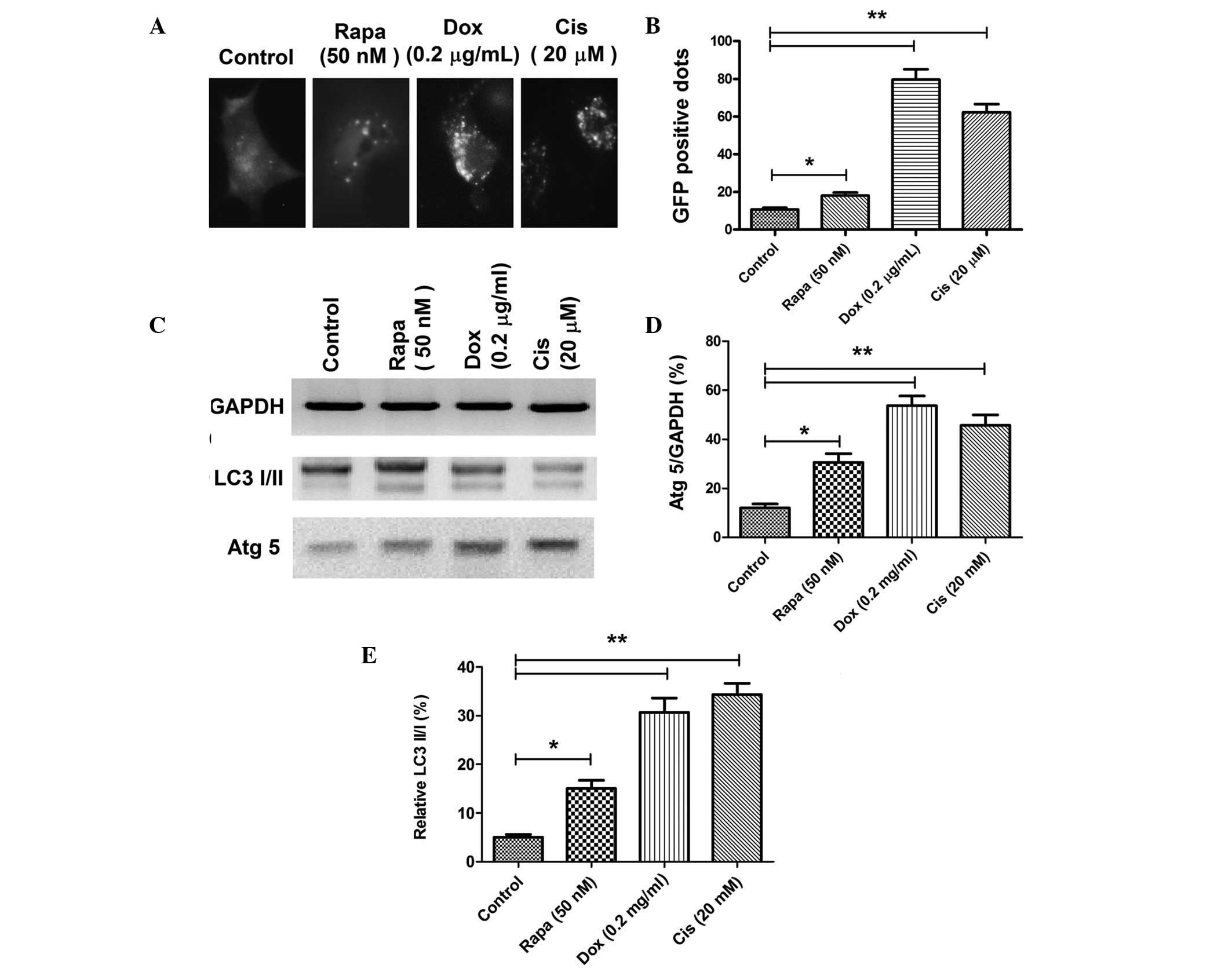
osteosarcoma cells following Dox or Cis treatment, autophagic
acidic vesicular organelles and LC3 punctas (31) were detected via a GFP-LC3 report
vector and fluorescence microscopy in the Saos-2 osteosarcoma cell
line. The conversion between LC3-I and LC3-II, as well as Atg5
expression, one of the autophagy-related gene products, was then
quantified by western blot analysis. Accumulation of LC3 punctas in
osteosarcoma cells was significantly higher in the Dox (0.2 μg/ml)
or Cis (20 μM) treatment groups (Fig.
