Introduction
As a severe central nervous system disease, spinal
cord injury (SCI) may lead to the complete or incomplete loss of
motor and sensory functions (1).
Damages caused by SCI can be divided into two phases, namely the
primary and secondary phases, which include spinal cord blood flow
reduction, excessive inflammatory response and neuron apoptosis
(2,3). Approximately 250,000-500,000
individuals are reported to suffer from SCI worldwide each year
(4). Although various therapeutic
strategies have been applied for SCI treatment, including
methylprednisolone administration and cell transplantation, there
is currently no effective therapeutic method for this injury
(5,6). Thus, it is urgent to develop a novel
and effective therapeutic method for SCI.
MicroRNAs (miRNAs or miRs), a family of endogenous
small no-coding RNA molecules with a length of 18–22 nucleotides,
are widely expressed in eukaryotes and serve important roles in
gene regulation by binding to the 3′-untranslated region (3′UTR) of
their target genes (7). Evidence has
demonstrated that miRNAs are involved in many developmental and
cellular processes in eukaryotic organisms (8,9). Due to
their key roles in the regulation of gene expression, cell
differentiation, proliferation and apoptosis, miRNAs have been
observed to participate in various neurological diseases (10,11).
Furthermore, an increasing number of studies have suggested that
miRNA serve an important role in the development of SCI (12–14).
miR-219-5p has been identified as a tumor suppressor
in several types of cancer, including colorectal cancer, gastric
cancer, papillary thyroid carcinoma and hepatocellular (15–18). In
addition, a previous study has revealed a high expression of
miR-219-5p in SCI (19); however,
the exact role of miR-219-5p in SCI remains unclear. Therefore, the
present study aimed to investigate the role of miR-219-5p in SCI
and to further examine the underlying molecular mechanism.
Materials and methods
Animals and establishment of an SCI
model in mice and neurons
Healthy adult male ICR mice (Sino-British SIPPR/BK
Lab Animal Ltd., Shanghai, China) weighing ~30 g (6 weeks of age)
were fed under a controlled environment, and provided with free
access to standard rodent chow and water. Mice were maintained
under a 12-h light/dark cycle, and the room temperature and
relative humidity were set at 25±3°C and 60±15%, respectively. All
experiments were performed in accordance with ethical standards of
the Third Hospital of Hebei Medical University (Shijiazhuang,
China), and were approved by the Ethics Committee Review Board of
this institution.
Mice were randomly divided into two groups (n=10 per
group), including the sham and SCI groups. The SCI model was
established as previously described (20). Briefly, the mice were anesthetized by
intraperitoneally injection with 10% chloral hydrate (30 mg/kg).
Following anesthetization, the mice were placed on table at a prone
position and an incision along the spine was made across the skin,
subcutis and muscle. Subsequently, a thoracic (T) 11-lumbar (L) 1
laminectomy was performed to expose the spinal cord. Following L1
laminectomy, the contusion injury was extended to the T11 spinal
cord. Subsequent to the contusion surgery, the skin was immediately
sutured. Mice in the sham group received a dorsal laminectomy only.
Mice were kept warm and allowed to recover from the anesthesia. The
majority of SCI mice presented flaccid paralysis in the lower
extremities, and other SCI mice displayed spastic symptoms. Mice
presenting with flaccid paralysis were used in subsequent
experiments in the present study.
Following the sacrifice of mice, the spinal cord at
L4-6 from SCI mice and the control mice was isolated, and the
tissue was then digested with 0.125% trypsin containing 0.02% EDTA
at 37°C for 20 min. Next, Dulbecco's modified Eagle medium was
added to stop the digestion. The tissues were then used for
miR-219-5p detection using reverse transcription-quantitative
polymerase chain reaction (RT-qPCR), and for neuron extraction and
purification. Neurons were isolated and purified from the spinal
cords from the sham mice as previously described (21). Neurons
(3×104/cm2) were plated into 35-mm petri
plates coated with polylysine. Subsequently, 2 ml neurobasal
culture medium (Invitrogen; Thermo Fisher Scientific, Inc.)
supplemented with glutamine, B27 and penicillin/streptomycin was
added to the plates, and the neurons were grown in an incubator at
37°C with 5% CO2. An SCI model in neurons was
established by scratch according to previous study (22), and neurons without any treatment were
used as the control group. At 24 h after scratching, neurons were
harvested for subsequent analysis.
