Introduction
Osteosarcoma (OS) is a bone tumor that typically
affects children and adolescents <20 years old. OS is
characterized by lung metastasis and poor prognosis, posing a
serious threat to health (1). In
recent years, research into the molecular changes associated with
cancer has received increasing attention (2). The identification of valuable novel
biomarkers and potential molecular-level treatment approaches for
OS are expected to improve future diagnosis and treatment (3).
Metastasis-associated protein 1 (MTA1) is a
chromatin remodeling factor that forms part of the nucleosome
remodeling and histone deacetylase (NuRD) complex, along with MTA2
and MTA3 and is crucial for gene expression, cell survival and
promoting the hypoxic response as an oncogene upregulated in human
cancer (4). A previous study
demonstrated that MTA1 promotes non-small-cell lung cancer cell
epithelial-mesenchymal transition and metastasis (5). Kim and Park (6) also reported the high-level expression
of MTA1 in OS.
MicroRNAs (miRNA/miRs) are a group of endogenous
small (19–25-nucleotide) non-coding RNAs. Differential expression
of miRNAs has been described in almost every type of tumor, and so
miRNAs have potential as diagnostic or prognostic markers (7,8). In the
present study, it was observed that miR-183 functioned as a tumor
suppressorand served a significant role in OS cell proliferation,
migration and apoptosis by binding to a target gene. MiRNAs
post-transcriptionally silence complementary target genes and
increasing evidence suggests that miR-183 functions as a tumor
suppressor (9). For example, miR-183
has been reported to function as an oncogene to suppress
neuropathic pain and the expression of the
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor
(10).
The aim of the present study was to investigate the
effects of miR-183 on cellular processes, including metastasis,
invasion and apoptosis and to evaluate the mechanisms by which
miR-183 regulates its direct target MTA1 in MG63 cells.
Materials and methods
Tumor sample collection
Paired OS tumor tissues and adjacent normal tissues
were obtained from patients (n=25), including 14 male and 11 female
(mean age 16 years; range, 6–58 years) who were treated at the
First Affiliated Hospital of Zhengzhou University (Henan, China)
between September 2014 and June 2016, in accordance with an
institutionally approved protocol. All samples were obtained from
primary OS patients who were not subjected to chemotherapy or
radiotherapy and were not suffering from any other diseases. All
patients were diagnosed with OS on the basis of clinicopathological
findings. All tissues were flash frozen in liquid nitrogen at the
time of surgery and stored at −80°C until use. All procedures were
conducted in accordance with the principles of the Declaration of
Helsinki. Informed consent was obtained from patients themselves or
their families and the procedures were approved by the ethics
committee of Zhengzhou University.
Cell lines and cell culture
The normal human osteoblastic cell line hFOB 1.19
and the human OS cell line MG63 were purchased from the Cell
Culture Center of the Chinese Academy of Medical Sciences (Beijing,
China). MG63 cells were maintained in RPMI 1640 medium (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10%
fetal bovine serum (FBS; Sigma-Aldrich, USA), 100 U/ml penicillin
and 100 µg/ml streptomycin at 37°C in a humidified incubator
containing 5% CO2. hFOB 1.19 cells were grown in
Dulbecco's modified Eagle's medium/F12 (1:1; Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS, 100 U/ml penicillin
and 100 µg/ml streptomycin at 37°C in a humidified incubator
containing 5% CO2.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was purified from tissue samples and
cultured cells using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. RT was
performed at 42°C for 2 min, 37°C for 15 min and 85°C for 5 sec
using a Takara RNA PCR kit (Takara Biotechnology Co., Ltd., Dalian,
China) according to the manufacturer's protocol. qPCR was performed
using SYBR Premix Ex TaqII (Takara Biotechnology Co., Ltd.) and an
Applied Biosystems 7500 Real-time PCR System (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. All
reactions were performed in triplicate and thermocycling conditions
were as follows: 95°C for 30 sec followed by 40 cycles of 95°C for
5 sec and 60°C for 30 sec, with a final dissociation stage for 1h
at4°C. U6 small nuclear RNA and β-actin mRNA were used as internal
controls. The relative level of miRNA expression and mRNA
expression was calculated using the 2−ΔΔCq method
(11). The primers used were
designed and synthesized by Sangon Biotech Co., Ltd. (Shanghai,
China) and are listed in Table
I.
 | Table I.Primers used for reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Primers used for reverse
transcription-quantitative polymerase chain reaction.
| Gene | Direction | Sequence (5′-3′) |
|---|
| U6 | Forward |
TCCGATCGTGAAGCGTTC |
|
| Reverse | GTGCAGGGTCCGAGGT |
| β-actin | Forward |
ACATCTCCAGCACCCTCATC |
|
| Reverse |
TAGATGGGCACAGTGTGGGTG |
| Human
microRNA-183 | Forward |
CTGCGAGTATGGCACTGGTAGAA |
|
| Reverse |
ATCCAGTGCAGGGTCCGAGG |
| Metastasis-associated
protein 1 | Forward |
CGGATTCTCCATTTCCTCTTC |
|
| Reverse |
ACATCTCCAGCACCCTCATC |
miRNA transfection
ThemiR-183 mimic and negative control miRNA used in
the current study were obtained from Shanghai GenePharma Co., Ltd.