1A); there were more GFP-positive dots (autophagic vesicles) in
the Dox- or Cis-treated Saos-2 cells when compared with the control
Saos-2 cells (P<0.05; Fig. 1B).
In addition, immunoblot analysis revealed significantly higher
conversion levels of LC3-I to LC3-II and a higher level of Atg5
expression in the osteosarcoma cells that had been treated with 0.2
μg/ml Dox or 20 μM Cis (Fig.
1C–E). These observations indicated that chemotherapy in
osteosarcoma cells induces autophagy.
miR-155 expression increases in
osteosarcoma cells following treatment with chemotherapy
agents
The role of miRNAs in chemotherapy-induced autophagy
of cancer cells remains unknown. To screen possible miRNAs that may
be important for anticancer drug-induced autophagy in osteosarcoma
cells, miRNA expression levels were analyzed by microarray in
osteosarcoma cells following treatment with Dox (data not shown).
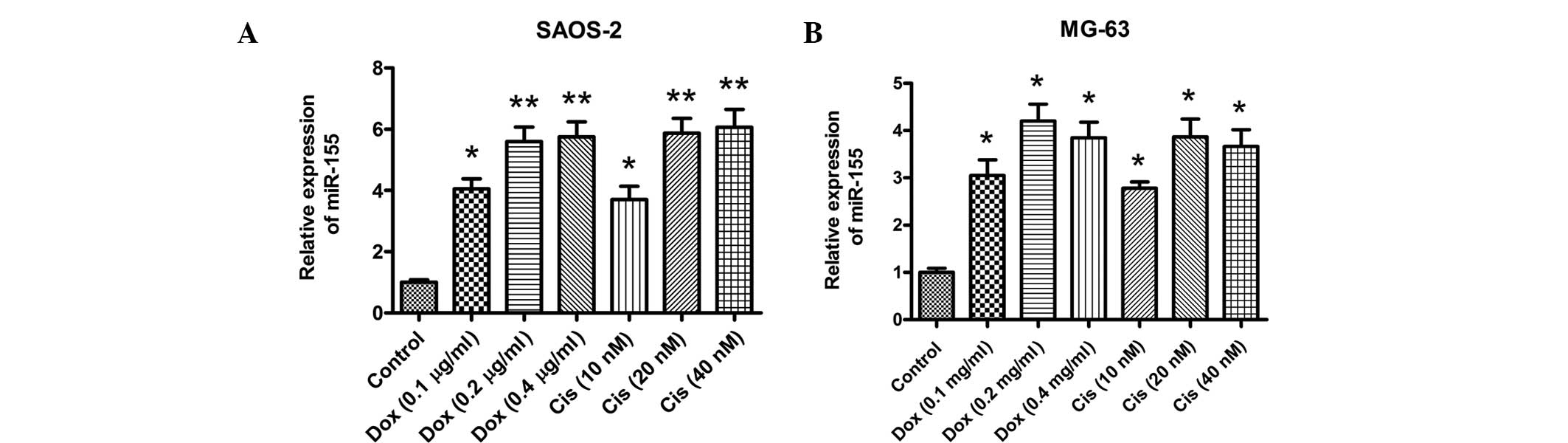
miR-155 was demonstrated to be the most highly-expressed miRNA.
Thus, the expression level of miR-155 was quantified in Saos-2 and
MG-63 cells following treatment with Dox or Cis. The results
indicated that treatment with 0.2 μg/ml Dox or 20 μM Cis
significantly upregulated the miR-155 expression levels in the two
cell lines. A qPCR assay demonstrated that significantly higher
expression levels of miR-155 were induced in Saos-2 or MG-63 cells
following Dox or Cis treatment (Fig.