Cell transfection
A transfection assay was performed using
Lipofectamine 2000 regent (Thermo Fisher Scientific, Inc., Waltham,
MA, USA). Neurons were transiently transfected with miR-219-5p
inhibitors (GenePharma, Shanghai, China), the negative control of
miR-219-5p inhibitors (NC) (GenePharma), small interfering (si)RNA
(si)-liver receptor homolog-1 (LRH-1) (Santa Cruz Biotechnology,
Inc., Dallas, CA, USA), control siRNA (the control of LRH-1 siRNA)
(Santa Cruz Biotechnology, Inc.) or miR-219-5p inhibitor + LRH-1
siRNA (in+si-LRH-1) using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according with the
manufacturer's protocol. Cells without any treatment were used as
the control (Con) group. The transfection efficiency was determined
by RT-qPCR.
MTT assay
At 48 h after cell transfection, an MTT assay was
performed to investigate the neuron viability. Briefly, the neurons
were seeded into 96-well plates (Costar; Corning Incorporated,
Corning, NY, USA) at a density of 2.0×103 cells per well
and incubated for ~24 h before treatment. A total of 24 h later, 20
µl MTT (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) solution at
a concentration of 5 mg/ml was added to each well, and then
incubated for a further 4 h at 37°C. Finally, the cell
proliferation ability was assessed by measuring the optical density
at 490 nm using a microplate reader (Thermo Fisher Scientific,
Inc.).
Apoptosis analysis assay
An Annexin V-FITC Early Apoptosis Detection Kit
(cat. no. 6592; Cell Signaling Technology, Inc., Danvers, MA, USA)
was used to detect cell apoptosis. Briefly, at 48 h after cell
transfection, the neurons were harvested with trypsin, re-suspended
in Annexin V-FITC/propidium iodide, and then incubated for ~15 min
at room temperature in the dark. A BD FACSCelesta™ flow
cytometer (BD Biosciences, Franklin Lakes, NJ, USA) was
subsequently conducted to assess the apoptotic rate of cells in
different groups in line with the instrument's operating protocol.
Data were analyzed using version 2.5 WinMDI (Purdue University
Cytometry Laboratories; http://www.cyto.purdue.edu/flowcyt/software/Catalog.htm).
Bioinformatics and dual-luciferase
reporter analyses
Bioinformatics prediction was performed to identify
potential target genes of miR-219-5p, including LRH-1, which were
selected using miRNA target site prediction software (http://www.microrna.org).
In order to examine whether miR-219-5p targets the
3′UTR of LRH-1, a cDNA fragment of the LRH-1-3′UTR mRNA containing
the seed sequence of the wild-type (WT) miR-219-5p binding site or
a mutated (MUT) binding site of the 3′UTR sequence was cloned into
the pmirGLO dual-luciferase vector (Promega Corp., Madison, WI,
USA). The psiCHECK-2 reporter plasmid (Sangon Biotech Co., Ltd.,
Shanghai, China) vectors were termed LRH-1-3′UTR-WT and
LRH-1-3′UTR-MUT, respectively. Subsequently, neurons were seeded
into a 24-well plate (5×104 cells/well), and
co-transfected with LRH-1-3′UTR-WT or LRH-1-3′UTR-MUT and with
miR-219-5p mimic or its control (mimic control) vector (GenePharma,
Shanghai, China). The luciferase activity was then analyzed using a
Dual-Luciferase Reporter Assay kit (Promega Corp.) following the
manufacturer's protocols, and normalized to Renilla
luciferase activity. Each experiment was repeated at least three
times.
RT-qPCR analysis
Total RNA from the spinal cord tissue and neurons
was extracted using the TRIzol reagent (Thermo Fisher Scientific,
Inc.) as per the manufacturer's protocol. The RevertAid First
Strand cDNA synthesis kit (Fermentas; Thermo Fisher Scientific,
Inc.) was used to reverse transcribe the total RNA into cDNA. qPCR
was subsequently performed using the SYBR Green qRCR Mix (Toyobo
Life Science, Osaka, Japan). U6 was used as an internal reference
for the determination of miRNA expression, while GAPDH served as
the internal reference for the detection of mRNA expression. The
following primer sequences were synthesized by GenScript
(Piscataway, NJ, USA): LRH-1, forward 5′-GCACGGACTTACACCTATTGTG-3′
and reverse 5′-TGTCAATTTGGCAGTTCTGG-3′; β-catenin, forward
5′-AACAGGGTCTGGGACATTAGTC-3′ and reverse
5′-CGAAAGCCAATCAAACACAAAC-3′; c-Myc, forward
5′-CACCAGCAGCGACTCTGA-3′ and 5′-GATCCAGACTCTGACCTTTTGC-3′; cyclin
D1, forward 5′-AACTACCTGGACCGCTTCCT-3′ and reverse
5′-CCACTTGAGCTTGTTCACCA-3′; GAPDH, forward
5′-GAAATCCCATCACCATCTTCCAGG-3′ and reverse
5′-GAGCCCCAGCCTTCTCCATG-3′; miR-219-5p, forward
5′-ACACTCCAGCTGGGTGATTGTCCAAACGCAAT-3′ and reverse
5′-CTCAACTGGTGTCGTGGA-3′; and U6, forward
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse
5′-CGCTTCACGAATTTGCGTGTCAT-3′. The following thermocycling
conditions were performed: 95°C for 5 min, followed by 40 cycles of
denaturation at 95°C for 15 sec and annealing/elongation at 60°C
for 30 sec. Relative gene expression was calculated by using the
2−ΔΔCq method (23). All
experiments were repeated at least three times.
