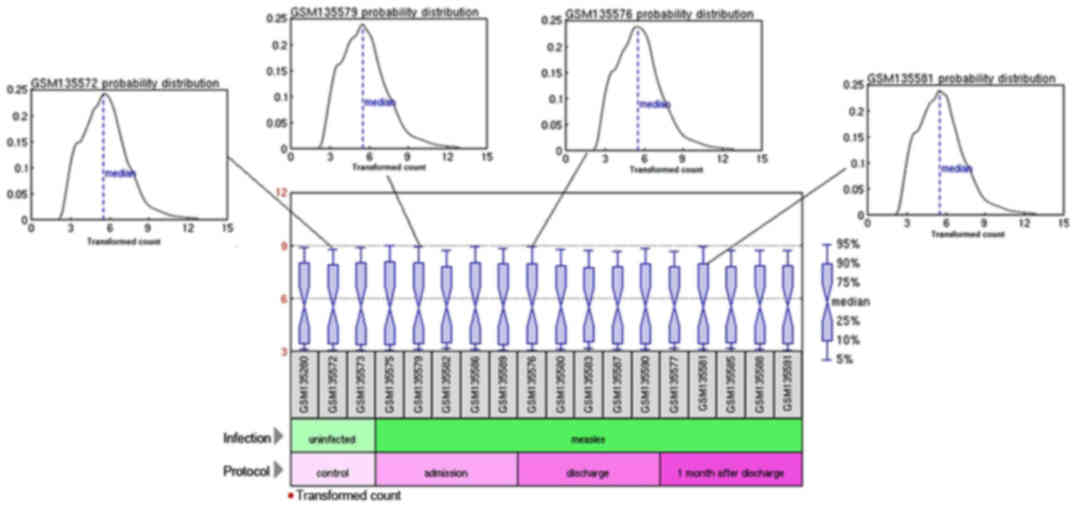
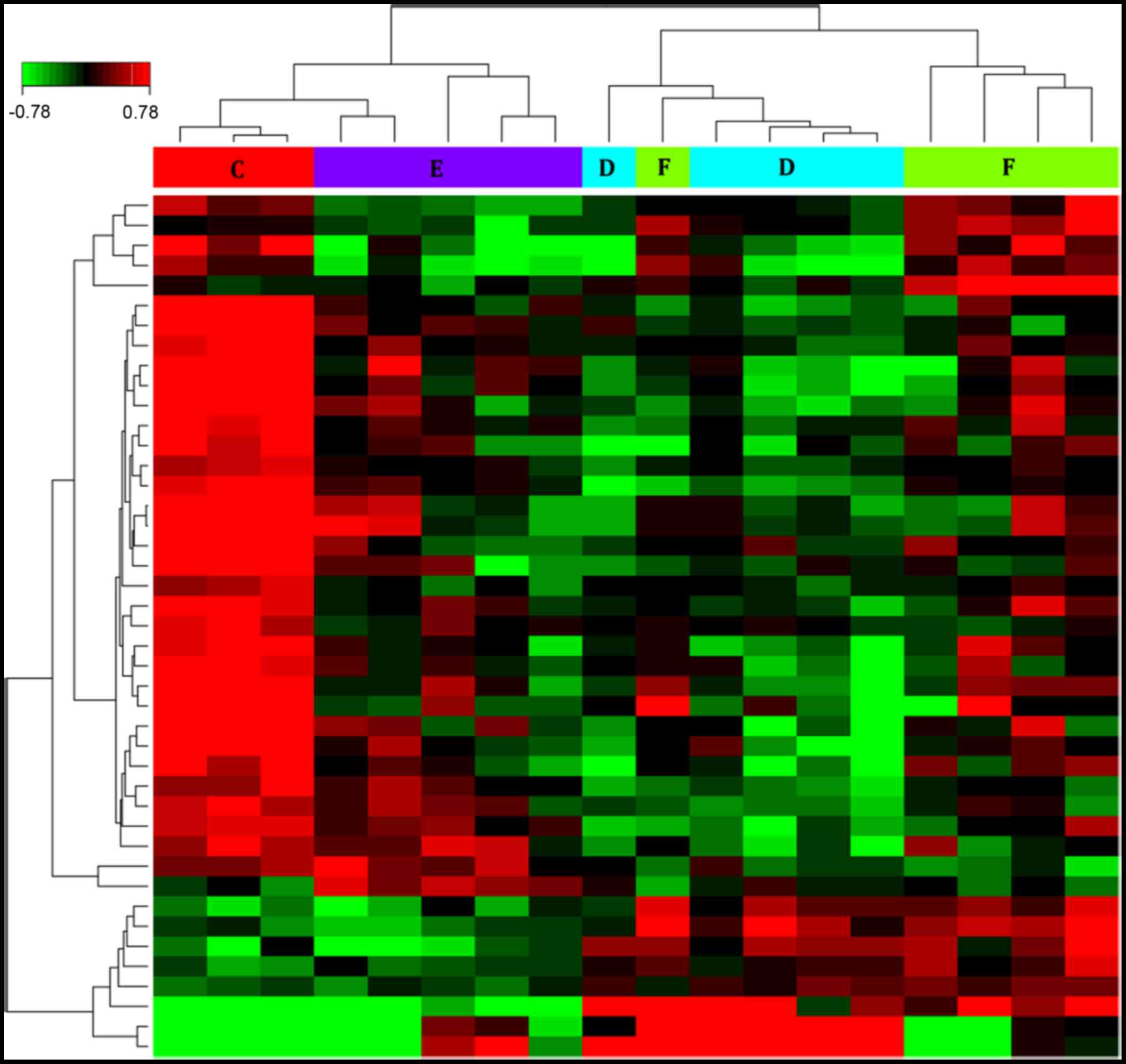
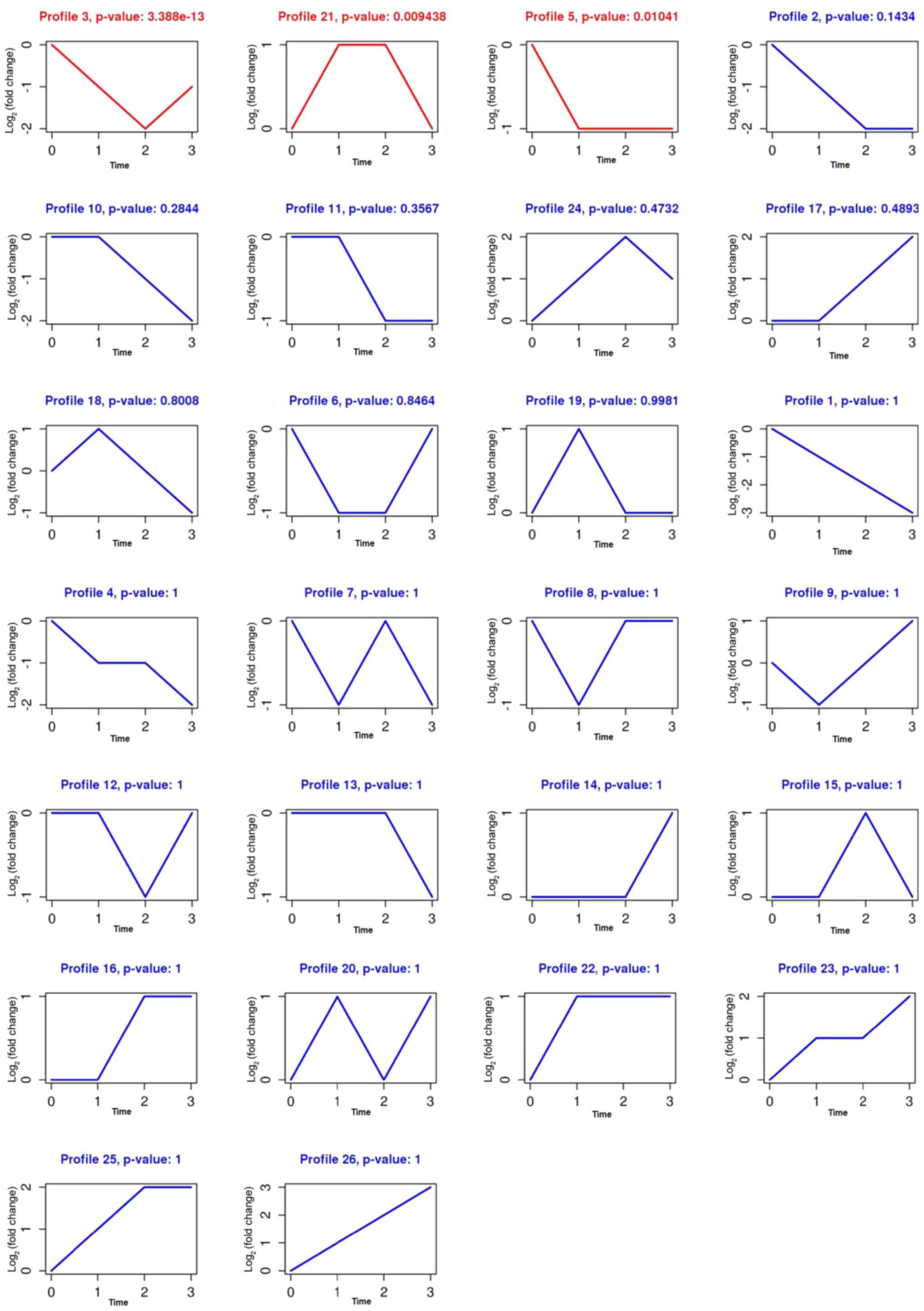