2A and B). Therefore, miR-155 expression is induced in
vitro during anticancer drug therapy in osteosarcoma cells.
Overexpression of miR-155 ameliorates the
anticancer drug-induced cell proliferation decrease
To determine the possible effect of miR-155 on
osteosarcoma cell proliferation, the proliferation of Saos-2 or
MG-63 cells that had been treated with Dox and transfected with
miR-155 mimics was determined using a CCK-8 assay. Transfection
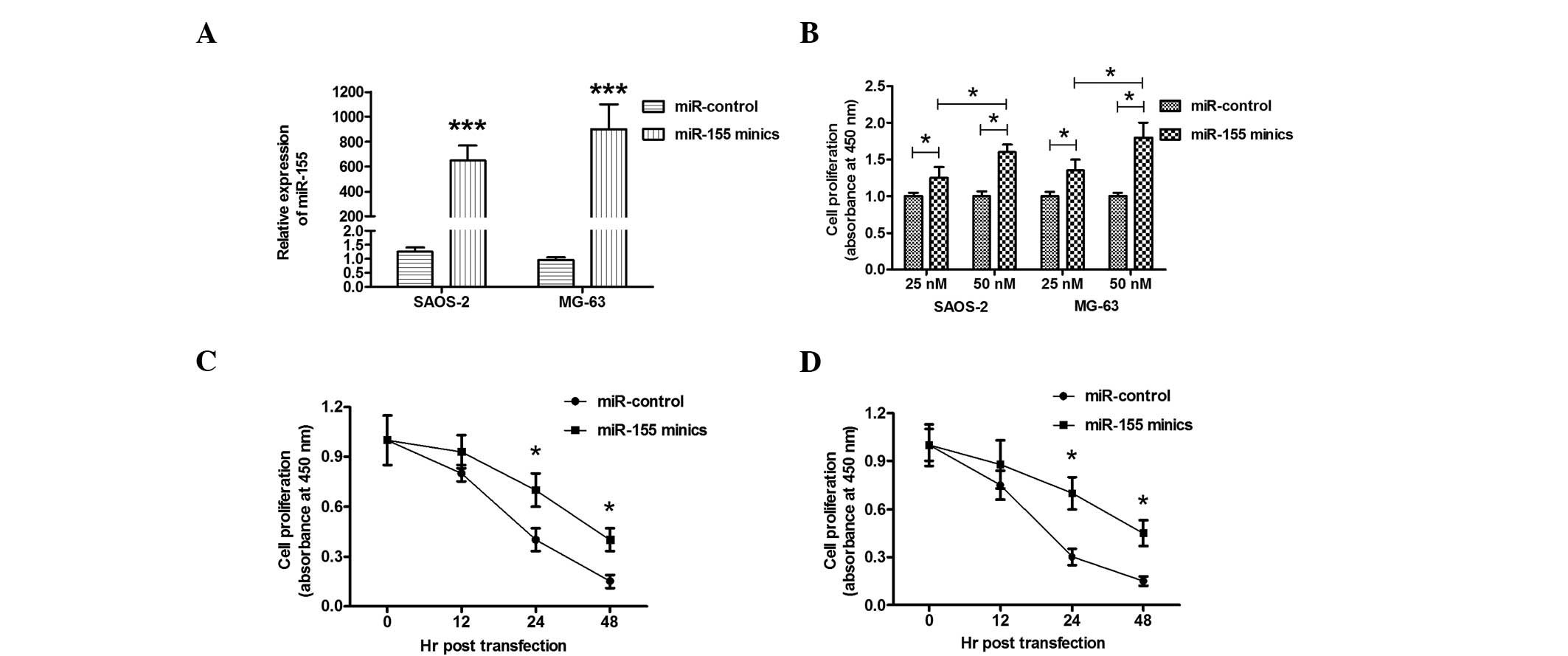
with miR-155 mimics significantly upregulated the miR-155 level in
Saos-2 or MG-63 cells (Fig. 3A).