Western blot analysis
To collect the total cell protein, neurons were
lysed on ice with the radioimmunoprecipitation assay lysis buffer
(Auragene Bioscience, Changsha, China). Protein concentration was
determined using the BCA assay kit (Beyotime Institute of
Biotechnology). Protein samples were separated by 12% SDS-PAGE, and
then transferred onto nitrocellulose membranes. Subsequent to
blocking with 5% non-fat milk at room temperature for 1.5 h, the
membranes were incubated overnight at 4°C with primary antibodies
against LRH-1 (cat no. #12800), β-catenin (cat no. #8480), Cyclin
D1 (cat no. #2978), c-Myc (cat no. #13978) and β-actin (cat no.
4970) (All dilutions, 1:1,000; Cell Signaling Technology, Inc.,
Danvers, MA, USA). Next, the membranes were incubated with
anti-rabbit immunoglobulin G horseradish peroxidase-coupled
secondary antibody (cat no. 7074; 1:1,000 dilution; Cell Signaling
Technology, Inc.) at room temperature for ~1 h. Subsequent to
washing three times with Tris-buffered saline/Tween-20, the
membranes were stained with an enhanced chemiluminescent reagent
(Applygen Technologies, Inc., Beijing, China) according to the
manufacturer's protocol, and the western blot bands were observed
using a ChemiDoc XRS+ System (Bio-Rad Laboratories, Inc., Hercules,
CA, USA).
Statistical analysis
Data are presented as the mean ± standard deviation.
Statistical analyses were performed using SPSS version 16.0 (SPSS
Inc., Chicago, IL, USA). The variations between groups were
statistically examined using the Student's t-test or one-way
analysis of variance. P<0.05 was considered to be an indication
of a statistically significant difference.
Results
SCI upregulates the level of
miR-219-5p in the spinal cords of mice
The expression level of miR-219-5p in the spinal
cord at 1, 3, 7 and 14 days after SCI was detected using RT-qPCR.
The results revealed that the relative expression of miR-219 in the
sham rats exhibited no significant alterations between days 1 and
14 after SCI. However, compared with the sham group, the level of
miR-219-5p in the SCI rats was significantly increased at day 1
after SCI, and this increase was maintained until day 14
post-injury (Fig. 1A). Furthermore,
the level of miR-219-5p in the control and SCI model neurons were
also determined by RT-qPCR. The results demonstrated that, compared
with the control neurons, the level of miR-219-5p was significantly
upregulated in the SCI model neurons (Fig. 1B). The data indicated that the level
of miR-219-5p was upregulated in spinal cords.
LRH-1 is a target gene of
miR-219-5p
An miRNA target site prediction software was used to
predict the target genes of miR-219-5p. In total, ~4,300 target
genes were identified to be the potential target genes of
miR-219-5p, including LRH-1 (Fig.
2A). Based on our previous studies and the literature analysis,
it was determined that LRH-1 is involved in a variety of biological
progress, and regulates cell proliferation, apoptosis and cell
cycle by modulating the Wnt/β-catenin and p53 signaling pathways
(16,24,25).
However, whether this gene is involved in SCI remains unclear.
Thus, LRH-1 was selected for further investigation in the present
study.
Subsequently, a dual-luciferase reporter assay was
conducted to confirm whether miR-219-5p directly targets LRH-1. As
shown in Fig. 2B, compared with the
control (miR-C) group, the luciferase activity of cells transfected
with the LRH-1-3′UTR-WT vector was significantly reduced
(P<0.01). However, no evident difference in fluorescence was
observed between the LRH-1-3′UTR-MUT and the control groups.