(Shanghai, China). MG63 cells were seeded in 24-well plates
(2×105 cells/well) and grown to ~80% confluence as
described above. Cells were transiently transfected using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. The
following groups of cells were prepared: Cells treated with vehicle
only (control group); cells transfected with a scramble miR-183
sequence [negative control (NC) group]; and cells transfected with
miR-183 mimic (miR-183 group). The transfection efficiency of
miR-183 mimic was tested using RT-qPCR as described above, after
1–2 days and cells were used for further experiments.
Cell Counting Kit-8 (CCK-8) cell
viability assay
Transfected MG63 cells were seeded in 96-well plates
(5×103 cells/well, five replicate wells per group),
cultured for 1–2 days and subsequently subjected to a cell
viability assay using a CCK-8 (Dojindo Molecular Technologies,
Inc., Kumamoto, Japan) to detect viable cells on days 1–5. In
brief, cells were incubated with 10 µl CCK-8 solution for 48 h at
37°C in a humidified atmosphere containing 5% CO2 until
a visible color conversion occurred. Absorbance values were
measured at 490 nm and proliferation curves were plotted.
Transwell invasion assay
Following 48 h of culture, transfected cells were
collected and resuspended in RPMI 1640 medium at 2×105
cells/ml. The cell suspension (200 µl) was transferred to the upper
chamber of a Transwell Permeable Support device (Costar; Corning
Incorporated, Corning, NY, USA) with a membrane insert containing
8-µm pores and incubated at 37°C in a humidified incubator
containing 5% CO2. RPMI 1640 culture medium (500 µl)
containing 10% FBS was added to the lower chamber of the Transwell
device. Following 24 or 48 h of incubation, cells on the upper
surface of the membrane were removed using a cotton swab. The cells
that had migrated to the lower surface of the membrane were fixed
with 4% paraformaldehyde for 15 min at room temperature, washed
three times with PBS and stained with hematoxylin for 15 min at
room temperature. Cells were subsequently mounted and air-dried and
the number of cells that had invaded the lower surface of the
membrane was counted in three randomly selected fields under a
light microscopic at ×200 magnification.
Scratch-wound healing assay
At 24 h following transfection, cells were seeded in
6-well plates (2×103 cells/well) and allowed to grow
overnight to 80–90% confluence at 37°C. The cell layer was then
scratched using a sterile 10-µl pipette tip. The wounded monolayer
was washed twice with PBS and fresh serum-free culture medium was
added. At 24 and 36 h after wounding, micrographs were captured
with a light microscope (original magnification, ×100). All
experiments were repeated in triplicate.
Apoptosis assay
Fluorescence-activated cell sorting assays were
conducted to assess apoptosis. At 36 h following transfection,
cells were harvested and diluted in 500 µl PBS (1×106
cells/ml; Shanghai BestBio Biotechnology, Shanghai, China) and
subsequently stained with Annexin V-fluorescein isothiocyanate and
propidium iodide (Beyotime Institute of Biosciences, Haimen, China)
at room temperature in the dark for 15 min. A calibrated
FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ,
USA) equipped with Cell Quest software 5.1 (BD Biosciences) was
used to analyze the cells.
To further analyze apoptosis, a TUNEL assay was
performed. Following 36 h of culture in a 6-well plate (as above),
transfected cells were subjected to a Dead End™ Fluorometric TUNEL
System (Promega Corp., Madison, WI, USA) assay in accordance with
the manufacturer's protocol to detect apoptotic cells. Briefly,
cells were washed twice with PBS, fixed with 4% paraformaldehyde
for 20 min at room temperature and treated with 50 µl of the TUNEL
reaction solution, 50 µl of the enzyme solution and 450 µl of the
labeling solution at 37°C in the dark for 1 h. The cells were then
observed by fluorescence microscopy at magnification, ×200 (Olympus
Corporation, Tokyo, Japan).
Bioinformatics analysis
Using starBase V2.0 (http://starbase.sysu.edu.cn/), TargetScan 7.1
(http://www.targetscan.org/) and
miRanda(http://www.microrna.org)bioinformatics
software, the target genes were identified. The results obtained
from the software packages were considered and the 3′-untranslated
region (3′-UTR) region of MTA1 mRNA was selected as the sequence
most likely to be complementary to miR-183.
Dual luciferase reporter assay
PCR was used to amplify the region of the MTA1 gene
that included putative binding sites for miR-183 from MG63 cells
with a Taq DNA Mix kit (Takara Biotechnology Co., Ltd.).
Overlap-extension PCR was used to amplify the mutant segment of the
MTA1 mRNA 3′UTR. The primers usedwere as follows: Wild type forward
5′TTTCTCGAGGCCGAGGAGTTGTCGTTTTTAGCTT3′, and wild type reverse,
5′TTCTCTAGACTTGAAAAGACAAGGGCCAACCCCG3′; mutantforward,
5′TTGAAGCACGGTATTTAAATTTTATTTTTATTACTTTTTTTGTAG3′ and
mutantreverse, 5′AATACCGTGCTTCAACAGAACACAACAAAACCTTAGGGCCCGGCC3′.