The elevated miR-155 expression in the Saos-2 or MG-63 cell lines
ameliorated the cell proliferation retardation that had been caused
by Dox in a dose-dependent manner, when compared with the miRNA
control (Fig. 3B). To further
observe the effect of miR-155 overexpression on cell proliferation,
the proliferation of Saos-2 or MG-63 cells was determined at
various time points following miR-155 mimics transfection. As shown
in Fig. 3C and D, miR-155 mimics
transfection resulted in a time-dependent amelioration of the cell
proliferation decrease in Saos-2 and MG-63 cells. Thus,
overexpression of miR-155 ameliorated the anticancer drug-induced
cell proliferation decrease in osteosarcoma cells.
Overexpression of miR-155 upregulates
anticancer drug-induced autophagy in osteosarcoma cells
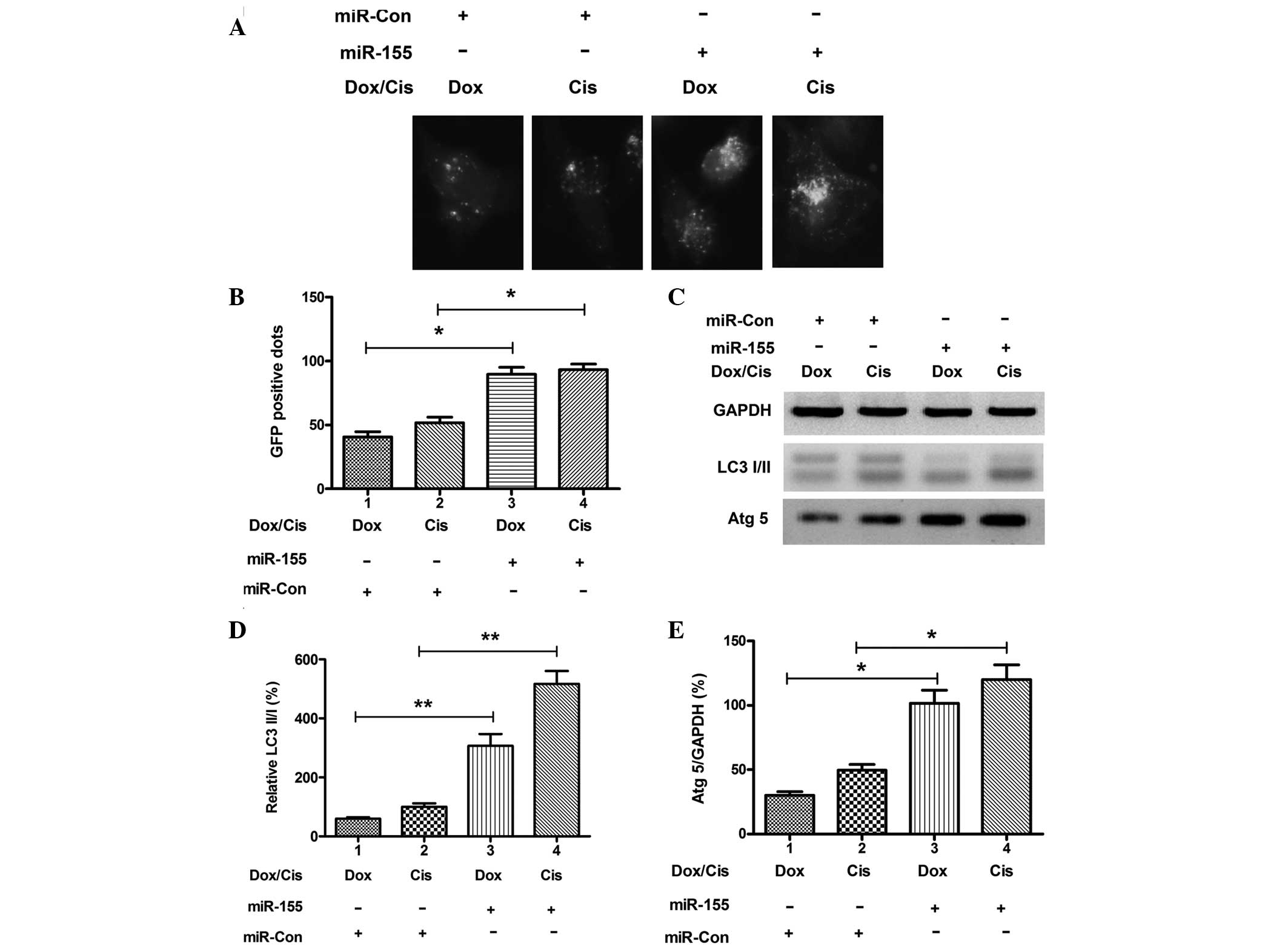
To determine the possible contribution of miR-155 to
autophagy in drug-treated osteosarcoma cells, miR-155 expression
was manipulated in Saos-2 cells via transfection with miR-155
mimics or miRNA control. Transfection with miR-155 mimics
significantly upregulated the miR-155 expression level in the cells
(Fig. 3A; P<0.001). The level
of autophagy was determined in Saos-2 cells following miR-155
mimics transfection. As shown in Fig.