To further determine whether miR-219-5p regulates
LRH-1 expression in neurons, the effect of an miR-219-5p inhibitor
on LRH-1 expression in neurons was investigated. Neurons were
transfected with miR-219-5p inhibitor, NC, LRH-1 siRNA, or control
siRNA. It was indicated that the miR-219-5p inhibitor significantly
enhanced the protein (Fig. 2C) and
mRNA (Fig. 2D) expression levels of
LRH-1 in neurons as compared with the control and the
NC-transfected cells. Additionally, the transfection efficiency was
determined by RT-qPCR (Fig. 3).
Taken together, the findings suggested that LRH-1 is a target gene
of miR-219-5p and it was negatively regulated by miR-219-5p.
 | Figure 3.Cell transfection efficiency was
determined by reverse transcription-quantitative polymerase chain
reaction. At 48 h after cell transfection, transfection efficiency
was determined by reverse transcription-quantitative polymerase
chain reaction. (A) Relative miR-219-5p and (B) LRH-1 expression
levels were determined. Con, control group cells without any
treatment; NC, cells transfected with the negative control of
miR-219-5p inhibitor; inhibitor, cells transfected with miR-219-5p
inhibitor; si-Con, cells transfected with control siRNA; siRNA,
cells transfected with LRH-1 siRNA. Data are displayed as the mean
± standard deviation. **P<0.01 vs. control group. miR, microRNA;
Con, control; NC, negative control; LRH-1, liver receptor
homolog-1; si, small interfering. |
miR-219-5p inhibitor rescues the
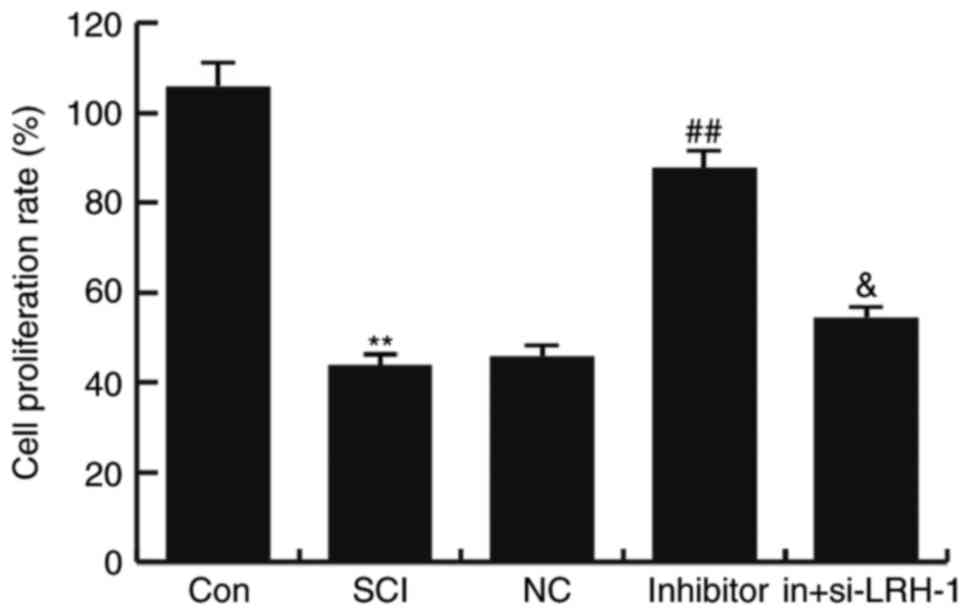
SCI-induced neuron activity inhibition
To detect the influence of miR-219-5p inhibitor on
neuron activity, the neuron viability was determined using an MTT
assay. The results demonstrated that, compared with the control
group (untreated control cells), the viability of neurons was
markedly reduced in the SCI group (P<0.01; Fig. 4). This decreased viability was
rescued by miR-219-5p inhibitor treatment, which significantly
increased the cell proliferation rate compared with the SCI group
(P<0.01). In addition, transfection with siRNA-LRH-1 for
knockdown of LRH-1 expression eliminated the increased neuron
viability caused by the miR-219-5p inhibitor (Fig. 4). These findings suggest that
miR-219-5p inhibitor could rescue SCI-induced neuron activity
inhibition.
miR-219-5p inhibitor inhibits the
SCI-induced neuronal apoptosis
To determine the effects of miR-219-5p on neuronal
apoptosis, the apoptosis of cells was analyzed by FCM assay.
Compared with the control group, the number of apoptotic cells was
significantly enhanced in the SCI group, while this SCI-induced
increase was then inhibited by miR-219-5p inhibitor treatment (both
P<0.01). Furthermore, it was observed that si-LRH-1 eliminated
the decreased neuron apoptosis caused by the miR-219-5p inhibitor,
and this change was statistically significant (P<0.05; Fig. 5). The data indicated that miR-219-5p
inhibitor could inhibit the SCI-induced neuronal apoptosis.