The PCR steps were as follows: Initial denaturation for 3 min at
95°C; 34 cycles of denaturation for 35 sec at 95°C, annealing for
35 sec at 55°C and elongation for 60 sec at 72°C; and a final
extension step for 60 min at 10°C. Amplified fragments were
inserted into a pmir-GLO plasmid (Promega Corp.) downstream of the
luciferase gene to create the recombinant vectors pGLO-MAT1-wt
(wild-type reporter vector) and pGLO-MTA1-mut (mutant reporter
vector). MG63 cells were cultured for 48 h in 96-well plates
(2×105 cells/ml) and the miRNA and reporter plasmid were
cotransfected into the cells using Lipofectamine® 2000
reagent according to the manufacturer's protocol. At 24 h following
transfection, luciferase activity was determined using a Dual
Luciferase Reporter Assay System (Promega Corp.) according to the
manufacturer's protocol.
Western blotting
Western blotting was performed to examine MTA1
protein expression levels in the OS tissue samples and MG63 cells.
Cells were incubated and lysed in radioimmunoprecipitation assay
buffer containing phenylmethanesulfonyl fluoride (Beyotime
Institute of Biotechnology) to extract the total protein, following
which the total protein concentration was quantified using a
Bicinchoninic Acid Protein Assay kit (Beyotime Institute of
Biotechnology). The procedure for western blotting was as follows:
Extracted proteins (10 µl per lane)were separated by 10% SDS-PAGE
and transferred to polyvinylidene difluoride membranes. The
membranes were subsequently blocked with 5% non-fat milk at room
temperature for 2 h, rinsed four times with Tris-buffered saline
containing Tween-20 (TBST) and incubated with primary antibodies
against MTA1 (cat. no. sc-373765; diluted 1:1,000) and β-actin
(cat. no. sc-130065; diluted 1:3,000) (both Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) overnight at 4°C. Membranes
were then washed in TBST four times and incubated with horseradish
peroxidase-conjugatedgoat anti-mouse secondary immunoglobulin G
(cat. no. sc-2005; diluted 1:2,500; Santa Cruz Biotechnology, Inc.)
for 2 h at room temperature. The membranes were then washed four
times (5 min per wash) with TBST and signals from immunoreactive