4A and B, there were more GFP-positive dots (LC3 punctas) in
the Saos-2 cells that had been transfected with miR-155 mimics when
compared with transfection with miRNA control (P<0.05). In
addition, significantly higher conversion levels of LC3-I to LC3-II
and high expression levels of Atg5 were also confirmed in the
osteosarcoma cells transfected with miR-155 mimics (P<0.01 and
P<0.05 respectively; Fig.
4C–E). These results confirm that overexpression of miR-155
contributes to anticancer drug-induced autophagy in osteosarcoma
cells.
Discussions
Chemoresistance to anticancer therapeutic drugs is a
common occurrence and contributes to cancer mortality, as it often
leads to failure in the blockage of disease progression. A number
of studies have evaluated the mechanisms of resistance and the
biological factors involved (32).
Autophagy is an intracellular self-protective mechanism that
prevents the toxic accumulation of damaged components, but also
recycles the degraded components to sustain metabolic homeostasis.
Upregulated autophagy has been found in a wide variety of cancer
cells faced with metabolic and therapeutic stress, and has been
shown to contribute to the chemotherapy resistance of various types
of tumors (4,33). Blocking cancer cell autophagy is
emerging as a novel approach to enhance the efficiency of
chemotherapy in cancer treatment (8,9). The
results of the present study demonstrated that treatment with Dox
or Cis caused the activation of autophagy in osteosarcoma cells,
which enhanced autophagy and facilitated the survival of tumor
cells under these drug treatments. The results from the in
vitro experiments demonstrated that miR-155 expression was also
enhanced in osteosarcoma cells following anticancer treatment, and
the upregulated miR-155 expression mediated the autophagy.
Tight control of autophagy is essential for normal
or tumor cells to survive, and recent advances in this field have
begun to unveil the molecular mechanisms underlying autophagy
regulation (34). Within the past
decade, genetic screening in yeast has identified a large family of
core autophagy regulators, the Atg-related genes, a number of which
have known orthologs in mammalian cells that serve to coordinately
regulate the stepwise progression of this degradation pathway
(35,36). In addition, a diverse and complex
network of upstream signaling pathways contribute to autophagy
regulation, including the phosphatidylinositol 3-kinase, RAS and
AMP-activated protein kinase pathways, a number of which converge
at mammalian target of rapamycin complex 1, a key negative
regulator of autophagy signaling (37,38).
There is much evidence that miRNA molecules are differentially
expressed in human cancers, in which they function as tumor
suppressors or oncogenes, with their oncogenic regulation spanning
from initiation, progression to metastasis and treatment
sensitivity. Notably, miRNAs can regulate a multitude of targets
and biological networks in autophagy (39,40).
A previous study indicated clear roles of miRNAs in autophagy
induction, autophagic vesicle nucleation, autophagic vesicle
elongation and vesicle fusion to lysosomes (39). The present study confirmed that
during treatment with Dox or Cis in osteosarcoma cells, miR-155
expression was strongly induced. The increased miR-155 expression
facilitated tumor cell proliferation via upregulating autophagy,
thus, facilitated the resistance of osteosarcoma cells to Dox or
Cis. However, the details underlying the mechanism of
miR-155-mediated autophagic resistance in osteosarcoma cells
requires further study.