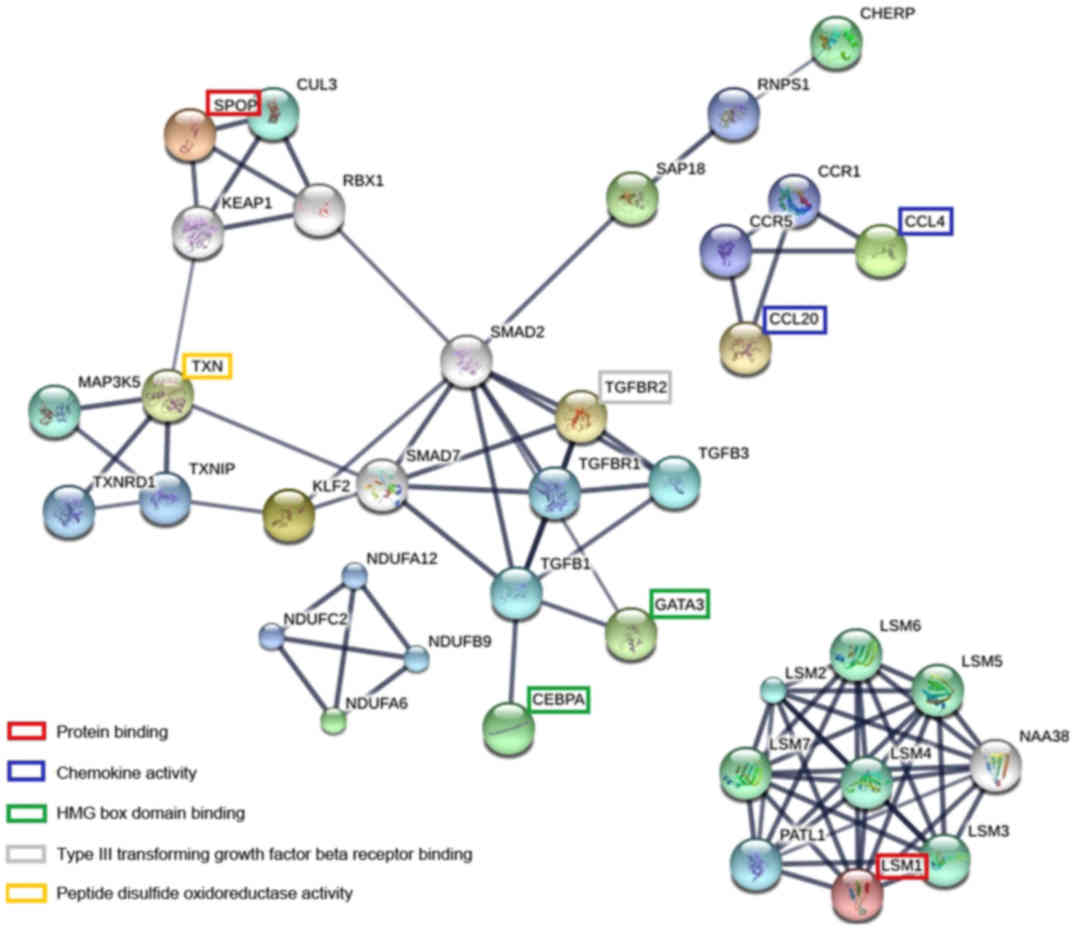
|
1
|
World Health Organization, . Measles Fact
Sheets. 2016.http://www.who.int/mediacentre/factsheets/fs286/en/September
29–2017
|
|
2
|
Centers for Disease Control and Prevention
(CDC): Measles-United States, 2011. MMWR Morb Mortal Wkly. Rep.
