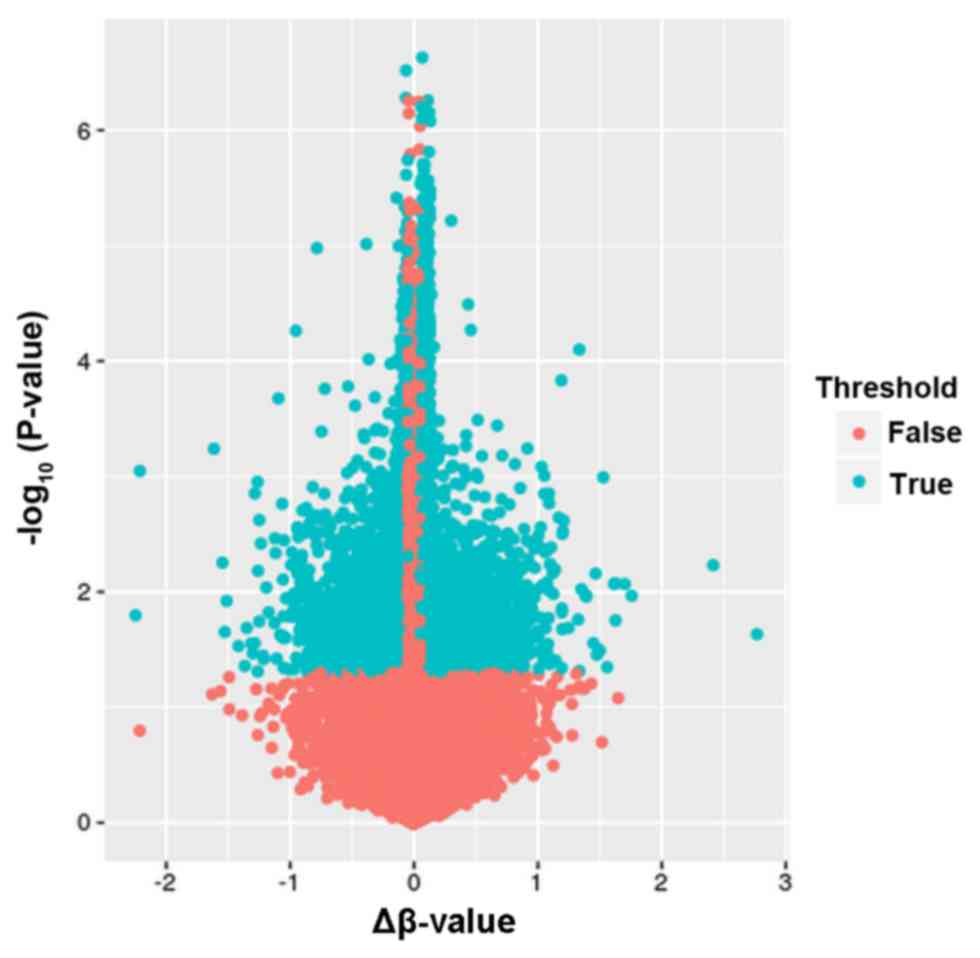
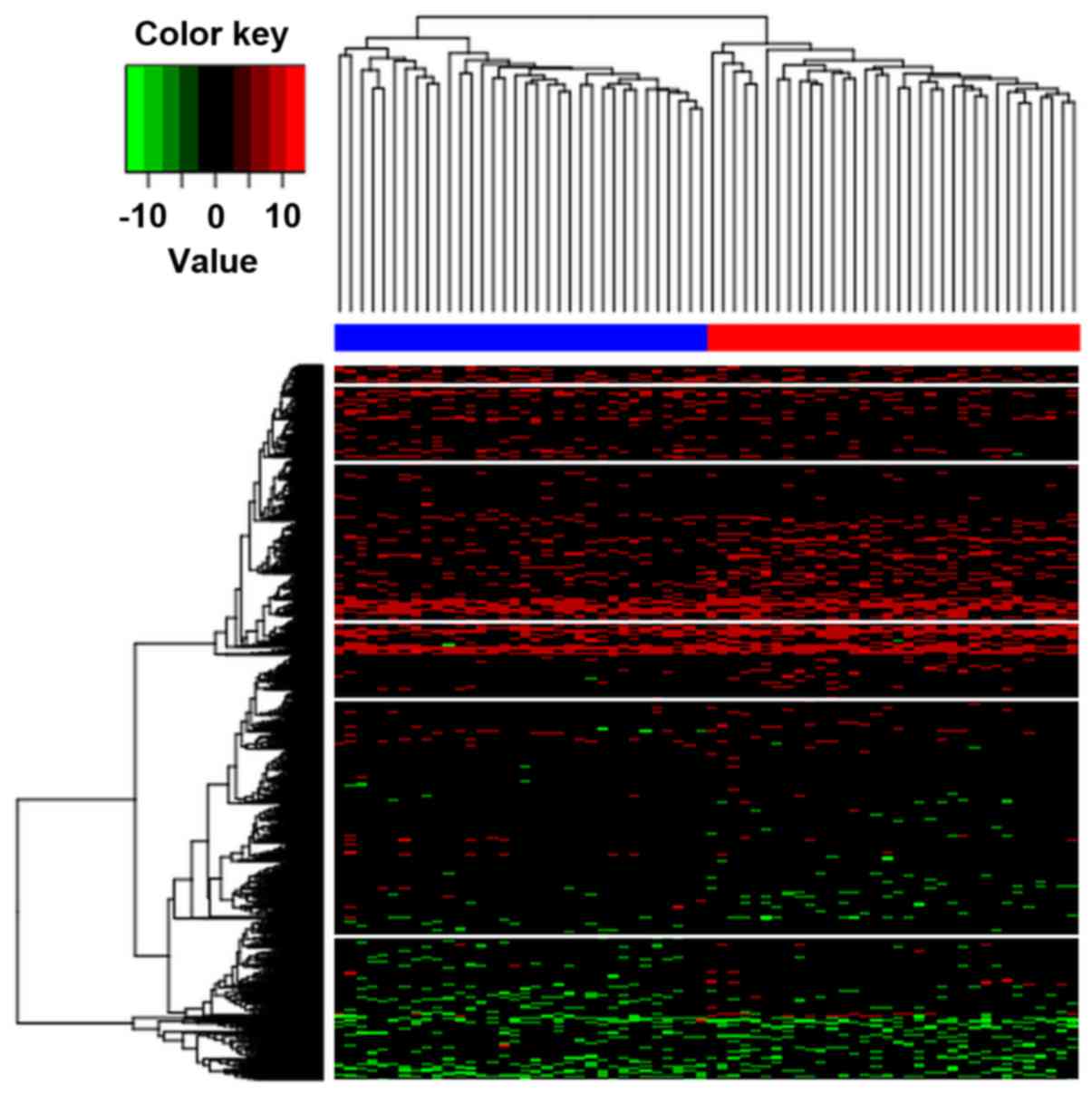
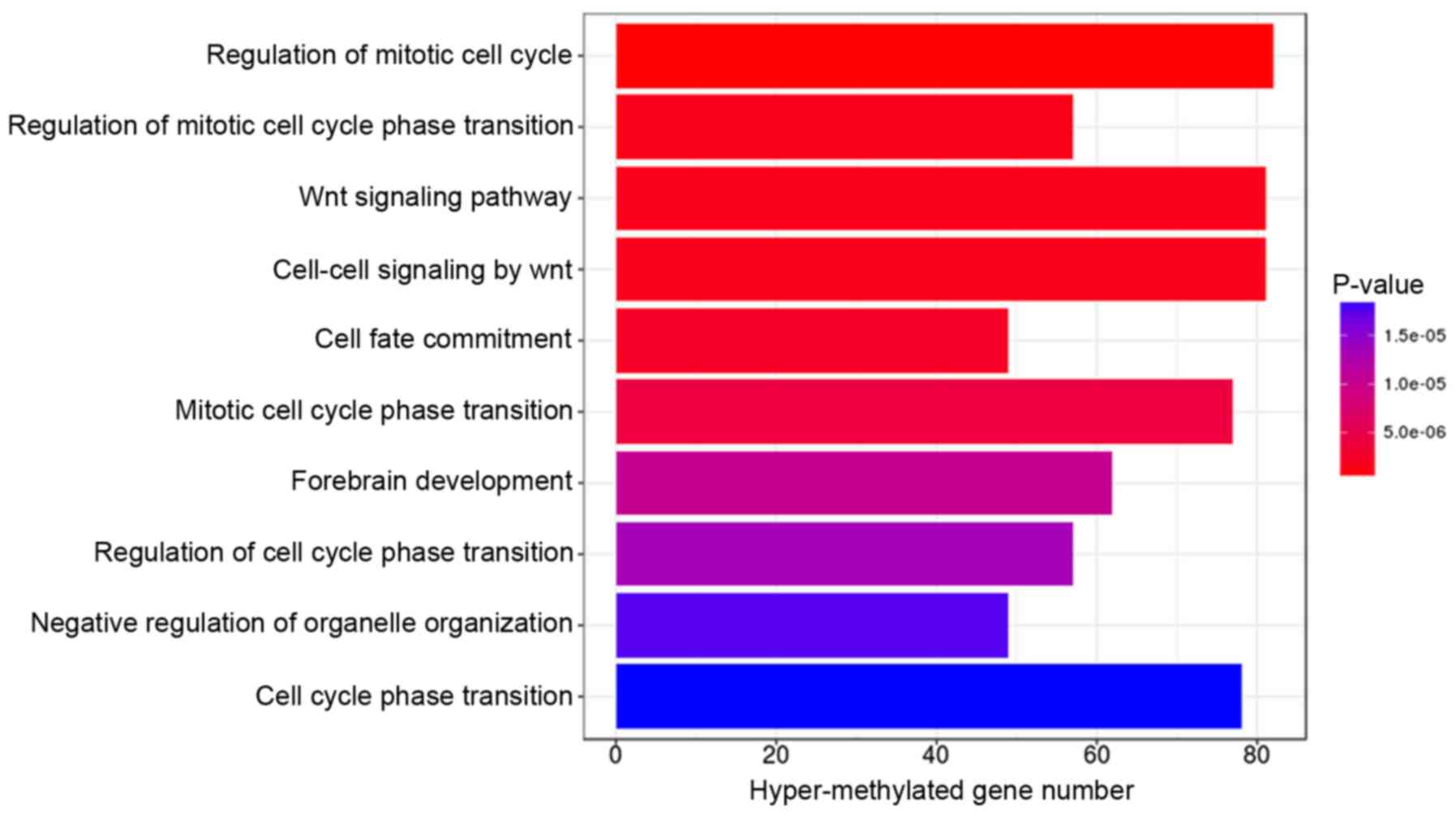
|
1
|
Wilson RS, Segawa E, Boyle PA, Anagnos SE,
Hizel LP and Bennett DA: The natural history of cognitive decline
in Alzheimer's disease. Psychol Aging. 27:1008–1017. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Association As: 2017 Alzheimer's disease
facts and figures. Alzheimers Dement. 13:325–37310. 2017.
View Article : Google Scholar
|
|
3
|
Selkoe DJ: Translating cell biology into
therapeutic advances in Alzheimer's disease. Nature. 399(6738
Suppl): A23–A31. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Migliore L and Coppedè F: Genetics,
environmental factors and the emerging role of epigenetics in
neurodegenerative diseases. Mutat Res. 667:82–97. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Iatrou A, Kenis G, Rutten BP, Lunnon K and
van den Hove DL: Epigenetic dysregulation of brainstem nuclei in
the pathogenesis of Alzheimer's disease: Looking in the correct
place at the right time? Cell Mol Life Sci. 74:509–523. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gatz M, Reynolds CA, Fratiglioni L,
Johansson B, Mortimer JA, Berg S, Fiske A and Pedersen NL: Role of
genes and environments for explaining Alzheimer disease. Arch Gen
Psychiatry. 63:168–174. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bird A: Perceptions of epigenetics.