In conclusion, the present study has demonstrated
that anticancer drug treatment upregulates miR-155 expression in
osteosarcoma cells. Overexpression of miR-155 induces the
activation of autophagy, which promotes tumor cell survival and
chemoresistance. These observations reveal a novel role for miR-155
in chemotherapy resistance during the treatment of
osteosarcoma.
References
|
1
|
Ottaviani G and Jaffe N: The epidemiology
of osteosarcoma. Cancer Treat Res. 152:3–13. 2009. View Article : Google Scholar
|
|
2
|
Wittig JC, Bickels J, Priebat D, et al:
Osteosarcoma: a multidisciplinary approach to diagnosis and
treatment. Am Fam Physician. 65:1123–1132. 2002.PubMed/NCBI
|
|
3
|
Rosen G, Caparros B, Groshen S, et al:
Primary osteogenic sarcoma of the femur: a model for the use of
preoperative chemotherapy in high risk malignant tumors. Cancer
Invest. 2:181–192. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang ZJ, Chee CE, Huang S and Sinicrope
FA: The role of autophagy in cancer: therapeutic implications. Mol
Cancer Ther. 10:1533–1541. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Provisor AJ, Ettinger LJ, Nachman JB, et
al: Treatment of nonmetastatic osteosarcoma of the extremity with
preoperative and postoperative chemotherapy: a report from the
Children’s Cancer Group. J Clin Oncol. 15:76–84. 1997.
|
|
6
|
Goorin AM, Schwartzentruber DJ, Devidas M,
et al; Pediatric Oncology Group. Presurgical chemotherapy compared
with immediate surgery and adjuvant chemotherapy for nonmetastatic
osteosarcoma: Pediatric Oncology Group Study POG-8651. J Clin
Oncol. 21:1574–1580. 2003. View Article : Google Scholar
|
|
7
|
Huang J, Ni J, Liu K, et al: HMGB1
promotes drug resistance in osteosarcoma. Cancer Res. 72:230–238.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Amaravadi RK, Lippincott-Schwartz J, Yin
XM, et al: Principles and current strategies for targeting
autophagy for cancer treatment. Clin Cancer Res. 17:654–666. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Livesey KM, Tang D, Zeh HJ and Lotze MT:
Autophagy inhibition in combination cancer treatment. Curr Opin
Investig Drugs. 10:1269–1279. 2009.PubMed/NCBI
|
|
10
|
Kang R, Tang D, Schapiro NE, et al: The
receptor for advanced glycation end products (RAGE) sustains
autophagy and limits apoptosis, promoting pancreatic tumor cell
survival. Cell Death Differ. 17:666–676. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mizushima N, Levine B, Cuervo AM and
Klionsky DJ: Autophagy fights disease through cellular
self-digestion. Nature. 451:1069–1075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
White E and DiPaola RS: The double-edged
sword of autophagy modulation in cancer. Clin Cancer Res.
15:5308–5316. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mathew R, Karantza-Wadsworth V and White
E: Role of autophagy in cancer. Nat Rev Cancer. 7:961–967. 2007.
View Article : Google Scholar
|
|
14
|
Tang D, Kang R, Livesey KM, et al:
Endogenous HMGB1 regulates autophagy. J Cell Biol. 190:881–892.