bands were quantified with a chemiluminescence detection kit (GE
Healthcare Life Sciences, Little Chalfont, UK). AlphaView3.3.0
software (ProteinSimple; Bio-Techne, Minneapolis, MN, USA) was used
for densitometry analysis. β-actin was used as an internal
reference to confirm equal protein loading.
Statistical analysis
All statistical analyses including correlation
analysis were performed using SPSS 19.0 software (IBM Corp.,
Armonk, NY, USA) and data are presented as the mean ± standard
deviation. The Student's t-test, Pearson correlation analysis,
one-way analysis of variancewith Dunnett's post hoc test were used
to evaluate differences between groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-183 and MTA1 expression in human
OS tissues and cell lines
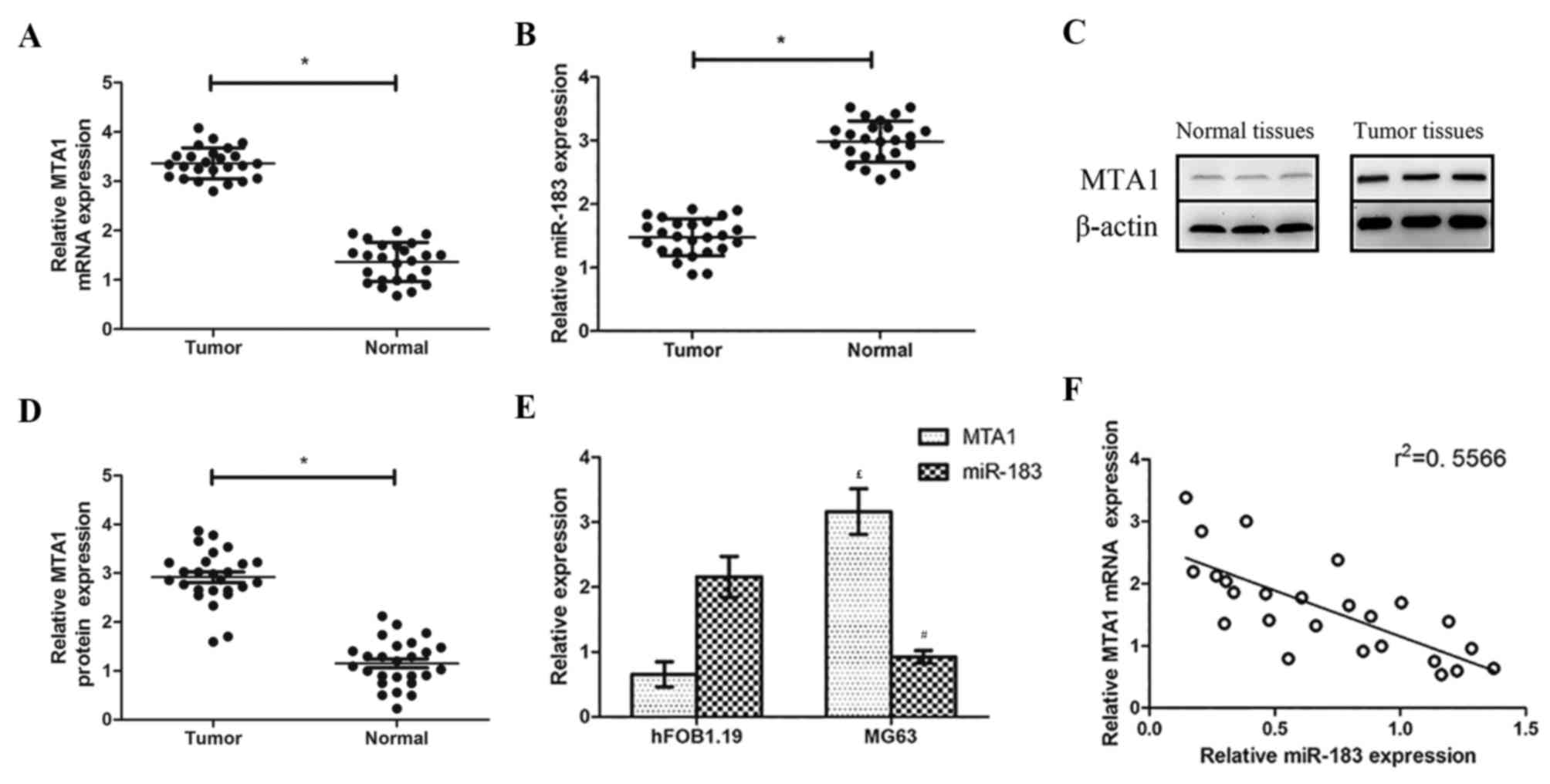
RT-qPCR analysis revealed that MTA1 mRNA and MTA1
protein levels were higher in OS tissues compared with normal
adjacent tissues, whereas relative miR-183 levels were lower (both
P<0.05; Fig. 1A and B). Western
blotting revealed that MTA1 was overexpressed in OS tissues
compared with the normal paracancerous tissues (Fig. 1C and D). The same patterns were
observed in the OS and normal cell lines via the same methods:
miR-183 expression was significantly lower in MG63 cells compared
with their expression levels in hFOB 1.19 cells, while MTA1 mRNA
expression was higher (both P<0.05; Fig. 1E). A negative correlation was also
observed between miR-183 and MTA1 expression (Fig. 1F).
miR-183 overexpression impairs
proliferation, invasion and migration in MG63 cells in vitro
To determine the potential functional role of
miR-183 in OS cell proliferation, migration and invasion, CCK-8,
Transwell and scratch-wound healing assays were performed to
examine the differences between cells prior to and following
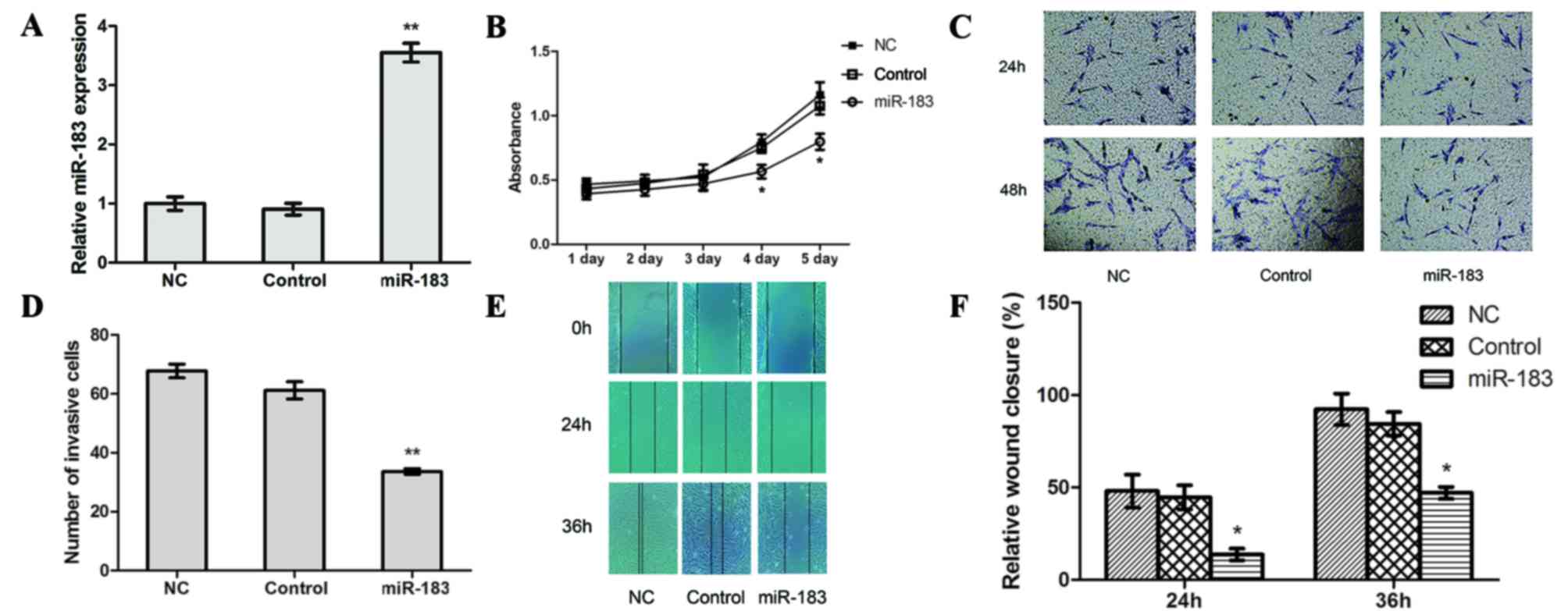
transfection with an miR-183 mimic. The results of RT-qPCR revealed
that the transfection efficiency of miR-183 mimic-transfected cells
was significantly higher compared with control and NC cells
(P<0.01; Fig. 2A), whereas no
significant differences were observed between the control and NC
cells.