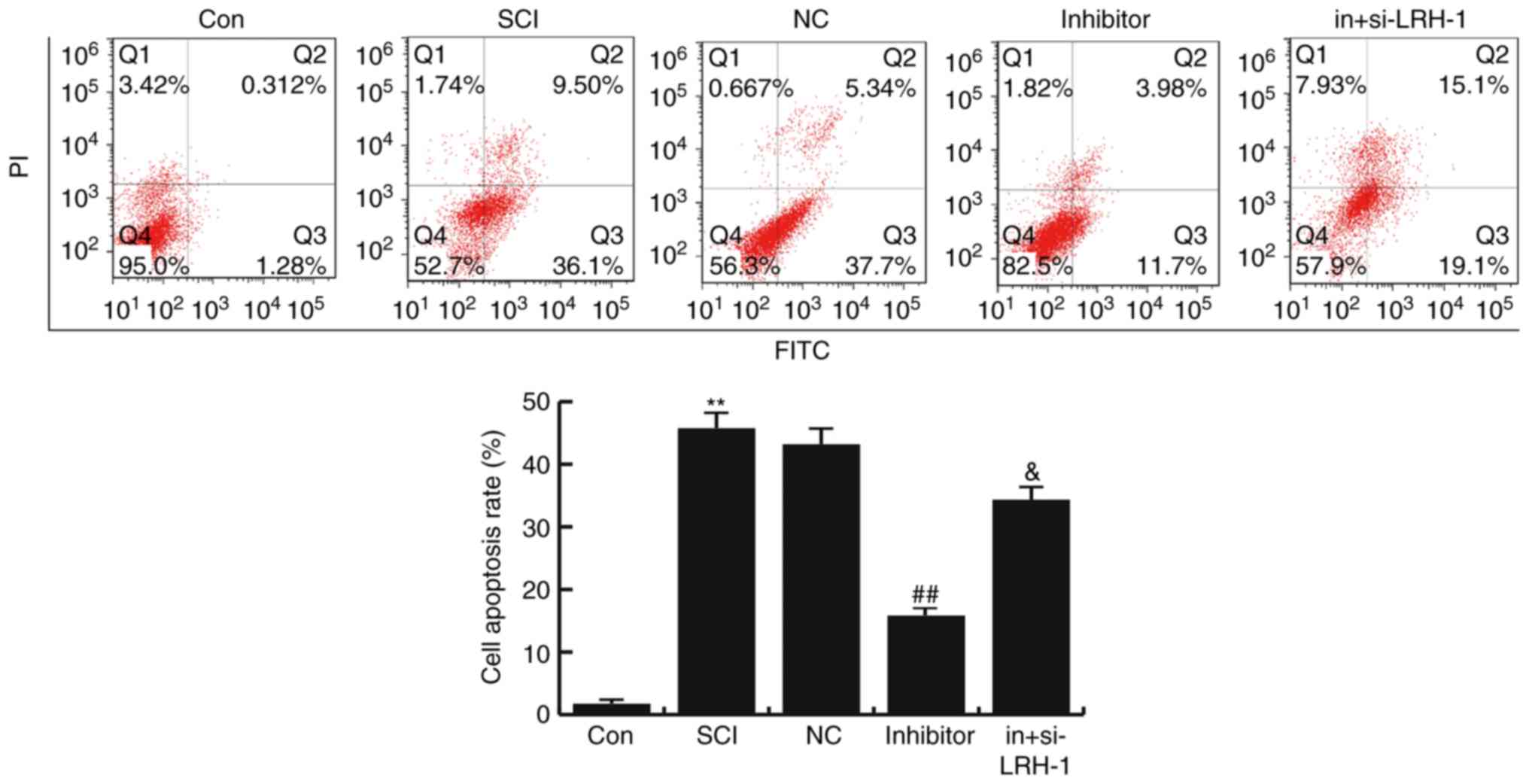 | Figure 5.Effects of miR-219-5p on neuronal
apoptosis. At 48 h after cell transfection, neuronal apoptosis was
detected by flow cytometry, and the cell apoptosis rate was
calculated according to Q3 and Q2, which refer to the early and
late apoptosis rates, respectively. Data are displayed as the mean
± standard deviation. **P<0.01 vs. Con group;
##P<0.01 vs. SCI group; &P<0.05 vs.