61:253–257. 2012.
|
|
3
|
World Health Organization: Measles-Rubella
Bulletin. 2017.http://iris.wpro.who.int/bitstream/handle/10665.1/13539/Measles-Rubella-Bulletin-2017-Vol-11-No-01.pdf?ua=1September
29–2017
|
|
4
|
No authors listed: Measles vaccines: WHO
position paper. Wkly Epidemiol Rec. 84:349–360. 2009.(In English,
French). PubMed/NCBI
|
|
5
|
Zheng J, Zhou Y, Wang H and Liang X: The
role of the China experts advisory committee on immunization
program. Vaccine. 28 Suppl 1:A84–A87. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang RQ, Li HB, Li FY, Han LX and Xiong
YM: Epidemiological characteristics of measles from 2000 to2014:
Results of a measles catch-up vaccination campaign in Xianyang,
China. J Infect Public Health. 10:624–629. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li L, Yu WZ, Shui TJ, Ma C, Wen N and
Liang XF: Analysis on epidemiological characteristics of age
distribution of measles in China during 2003–2006. Chin J Vaccines
Immun. 13:101–105. 2007.
|
|
8
|
Wang Z, Yan R, He H, Li Q, Chen G, Yang S
and Chen E: Difficulties in eliminating measles and controlling
rubella and mumps: A Cross-sectional study of a first measles and
rubella vaccination and a second measles, mumps, and rubella
vaccination. PLoS One. 9:e893612014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu Y, Lu P, Hu Y, Wang Z, Deng X, Ma F,
Tao H, Jia C, Ding X, Yang H, et al: Cross-sectional surveys of
measles antibodies in the Jiangsu Province of China from 2008 to
2010: The effect of high coverage with two doses of measles vaccine
among children. PLoS One. 8:e667712013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zilliox MJ, Moss WJ and Griffin DE: Gene
expression Changes in Peripheral blood mononuclear cells during
measles virus infection. Clin Vaccine Immunol. 14:918–923. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xiao S, Mo D, Wang Q, Jia J, Qin L, Yu X,
Niu Y, Zhao X, Liu X and Chen Y: Aberrant host immune response
induced by highly virulent PRRSV identified by digital gene
expression tag profiling. BMC Genomics. 11:5442010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Honda M, Kaneko S, Kawai H, Shirota Y and
Kobayashi K: Differential gene expression between chronic hepatitis
B and C hepatic lesion. Gastroenterology. 120:955–966. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sankaran S, Guadalupe M, Reay E, George
MD, Flamm J, Prindiville T and Dandekar S: Gut mucosal T cell
responses and gene expression correlate with protection against
disease in long-term HIV-1-infected nonprogressors. Proc Natl Acad
Sci USA. 102:9860–9865. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu SY, Hu YW, Liu XY, Xiong W, Zhou ZT and
Yuan ZH: Gene expression profiles in peripheral blood mononuclear
cells of SARS patients. World J Gastroenterol. 11:5037–5043. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rota PA, Liffick SL, Rota JS, Katz RS,
Redd S, Papania M and Bellini WJ: Molecular epidemiology of measles
viruses in the United States, 1997–2001. Emerg Infect Dis.