The results of a CCK-8 assay revealed a lower
absorption value in miR-183 mimic-transfected cells compared with
control and NC cells (P<0.05; Fig.
2B), which suggests that high miR-183 expression may suppress
OS cell proliferation. In the Transwell assay, the number of cells
that crossed the basement membrane was significantly lower in
miR-183 mimic-transfected cells (33.5924±0.9857) compared with the
control (61.1830±2.9217) and NC (67.7391±2.3318) cells (P<0.01;
Fig. 2C and D). This suggests that
increased miR-183 expression inhibits OS cell invasion.
Furthermore, the scratch-wound healing assay revealed that the
relative extent of migration of miR-183 mimic-transfected cells was
significantly lower compared with the control and NC cells
(P<0.05; Fig. 2E and F), which
suggests that miR-183 overexpression impairs OS cell migration.
miR-183 overexpression promotes
apoptosis in OS cells in vitro
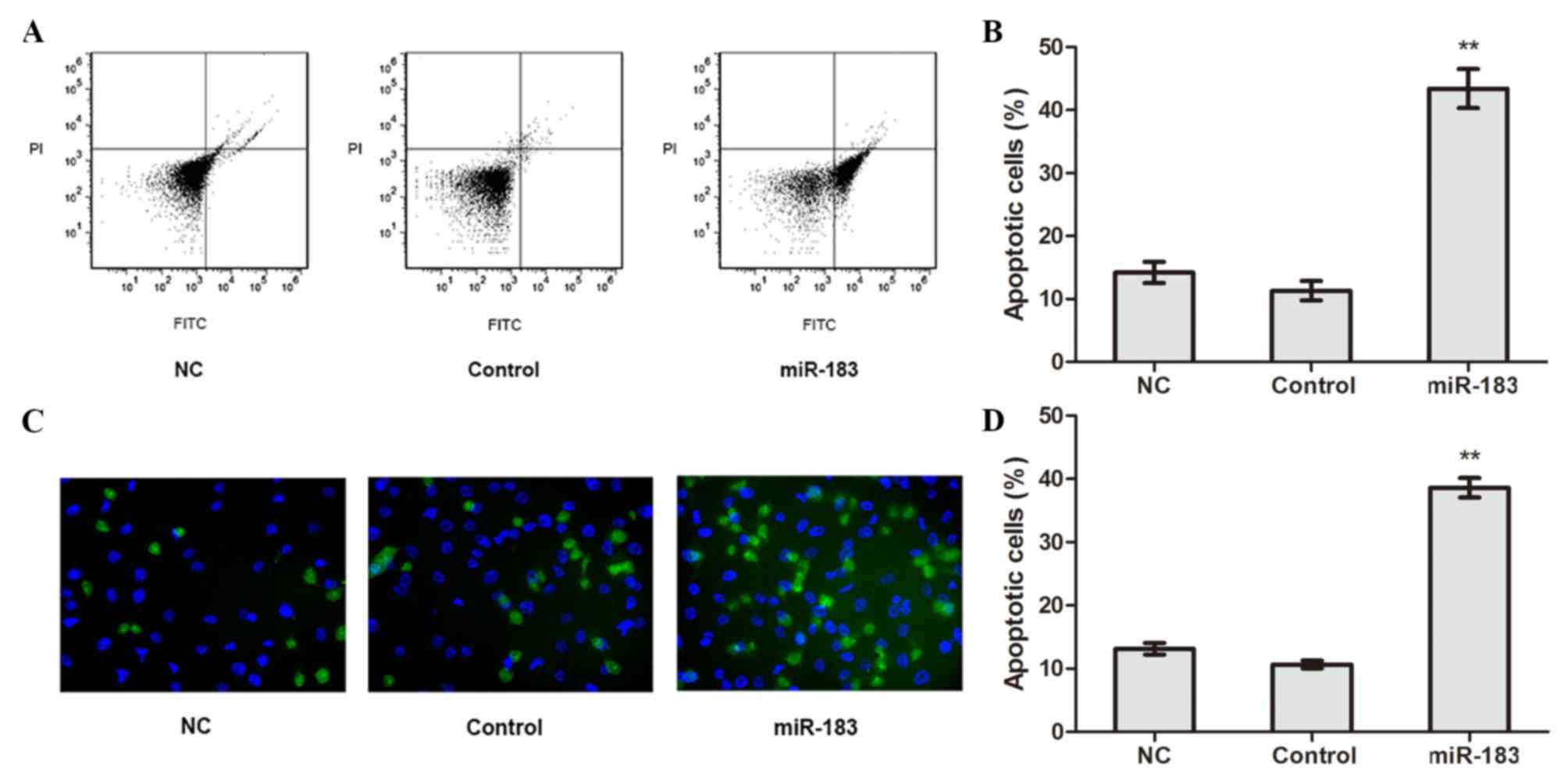
The apoptosis of transfected cells was detected
using flow cytometry. The results revealed that the apoptotic rate
of miR-183 mimic-transfected cells was significantly higher
compared with control and NC cells (P<0.01; Fig. 3A and B), whereas no significant
difference in apoptotic rate was observed between the control and
NC cells. TUNEL assay results confirmed the apoptotic rate of
miR-183 mimic-transfected cells was higher compared with control
and NC cells (P<0.01; Fig. 3C and
D). These results suggest that elevated miR-183 expression
promotes OS cell apoptosis.