inhibitor group. miR, microRNA; Con, control; SCI, spinal cond
injury; NC, negative control; in+si-LRH-1, miR-219-5p inhibitor +
siRNA-LRH-1; LRH-1, liver receptor homolog-1. |
miR-219-5p inhibitor rescues the
LRH-1/Wnt/β-catenin inhibition induced by SCI
To further determine the underlying molecular
mechanisms of the effect of miR-219-5p on SCI development, the
LRH-1/Wnt/β-catenin pathway was analyzed. The protein and mRNA
levels of LRH-1, β-catenin, Cyclin D1 and c-Myc in different groups
were detected using western blotting and RT-qPCR, respectively. As
illustrated in Fig. 5, the mRNA
levels of LRH-1, β-catenin, Cyclin D1 and c-Myc in SCI neurons were
markedly lower compared with the control group (P<0.05 or
P<0.01). However, the miR-219-5p inhibitor prevented this
SCI-induced reduction of LRH-1, β-catenin, Cyclin D1 and c-Myc
levels (P<0.05 or P<0.01; Fig.
6). In addition, LRH-1 gene silencing by siRNA transfection
eliminated the increased mRNA expression levels of LRH-1,
β-catenin, Cyclin D1 and c-Myc caused by miR-219-5p inhibitor in
the neurons (P<0.05; Fig. 6).
Similar results were obtained from western blot analysis.
MiR-219-5p inhibitor rescued the LRH-1/Wnt/β-catenin inhibition
induced by SCI.
 | Figure 6.Effects of miR-219-5p on the
LRH-1/Wnt/β-catenin signaling pathway. At 48 h after cell
transfection, the (A) LRH-1 protein was examined by western
blotting, while the (B) LRH-1, (C) β-catenin, (D) Cyclin D1 and (E)
c-Myc were determined by reverse transcription-quantitative
polymerase chain reaction. Data are displayed as the mean ±
standard deviation. *P<0.05 and **P<0.01 vs. control group;
#P<0.05 and ##P<0.01 vs. SCI group;
&P<0.05 and &&P<0.01 vs.
inhibitor group. miR, microRNA; LRH-1, liver receptor homolog-1;
Con, control; SCI, spinal cord injury; NC, negative control;
in+si-LRH-1, miR-219-5p inhibitor + siRNA-LRH-1. |
Discussion
SCI is considered to be a severe disease that
affects a great number of individuals worldwide. Previous evidence
has revealed that apoptosis and neuroplasticity contribute to the
functional defects in patients with SCI (26). Although the pathophysiological
processes of SCI have been extensively studied, there are currently
no effective treatments for patients with SCI (22).
miRNAs are small, non-coding RNAs that suppress mRNA
translation or induce mRNA degradation by binding to the 3′UTR of
mRNA targets. In recent years, the potential roles that miRNA may
serve in the development and progression of SCI have been reported
(27–33). To date, various miRNAs have been
observed to be abnormally expressed in SCI patients and to serve
critical roles in the development of SCI (27). Liu et al (28) reported that inhibition of miR-223
exerted a protective role in functional recovery, angiogenesis and
anti-apoptosis during SCI. In addition, Yang et al (29) suggested that downregulation of
miR-128 in murine microglial cells may contribute to the
development of neuropathic pain following SCI via the activation of
p38. Zhou et al (30) also
demonstrated that miR-199b relieved SCI, at least partly, through
regulating the IKKβ-NF-kB signaling pathway and influencing the
microglia function. miR-195 has been observed to be decreased
following SCI, and may protect rats from SCI (31). Furthermore, miR-208b participated in
SCI progression via modulating myostatin expression (32). Wang et al (33) also considered that miR-142-3p was a
key therapeutic target for repairing the sensory function in
SCI.
miR-219-5p, which has been widely investigated in
several cancer processes (15–18,34,35), was
reported to be highly expressed in SCI (19). However, to date, the exact role of
miR-219-5p in SCI remains unclear. Therefore, the present study
investigated the role of miR-219-5p in SCI using a mouse/neuron SCI
model. The results revealed that the miR-219-5p inhibitor resulted
in the recovery of SCI-induced neuron activity inhibition, as well
as inhibited the SCI-induced neuron apoptosis. In addition, the
study revealed that LRH-1 was a direct target of miR-219-5p. LRH-1
has been suggested as a coactivator of the Wnt/β-catenin signaling
pathway, and it can interact with transcription factor 4 and
β-catenin to promote the expression of c-Myc and cyclin D1/E1
(36,37). Therefore, β-catenin, cyclin D1 and
c-Myc were analyzed in the present study. The present results
suggested that miR-219-5p inhibitor was able to reverse the
inhibition of the LRH-1/Wnt/β-catenin pathway induced by SCI.