Nature. 447:396–398. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jones PA and Takai D: The role of DNA
methylation in mammalian epigenetics. Science. 293:1068–1070. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ziller MJ, Gu H, Müller F, Donaghey J,
Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein
BE, et al: Charting a dynamic DNA methylation landscape of the
human genome. Nature. 500:477–481. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Smith ZD and Meissner A: DNA methylation:
Roles in mammalian development. Nat Rev Genet. 14:204–220. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang J, Yu JT, Tan MS, Jiang T and Tan L:
Epigenetic mechanisms in Alzheimer's disease: Implications for
pathogenesis and therapy. Ageing Res Rev. 12:1024–1041. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bollati V, Galimberti D, Pergoli L, Valle
E Dalla, Barretta F, Cortini F, Scarpini E, Bertazzi PA and
Baccarelli A: DNA methylation in repetitive elements and Alzheimer
disease. Brain Behav Immun. 25:1078–1083. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bakulski KM, Dolinoy DC, Sartor MA,
Paulson HL, Konen JR, Lieberman AP, Albin RL, Hu H and Rozek LS:
Genome-wide DNA methylation differences between late-onset
Alzheimer's disease and cognitively normal controls in human
frontal cortex. J Alzheimers Dis. 29:571–588. 2012.PubMed/NCBI
|
|
14
|
Hannum G, Guinney J, Zhao L, Zhang L,
Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, et al:
Genome-wide methylation profiles reveal quantitative views of human
aging rates. Mol Cell. 49:359–367. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bussière T, Gold G, Kövari E,
Giannakopoulos P, Bouras C, Perl DP, Morrison JH and Hof PR:
Stereologic analysis of neurofibrillary tangle formation in
prefrontal cortex area 9 in aging and Alzheimer's disease.
Neuroscience. 117:577–592. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
von Gunten A, Kövari E, Rivara CB, Bouras
C, Hof PR and Giannakopoulos P: Stereologic analysis of hippocampal
Alzheimer's disease pathology in the oldest-old: Evidence for
sparing of the entorhinal cortex and CA1 field. Exp Neurol.
193:198–206. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ginsberg SD, Hemby SE, Lee VM, Eberwine JH
and Trojanowski JQ: Expression profile of transcripts in
Alzheimer's disease tangle-bearing CA1 neurons. Ann Neurol.
48:77–87. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Haroutunian V, Katsel P and Schmeidler J:
Transcriptional vulnerability of brain regions in Alzheimer's
disease and dementia. Neurobiol Aging. 30:561–573. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Watson CT, Roussos P, Garg P, Ho DJ, Azam
N, Katsel PL, Haroutunian V and Sharp AJ: Genome-wide DNA
methylation profiling in the superior temporal gyrus reveals
epigenetic signatures associated with Alzheimer's disease. Genome
Med. 8:52016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huynh JL, Garg P, Thin TH, Yoo S, Dutta R,
Trapp BD, Haroutunian V, Zhu J, Donovan MJ, Sharp AJ and Casaccia
P: Epigenome-wide differences in pathology-free regions of multiple
sclerosis-affected brains. Nat Neurosci. 17:121–130. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Du P, Kibbe WA and Lin SM: lumi: A
pipeline for processing illumina microarray. Bioinformatics.
24:1547–1548. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Teschendorff AE, Marabita F, Lechner M,
Bartlett T, Tegner J, Gomez-Cabrero D and Beck S: A beta-mixture
quantile normalization method for correcting probe design bias in
Illumina Infinium 450 k DNA methylation data. Bioinformatics.
29:189–196. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Stadler MB, Murr R, Burger L, Ivanek R,
Lienert F, Schöler A, van Nimwegen E, Wirbelauer C, Oakeley EJ,
Gaidatzis D, et al: DNA-binding factors shape the mouse methylome
at distal regulatory regions. Nature. 480:490–495. 2011.PubMed/NCBI
|
|
24
|
Finer S, Mathews C, Lowe R, Smart M,
Hillman S, Foo L, Sinha A, Williams D, Rakyan VK and Hitman GA:
Maternal gestational diabetes is associated with genome-wide DNA
methylation variation in placenta and cord blood of exposed
offspring. Hum Mol Genet. 24:3021–3029. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sturn A, Quackenbush J and Trajanoski Z:
Genesis: Cluster analysis of microarray data. Bioinformatics.