miR-183 binds directly to MTA1 and
there is a negative association between miR-183 and MTA1 expression
in OS cells
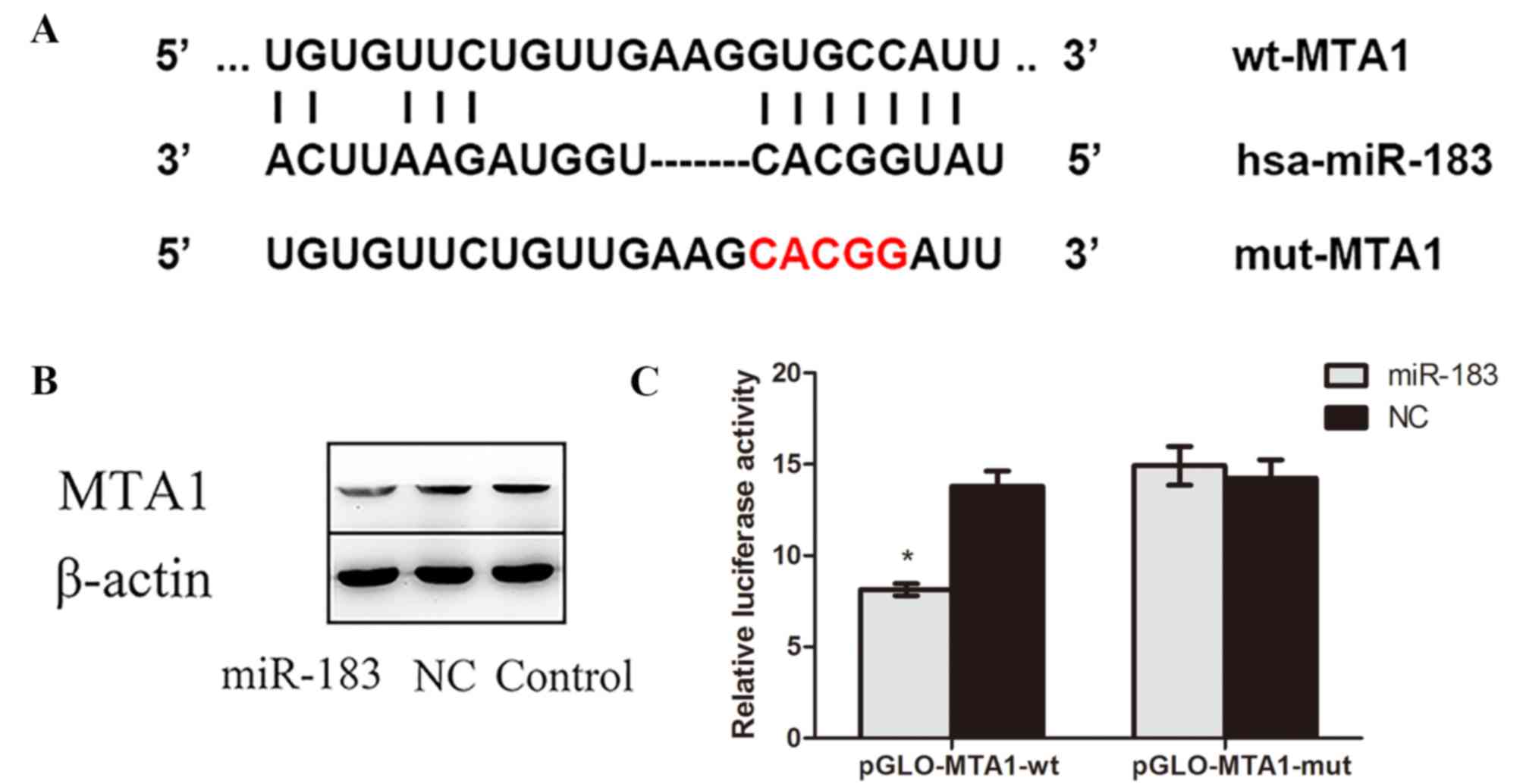
The complementary sites of MTA1 and miR-183
predicted by bioinformatics software, as well as the mutant
sequences of MTA1, are presented in Fig.
4A. The results of western blotting demonstrated that MTA1
protein expression was lower in miR-183 mimic-transfected cells
compared with control and NC cells (Fig.
4B), confirming that miR-183 targets MTA1. Furthermore, a dual
luciferase reporter assay indicated that luciferase activity in
miR-183 mimic and pmirGLO-MTA1-wtcotransfected OS cells was
significantly decreased compared with cells cotransfected with
scramble miR-183 pmirGLO-MTA1-mut (P<0.05; Fig. 4C). No significant difference in
luciferase activity was observed between cells cotransfected with
miR-183 negative control and pmirGLO-MTA1-wt and those transfected
with miR-183 negative control and pmirGLO-MTA1-mut. Collectively,
these results suggest that miR-183 targets the 3′-UTR of MTA1 and
negatively regulates its expression.