Moreover, LRH-1 silencing could eliminate the effects of miR-219-5p
inhibitor on SCI.
In conclusion, the present study suggested that the
miR-219-5p inhibitor served a protective role in SCI via regulating
the LRH-1/Wnt/β-catenin signaling pathway. Thus, miR-219-5p may be
used as a novel and potential therapeutic target for SCI treatment.
However, the role of miRNA-219-5p in SCI and its associated
mechanisms require further extensive research. In the future, it
should be also investigated whether miRNA-219-5p serves a role in
SCI by regulating other target genes, in order to provide a more
comprehensive theoretical basis for the clinical treatment of
SCI.
Acknowledgements
The authors would like to thank Dr Yujun Li
(Department of Spinal Surgery, the Second Hospital of Tangshan,
Tangshan, China) for the assistance in preparing the
manuscript.
References
|
1
|
Hulsebosch CE: Recent advances in
pathophysiology and treatment of spinal cord injury. Adv Physiol
Educ. 26:238–255. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dumont RJ, Okonkwo DO, Verma S, Hurlbert
RJ, Boulos PT, Ellegala DB and Dumont AS: Acute spinal cord injury,
part I: Pathophysiologic mechanisms. Clin Neuropharmacol.
24:254–264. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bareyre FM and Schwab ME: Inflammation,
degeneration and regeneration in the injured spinal cord: Insights
from DNA microarrays. Trends Neurosci. 26:555–563. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
W.H.O., . Spinal Cord Injury Fact Sheet N
384. 2013.
|
|
5
|
Ozdemir M, Attar A and Kuzu I:
Regenerative treatment in spinal cord injury. Curr Stem Cell Res
Ther. 7:364–369. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pereira JE, Costa LM, Cabrita AM, Couto
PA, Filipe VM, Magalhães LG, Fornaro M, Di Scipio F, Geuna S,
Maurício AC and Varejão AS: Methylprednisolone fails to improve
functional and histological outcome following spinal cord injury in
rats. Exp Neurol. 220:71–81. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Winter J, Jung S, Keller S, Gregory RI and
Diederichs S: Many roads to maturity: microRNA biogenesis pathways
and their regulation. Nat Cell Biol. 11:228–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang W, Kwon EJ and Tsai LH: MicroRNAs in
learning, memory, and neurological diseases. Learn Mem. 19:359–368.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rao P, Benito E and Fischer A: MicroRNAs
as biomarkers for CNS disease. Front Mol Neurosci. 6:392013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dong J, Lu M, He X, Xu J, Qin J, Cheng Z,
Liang B, Wang D and Li H: Identifying the role of microRNAs in
spinal cord injury. Neurol Sci. 35:1663–1671. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ning B, Gao L, Liu RH, Liu Y, Zhang NS and
Chen ZY: microRNAs in spinal cord injury: Potential roles and
therapeutic implications. Int J Biol Sci. 10:997–1006. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nieto-Diaz M, Esteban FJ, Reigada D,
Muñoz-Galdeano T, Yunta M, Caballero-López M, Navarro-Ruiz R, Del
Águila A and Maza RM: microRNA dysregulation in spinal cord injury:
Causes, consequences and therapeutics. Front Cell Neurosci.
8:532014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Q, Zhu L, Jiang Y, Xu J, Wang F and
He Z: miR-219-5p suppresses the proliferation and invasion of
colorectal cancer cells by targeting calcyphosin. Oncol Lett.
13:1319–1324. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li C, Dong J, Han Z and Zhang K:
MicroRNA-219-5p represses the proliferation, migration and invasion
of gastric cancer cells by targeting the LRH-1/Wnt/β-catenin
signaling pathway. Oncol Res. 25:617–627. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang C, Cai Z, Huang M, Mao C, Zhang Q,
Lin Y, Zhang X, Tang B, Chen Y, Wang X, et al: miR-219-5p modulates
cell growth of papillary thyroid carcinoma by targeting estrogen
receptor α. J Clin Endocrinol Metab. 100:E204–E213. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang N, Lin J, Ruan J, Su N, Qing R, Liu
F, He B, Lv C, Zheng D and Luo R: MiR-219-5p inhibits
hepatocellular carcinoma cell proliferation by targeting
glypican-3. FEBS Lett. 586:884–891. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hachisuka S, Kamei N, Ujigo S, Miyaki S,
Yasunaga Y and Ochi M: Circulating microRNAs as biomarkers for
evaluating the severity of acute spinal cord injury. Spinal Cord.