2010. View Article : Google Scholar
|
|
15
|
Tang D, Kang R, Livesey KM, et al:
High-mobility group box 1 is essential for mitochondrial quality
control. Cell Metab. 13:701–711. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu L, Yang M, Kang R, et al:
HMGB1-induced autophagy promotes chemotherapy resistance in
leukemia cells. Leukemia. 25:23–31. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tang D, Kang R, Cheh CW, et al: HMGB1
release and redox regulates autophagy and apoptosis in cancer
cells. Oncogene. 29:5299–5310. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ambros V: MicroRNA pathways in flies and
worms: growth, death, fat, stress, and timing. Cell. 113:673–676.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Brennecke J, Hipfner DR, Stark A, Russell
RB and Cohen SM: Bantam encodes a developmentally regulated
microRNA that controls cell proliferation and regulates the
proapoptotic gene hid in Drosophila. Cell. 113:25–36. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zou Z, Wu L, Ding H, et al: MicroRNA-30a
sensitizes tumor cells to cis-platinum via suppressing beclin
1-mediated autophagy. J Biol Chem. 287:4148–4156. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hu H, Li S, Cui X, et al: The
overexpression of hypomethylated miR-663 induces chemotherapy
resistance in human breast cancer cells by targeting heparin
sulfate proteoglycan 2 (HSPG2). J Biol Chem. 288:10973–10985. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bourguignon LY, Wong G, Earle C and Chen
L: Hyaluronan-CD44v3 interaction with Oct4-Sox2-Nanog promotes
miR-302 expression leading to self-renewal, clonal formation, and
cisplatin resistance in cancer stem cells from head and neck
squamous cell carcinoma. J Biol Chem. 287:32800–32824. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kastl L, Brown I and Schofield AC:
miRNA-34a is associated with docetaxel resistance in human breast
cancer cells. Breast Cancer Res Treat. 131:445–454. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tao J, Lu Q, Wu D, et al: microRNA-21
modulates cell proliferation and sensitivity to doxorubicin in
bladder cancer cells. Oncol Rep. 25:1721–1729. 2011.PubMed/NCBI
|
|
25
|
Song B, Wang Y, Xi Y, et al: Mechanism of
chemoresistance mediated by miR-140 in human osteosarcoma and colon
cancer cells. Oncogene. 28:4065–4074. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mukhopadhyay P, Pacher P and Das DK:
MicroRNA signatures of resveratrol in the ischemic heart. Ann NY
Acad Sci. 1215:109–116. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhu H, Wu H, Liu X, et al: Regulation of
autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer
cells. Autophagy. 5:816–823. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Brest P, Lapaquette P, Souidi M, et al: A
synonymous variant in IRGM alters a binding site for miR-196 and
causes deregulation of IRGM-dependent xenophagy in Crohn’s disease.
Nat Genet. 43:242–245. 2011.PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−delta delta C(T)) method. Methods. 25:402–408. 2001.
|
|
30
|
Kim SW, Li Z, Moore PS, et al: A sensitive
non-radioactive northern blot method to detect small RNAs. Nucleic
Acids Res. 38:e982010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Paglin S, Hollister T, Delohery T, et al:
A novel response of cancer cells to radiation involves autophagy
and formation of acidic vesicles. Cancer Res. 61:439–444.
2001.PubMed/NCBI
|
|
32
|
Rebucci M and Michiels C: Molecular
aspects of cancer cell resistance to chemotherapy. Biochemical
Pharmacology. 85:1219–1226. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rosenfeldt MT and Ryan KM: The multiple
roles of autophagy in cancer. Carcinogenesis. 32:955–963. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
He C and Klionsky DJ: Regulation
mechanisms and signaling pathways ofautophagy. Annu Rev Genet.
43:67–93. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xie Z and Klionsky DJ: Autophagosome
formation: core machinery and adaptations. Nat Cell Biol.
9:1102–1109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang Z and Klionsky DJ: Mammalian
autophagy: core molecular machinery and signaling regulation. Curr
Opin Cell Biol. 22:124–131. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
He C and Klionsky DJ: Regulation
mechanisms and signaling pathways of autophagy. Annu Rev Genet.
43:67–93. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang Z and Klionsky DJ: Eaten alive: a
history of macroautophagy. Nat Cell Biol. 12:814–822. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Frankel LB and Lund AH: MicroRNA
regulation of autophagy. Carcinogenesis. 33:2018–2025. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fu LL, Wen X, Bao JK and Liu B:
MicroRNA-modulated autophagic signaling networks in cancer. Int J
Biochem Cell Biol. 44:733–736. 2012. View Article : Google Scholar : PubMed/NCBI
|