Discussion
MTA1 has been established as a DNA damage response
protein and has been widely studied in a variety of types of cancer
(12). Tuncay Cagatay et al
(13) demonstrated that MTA1
enhances zing finger proteins SNAI1 and SNAI2 expression and
increases epithelial cadherin expression. Furthermore, they
reported that MTA1 overexpression promotes cell proliferation,
migration and invasion in colorectal cancer cells (13). In human non-small-cell lung carcinoma
and liver cancer cells, MTA1 has been reported to inhibit apoptosis
by regulating the NuRD complex, which mediates p53 deacetylation
(14,15). Furthermore, a meta-analysis of
several tumor types demonstrated that MTA1 may be used as a novel
indicator of poor prognosis in patients with tumors (16,17).
These findings suggest that MTA1 may be a potential target for
cancer therapy. In the present study, it was identified that MTA1
was significantlyupregulated in OS tissues and cells and served as
a potential oncogene in OS.
miRNAs silence target genes or induce mRNA
degradation by completely or incompletely binding to the 3′-UTR of
target mRNAs (18,19). Zhu et al (20) demonstrated that miR-183 downregulates
ezrin expression and inhibits the invasion and migration of OS.
Additionally, a number of studies have reported that miR-183
inhibits the function of transforming growth factor-β1 to induce
apoptosis in human hepatoma cells by targeting programmed cell
death 4 (21,22). Other studies have demonstrated that
miR-183 levels were abnormally low in a number of cancers,
including OS (23–27). In particular, a study by Wang et
al (28) identified low
expression of miR-183 and high expression of MTA1 in 29
nasopharyngeal carcinoma (NPC) tissues compared with 17 normal
nasopharyngeal epithelium tissues; cell experiments using NPC cell
lines compared with the human immortalized nasopharyngeal
epithelial cell line also obtained the same results. Furthermore,
Wang et al (28) reported
that miR-183 overexpression inhibits NPC cell proliferation and
migration, increases the rate of cellular apoptosis and improves
the cytotoxicity induced by the antitumor drug cisplatin by
targeting MTA1. These results were verified by a xenograft tumor
experiment in vivo (28). To
further confirm the reduction in miR-183 expression in OS, miR-183
expression levels in OS and paracancerous tissues were assessed in
the present study, as well as the OS cell line MG63 and the normal
osteoblast cell line hFOB1.19. The results obtained were in
agreement with previously published reports.
To the best of our knowledge, the effects of miR-183
binding to the 3′-UTR of MTA1 have not previously been studied. The
results of the present study demonstrate that MTA1 is a direct
target gene of miR-183 and that miR-183 negatively regulates the
expression of MTA1 by binding to the 3′-UTR. Additionally, low
miR-183 expression and high MTA1 expression may serve a role in the
pathogenesis of OS. The results of the present study also suggest
thatmiR-183 upregulation may reduce MTA1 expression, thereby
inhibiting proliferation, invasion and migration as well as
promoting apoptosis in OS cells. However, the present study is not
without limitations. The central hypothesis was tested in only one
cell line and so should be validated in more cell lines as well as
xenograft tumor experiments in future studies.
In conclusion, the present study is the first to
report that miR-183 regulates MTA1 gene expression in OS. These
results may provide a basis for the development of novel treatment
targets for the prevention and treatment of OS in the future.
Acknowledgements
The authors would like to thank Professor Li Yuebai
(Zhengzhou University, Zhengzhou, China) for their guidance and
support during the experimental project.
Funding
No fundingwas received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YL, XS and YX conceived the study, participated in
its design and coordination and helped to draft the manuscript. YW,
YZ and XL collected the samples. XS, YX, SZ, XL and YW performed
some of the experiments and wrote the manuscript. YL, XS, YW and XZ
performed the statistical analysis. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
All procedures were conducted in accordance with the
principles of the Declaration of Helsinki. Informed consent was
obtained from the patients or their families and the procedures
were approved by the Ethics Committee of Zhengzhou University.
Consent for publication
Informed consent was obtained from all individuals
included within the study for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Garimella R, Tadikonda P, Tawfik O,
Gunewardena S, Rowe P and Van Veldhuizen P: Vitamin D impacts the
expression of Runx2 Target Genes and Modulates Inflammation,
oxidative stress and membrane vesicle biogenesis gene networks in
143B osteosarcoma cells. Int J Mol Sci. 18:E6422017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mingardi J, Musazzi L, De Petro G and
Barbon A: miRNA editing: New insights into the fast control of gene
expression in health and disease. Mol Neurobiol. Feb 19–2018.(Epub
ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schadt EE: Molecular networks as sensors
and drivers of common human diseases. Nature. 461:218–223. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kaur E, Gupta S and Dutt S: Clinical
implications of MTA proteins in human cancer. Cancer Metastasis
Rev. 33:1017–1024. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li SH, Tian H, Yue WM, Li L, Li WJ, Chen
ZT, Hu WS, Zhu YC and Qi L: Overexpression of metastasis-associated
protein 1 is significantly correlated with tumor angiogenesis and
poor survival in patients with early-stage non-small cell lung
cancer. Ann Surg Oncol. 18:2048–2056. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim SS and Park YK: Significance of MTA1
in the molecular characterization of osteosarcoma. Cancer
Metastasis Rev. 33:981–991. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sadri D, Farhadi S, Shahabi Z and Sarshar
S: Expression of vascular endothelial growth factor in odontogenic
cysts: Is there any impression on clinical outcome. Open Dent J.