52:596–600. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jee MK, Jung JS, Choi JI, Jang JA, Kang
KS, Im YB and Kang SK: MicroRNA 486 is a potentially novel target
for the treatment of spinal cord injury. Brain. 135:1237–1252.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jaworski J and Sheng M: The growing role
of mTOR in neuronal development and plasticity. Mol Neurobiol.
34:205–219. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Z, Zhou L, Zheng X, Chen G, Pan R, Li
J and Liu W: Autophagy protects against PI3K/Akt/mTOR-mediated
apoptosis of spinal cord neuronsafter mechanical injury. Neurosci
Lett. 656:158–164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhai G, Song J, Shu T, Yan J, Jin X, He J
and Yin Z: LRH-1senses signaling from phosphatidylcholine to
regulate the expansion growth of digestive organs via synergy with
Wnt/β-catenin signaling in zebrafish. J Genet Genomics. 20:307–317.
2017. View Article : Google Scholar
|
|
25
|
Kramer HB, Lai CF, Patel H, Periyasamy M,
Lin ML, Feller SM, Fuller-Pace FV, Meek DW, Ali S and Buluwela L:
LRH-1 drives colon cancer cell growth by repressing the expression
of the CDKN1A gene in a p53-dependent manner. Nucleic Acids Res.
44:582–594. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Harkema SJ: Neural plasticity after human
spinal cord injury: Application of locomotor training to the
rehabilitation of walking. Neuroscientist. 7:455–468. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Martirosyan NL, Carotenuto A, Patel AA,
Kalani MY, Yagmurlu K, Lemole GM Jr, Preul MC and Theodore N: The
role of microRNA markers in the diagnosis, treatment and outcome
prediction of spinal cord injury. Front Surg. 3:562016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu D, Huang Y, Jia C, Li Y, Liang F and
Fu Q: Administration of antagomir-223 inhibits apoptosis, promotes
angiogenesis andfunctional recovery in rats with spinal cord
injury. Cell Mol Neurobiol. 35:483–491. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang Z, Xu J, Zhu R and Liu L:
Down-regulation of miRNA-128 contributes to neuropathic pain
following spinal cord injury via activation of P38. Med Sci Monit.
23:405–411. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou HJ, Wang LQ, Xu QS, Fan ZX, Zhu Y,
Jiang H, Zheng XJ, Ma YH and Zhan RY: Downregulation of miR-199b
promotes the acute spinal cord injury through IKKβ-NF-κB signaling
pathway activating microglial cells. Exp Cell Res. 349:60–67. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tao B and Shi K: Decreased miR-195
expression protects rats from spinal cord injury primarily by
targeting HIF-1α. Ann Clin Lab Sci. 46:49–53. 2016.PubMed/NCBI
|
|
32
|
Boon H, Sjögren RJ, Massart J, Egan B,
Kostovski E, Iversen PO, Hjeltnes N, Chibalin AV, Widegren U and
Zierath JR: MicroRNA-208b progressively declines after spinal cord
injury in humans and is inversely related to myostatin expression.
Physiol Rep. 3:pii e126222015. View Article : Google Scholar
|
|
33
|
Wang T, Yuan W, Liu Y, Zhang Y, Wang Z,
Chen X, Feng S, Xiu Y and Li W: miR-142-3p is a potential
therapeutic target for sensory function recovery of spinal cord
injury. Med Sci Monit. 21:2553–2556. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rao SA, Arimappamagan A, Pandey P, Santosh
V, Hegde AS, Chandramouli BA and Somasundaram K: miR-219-5p
inhibits receptor tyrosine kinase pathway by targeting EGFR in
glioblastoma. PLoS One. 8:e631642013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cheng J, Deng R, Zhang P, Wu C, Wu K, Shi
L, Liu X, Bai J, Deng M, Shuai X, et al: miR-219-5p plays a tumor
suppressive role in colon cancer by targeting oncogene Sall4. Oncol
Rep. 34:1923–1932. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nadolny C and Dong X: Liver receptor
homolog-1 (LRH-1): A potential therapeutic target for cancer.
Cancer Biol Ther. 16:997–1004. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Botrugno OA, Fayard E, Annicotte JS, Haby
C, Brennan T, Wendling O, Tanaka T, Kodama T, Thomas W, Auwerx J
and Schoonjans K: Synergy between LRH-1 and beta-catenin induces G1
cyclin-mediated cell proliferation. Mol Cell. 15:499–509. 2004.
View Article : Google Scholar : PubMed/NCBI
|