18:207–218. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Millan MJ: The epigenetic dimension of
Alzheimer's disease: Causal, consequence, or curiosity? Dialogues
Clin Neurosci. 16:373–393. 2014.PubMed/NCBI
|
|
27
|
Hebert LE, Bienias JL, Aggarwal NT, Wilson
RS, Bennett DA, Shah RC and Evans DA: Change in risk of Alzheimer
disease over time. Neurology. 75:786–791. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hernandez DG, Nalls MA, Gibbs JR, Arepalli
S, van der Brug M, Chong S, Moore M, Longo DL, Cookson MR, Traynor
BJ and Singleton AB: Distinct DNA methylation changes highly
correlated with chronological age in the human brain. Hum Mol
Genet. 20:1164–1172. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Probst AV, Dunleavy E and Almouzni G:
Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell
Biol. 10:192–206. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ortiz-Matamoros A, Salcedo-Tello P,
Avila-Muñoz E, Zepeda A and Arias C: Role of Wnt signaling in the
control of adult hippocampal functioning in health and disease:
Therapeutic implications. Curr Neuropharmacol. 11:465–476. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Arrázola MS, Silva-Alvarez C and Inestrosa
NC: How the Wnt signaling pathway protects from neurodegeneration:
The mitochondrial scenario. Front Cell Neurosci. 9:1662015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Riise J, Plath N, Pakkenberg B and
Parachikova A: Aberrant Wnt signaling pathway in medial temporal
lobe structures of Alzheimer's disease. J Neural Transm (Vienna).
122:1303–1318. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sinha A, Tamboli RS, Seth B, Kanhed AM,
Tiwari SK, Agarwal S, Nair S, Giridhar R, Chaturvedi RK and Yadav
MR: Neuroprotective role of novel triazine derivatives by
activating Wnt/β catenin signaling pathway in rodent models of
alzheimer's disease. Mol Neurobiol. 52:638–652. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
del Pino J, Ramos E, Aguilera OM,
Marco-Contelles J and Romero A: Wnt signaling pathway, a potential
target for Alzheimer's disease treatment, is activated by a novel
multitarget compound ASS234. CNS Neurosci Ther. 20:568–570. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tenayuca J, Cousins K, Yang S and Zhang L:
Computational modeling approach in probing the effects of cytosine
methylation on the transcription factor binding to DNA. Curr Top
Med Chem. 17:1778–1787. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Banovich NE, Lan X, McVicker G, van de
Geijn B, Degner JF, Blischak JD, Roux J, Pritchard JK and Gilad Y:
Methylation QTLs are associated with coordinated changes in
transcription factor binding, histone modifications and gene
expression levels. PLoS Genet. 10:e10046632014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Medvedeva YA, Khamis AM, Kulakovskiy IV,
Ba-Alawi W, Bhuyan MS, Kawaji H, Lassmann T, Harbers M, Forrest AR
and Bajic VB: FANTOM consortium: Effects of cytosine methylation on
transcription factor binding sites. BMC Genomics. 15:1192014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hu S, Wan J, Su Y, Song Q, Zeng Y, Nguyen
HN, Shin J, Cox E, Rho HS, Woodard C, et al: DNA methylation
presents distinct binding sites for human transcription factors.
Elife. 2:e007262013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Métivier R, Gallais R, Tiffoche C, Le
Péron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F
and Reid G: Cyclical DNA methylation of a transcriptionally active
promoter. Nature. 452:45–50. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pieper HC, Evert BO, Kaut O, Riederer PF,
Waha A and Wüllner U: Different methylation of the TNF-alpha
promoter in cortex and substantia nigra: Implications for selective
neuronal vulnerability. Neurobiol Dis. 32:521–527. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mastroeni D, Grover A, Delvaux E,
Whiteside C, Coleman PD and Rogers J: Epigenetic changes in
Alzheimer's disease: Decrements in DNA methylation. Neurobiol
Aging. 31:2025–2037. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mastroeni D, McKee A, Grover A, Rogers J
and Coleman PD: Epigenetic differences in cortical neurons from a
pair of monozygotic twins discordant for Alzheimer's disease. PLoS
One. 4:e66172009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chouliaras L, Mastroeni D, Delvaux E,
Grover A, Kenis G, Hof PR, Steinbusch HW, Coleman PD, Rutten BP and
van den Hove DL: Consistent decrease in global DNA methylation and
hydroxymethylation in the hippocampus of Alzheimer's disease
patients. Neurobiol Aging. 34:2091–2099. 2013. View Article : Google Scholar : PubMed/NCBI
|