8:902–908. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rota PA, Featherstone DA and Bellini WJ:
Molecular epidemiology of measles virus. Curr Top Microbiol
Immunol. 330:129–150. 2009.PubMed/NCBI
|
|
17
|
Riddell MA, Rota JS and Rota PA: Review of
the temporal and geographical distribution of measles virus
genotypes in the prevaccine and postvaccine eras. Virol J.
2:872005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Blattner M, Liu D, Robinson BD, Huang D,
Poliakov A, Gao D, Nataraj S, Deonarine LD, Augello MA, Sailer V,
et al: SPOP mutation drives prostate tumorigenesis in vivo through
coordinate regulation of PI3K/mTOR and AR signaling. Cancer Cell.
3:436–451. 2017. View Article : Google Scholar
|
|
19
|
Zhang L, Peng S, Dai X, Gan W, Nie X, Wei
W, Hu G and Guo J: Tumor suppressor SPOP ubiquitinates and degrades
EglN2 to compromise growth of prostate cancer cells. Cancer Lett.
390:11–20. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhi X, Tao J, Zhang L, Tao R, Ma L and Qin
J: Silencing speckle-type POZ protein by promoter hypermethylation
decreases cell apoptosis through upregulating Hedgehog signaling
pathway in colorectal cancer. Cell Death Dis. 7:e25692016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rump L, Mattey DL, Kehoe O and Middleton
J: An initial investigation into endothelial CC chemokine
expression in the human rheumatoid synovium. Cytokine. 97:133–140.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng Y, Han GW, Abagyan R, Wu B, Stevens
RC, Cherezov V, Kufareva I and Handel TM: Structure of CC Chemokine
receptor 5 with a potent chemokine antagonist reveals mechanisms of
chemokine recognition and molecular mimicry by HIV. Immunity.
46:1005–1017.e5. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Prabhakaran S, Rizk VT, Ma Z, Cheng CH,
Berglund AE, Coppola D, Khalil F, Mulé JJ and Soliman HH:
Evaluation of invasive breast cancer samples using a 12-chemokine
gene expression score: Correlation with clinical outcomes. Breast
Cancer Res. 19:712017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Arfsten DP, Schaeffer DJ and Muneny DC:
The effects of near ultraviolet radiation on the toxic effects of
polycyclic aromatic hydrocarbons in animals and plants: A review.
Ecotoxicol Environ Saf. 33:1–24. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Saunders CR, Ramesh A and Shockley DC:
Modulation of neurotoxic behavior in F-344 rats by temporal
disposition of benzo(a)pyrene. Toxicol Lett. 129:33–45. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yamaguchi K, Near R, Shneider A, Cui H, Ju
ST and Sherr DH: Fluoranthecene-induced apoptosis in murine T cell
hybridomas is independent of the aromatic hydrocarbon receptor.
Toxicol Appl Pharmacol. 139:144–152. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kang HG, Jeong SH, Cho MH and Cho JH:
Changes of biomarkers with oral exposure to benzo(a)pyrene,
phenanthrene and pyrene in rats. J Vet Sci. 8:361–368. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fries GF: A review of the significance of
animal food products as potential pathways of human exposures to
dioxins. J Anim Sci. 73:1639–1650. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kogevinas M: Human health effects of
dioxins: Cancer, reproductive and endocrine system effects. Hum
Reprod Update. 7:331–339. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Schug TT, Janesick A, Blumberg B and
Heindel JJ: Endocrine disrupting chemicals and disease
susceptibility. J Steroid Biochem Mol Biol. 127:204–215. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liem AK, Fürst P and Rappe C: Exposure of
populations to dioxins and related compounds. Food Addit Contam.
17:241–259. 2000. View Article : Google Scholar : PubMed/NCBI
|