10:752–759. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Matsumoto K, Xavier S, Chen J, Kida Y,
Lipphardt M, Ikeda R, Gevertz A, Caviris M, Hatzopoulos AK,
Kalajzic I, et al: Instructive role of the microenvironment in
preventing renal fibrosis. Stem Cells Translat Med. 6:992–1005.
2017. View Article : Google Scholar
|
|
9
|
Fan D, Wang Y, Qi P, Chen Y, Xu P, Yang X,
Jin X and Tian X: MicroRNA-183 functions as the tumor suppressor
via inhibiting cellular invasion and metastasis by targeting MMP-9
in cervical cancer. Gynecol Oncol. 141:166–174. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xie X, Ma L, Xi K, Zhang W and Fan D:
MicroRNA-183 suppresses neuropathic pain and expression of AMPA
receptors by targeting mTOR/VEGF signaling pathway. Cell Physiol
Biochem. 41:181–192. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li DQ, Pakala SB, Reddy SD, Ohshiro K,
Peng SH, Lian Y, Fu SW and Kumar R: Revelation of p53-independent
function of MTA1 in DNA damage response via modulation of the p21
WAF1-proliferating cell nuclear antigen pathway. J Biol Chem.
285:10044–10052. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cagatay Tuncay S, Cimen I, Savas B and
Banerjee S: MTA-1 expression is associated with metastasis and
epithelial to mesenchymal transition in colorectal cancer cells.
Tumour Biol. 34:1189–1204. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Moon HE, Cheon H and Lee MS:
Metastasis-associated protein 1 inhibits p53-induced apoptosis.
Oncol Rep. 18:1311–1314. 2007.PubMed/NCBI
|
|
15
|
Xue Y, Wong J, Moreno GT, Young MK, Côté J
and Wang W: NURD, a novel complex with both ATP-dependent
chromatin-remodeling and histone deacetylase activities. Mol Cell.
2:851–861. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Oliveto S, Mancino M, Manfrini N and Biffo
S: Role of microRNAs in translation regulation and cancer. World J
Biol Chem. 8:45–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang H, Zhu X, Li N, Li D, Sha Z, Zheng X
and Wang H: MiR-125a-3p targets MTA1 to suppress NSCLC cell
proliferation, migration, and invasion. Acta Biochim Biophys Sin
(Shanghai). 47:496–503. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cao JM, Li GZ, Han M, Xu HL and Huang KM:
MiR-30c-5p suppresses migration, invasion and epithelial to
mesenchymal transition of gastric cancer via targeting MTA1. Biomed
Pharmacother. 93:554–560. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Miao Y, Lu M, Yan Q, Li S and Feng Y:
Inhibition of proliferation, migration, and invasion by knockdown
of pyruvate kinase-M2 (PKM2) in ovarian cancer SKOV3 and OVCAR3
cells. Oncol Res. 24:463–475. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu J, Feng Y, Ke Z, Yang Z, Zhou J, Huang
X and Wang L: Down-regulation of miR-183 promotes migration and
invasion of osteosarcoma by targeting Ezrin. Am J Pathol.
180:2440–2451. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gu W, Gao T, Shen J, Sun Y, Zheng X, Wang
J, Ma J, Hu XY, Li J and Hu MJ: MicroRNA-183 inhibits apoptosis and
promotes proliferation and invasion of gastric cancer cells by
targeting PDCD4. Int J Clin Exp Med. 7:2519–2529. 2014.PubMed/NCBI
|
|
22
|
Li J, Fu H, Xu C, Tie Y, Xing R, Zhu J,
Qin Y, Sun Z and Zheng X: miR-183 inhibits TGF-beta1-induced
apoptosis by downregulation of PDCD4 expression in human
hepatocellular carcinoma cells. BMC Cancer. 10:3542010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cao LL, Xie JW, Lin Y, Zheng CH, Li P,
Wang JB, Lin JX, Lu J, Chen QY and Huang CM: miR-183 inhibits
invasion of gastric cancer by targeting Ezrin. Int J Clin Exp
Pathol. 7:5582–5594. 2014.PubMed/NCBI
|
|
24
|
Zhao H, Guo M, Zhao G, Ma Q, Ma B, Qiu X
and Fan Q: miR-183 inhibits the metastasis of osteosarcoma via
downregulation of the expression of Ezrin in F5M2 cells. Int J Mol
Med. 30:1013–1020. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xu L, Li Y, Yan D, He J and Liu D:
MicroRNA-183 inhibits gastric cancer proliferation and invasion via
directly targeting Bmi-1. Oncol Lett. 8:2345–2351. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang G, Mao W and Zheng S: MicroRNA-183
regulates Ezrin expression in lung cancer cells. FEBS Lett.
582:3663–3668. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang L, Quan H, Wang S, Li X and Che X:
MiR-183 promotes growth of non-small cell lung cancer cells through
FoxO1 inhibition. Tumour Biol. 36:8121–8126. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang G, Wang S and Li C: MiR-183
overexpression inhibits tumorigenesis and enhances DDP-induced
cytotoxicity by targeting MTA1 in nasopharyngeal carcinoma. Tumour
Biol. 39:10104283177038252017.PubMed/NCBI
|