Introduction
Osteosarcoma (OS) is the most common form of primary
bone malignancy that frequently affects children, adolescents and
young adults, with ~3,260 cases in the United States reported in
2017(1). Although progress has been
made in the treatment of OS, including surgery and chemotherapy,
the five-year survival rate remains relatively low in OS patients
(2). Numerous studies indicate that
epigenetic changes including non-coding RNA signatures, DNA
methylation and histone modifications are present in all human
malignancies and can facilitate cancer development and progression
(3,4).
MicroRNAs (miRNAs) have attracted major interest due
to their ability to affect a wide range of fundamental biological
processes such as proliferation, invasion, differentiation and
apoptosis (5). Studies into
OS-specific miRNAs have identified their potential in the
prevention and management of OS (6,7). Based
on the latest version of miRBase, ~3,700 miRNAs loci were annotated
in human tissues (8). Bioinformatic
predictions suggest that 30-60% of the protein-coding genes are
regulated by miRNAs (9). MiRNAs
cause mRNA degradation and post-transcriptional downregulation by
binding to the 3'untranslated region (UTR) of their target genes
(10). miR-106b-5p, a member of the
miR-106b-25 cluster, is mapped to human chromosome 7q21 (Chr7q21)
locus and has been reported to be amplified in several types of
human malignancies, including renal cell carcinoma (11), hepatocellular carcinoma (12), glioma (13), melanoma (14) and non-small cell lung cancer
(15). The deleterious effects of
miRNA dysregulation on malignant behaviors appear to be strongly
associated with tumorigenesis and cell cycle progression (12-15).
However, current understanding regarding the role of miR-106b-5p in
OS development and progression remains limited.
Cyclin-dependent kinase (CDK) represents a family of
proline-directed serine/threonine kinases with important regulatory
roles in modulating cell division in response to extrinsic and
intrinsic signaling events (16).
CDK inhibitor 1A (CDKN1A) is considered as a critical and universal
CDK-interacting protein, which binds to CDKs and/or its subunits
(17). There is clear evidence that
CDKN1A is involved in cell cycle progression, proliferation,
survival, motility and senescence (18,19).
CDKN1A expression has been previously found to be downregulated in
numerous human malignancies, the restoration of which has been
observed to attenuate metastasizes in vivo (19,20). To
date, a number of studies have identified that some miRNAs are
associated with the expression of CDKN1A, including miR-93(21), miR-130a (22), miR-519d (23) and miR-4295(24). In OS, knockdown of miR-95-3p has been
shown to inhibit cell growth by epigenetically regulating CDKN1A
(25).
The purpose of the present study was to explore the
biological role of miR-106b-5p in OS and identify the critical
tumor-suppressed targets of miR-106b-5p. To the best of our
knowledge, the current study first revealed that CDKN1A was a
direct target of miR-106b-5p in OS, which will establish the
miR-106b-5p/CDKN1A axis in the development and progression of
OS.
Materials and methods
Patients and tumor specimens
A total of 18 pairs of fresh surgically resected OS
tissue and adjacent bone tissue, 5 cm from the edge of tumor site,
were obtained from OS patients (age range, 13-68 years; sex, 12
females and 6 males) after diagnosis by experienced pathologists
between March 2015 and September 2017 at the Jingzhou Traditional
Chinese Medicine Hospital (Hubei, China). All collected tissues
were immediately frozen in liquid nitrogen. The present study was
approved by the Ethics Committee of Jingzhou Traditional Chinese
Medicine Hospital and all patients provided their written informed
consent.
Cell culture
Human OS cell lines (Saos-2, MG-63, SW1353 and
U2OS), osteoblast cell line hFOB 1.19 and embryonic kidney cell
line 293T were purchased from the American Type Culture Collection.
All cell lines were cultured in RPMI-1640 medium (Gibco; Thermo
Fisher Scientific, Inc.) with 10% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc.), 100 U/ml penicillin and streptomycin
(Invitrogen; Thermo Fisher Scientific, Inc.). Samples were
maintained in a humidified atmosphere containing 5% CO2
at 37˚C.
Oligonucleotides and cell
transfection
Oligonucleotides, including miR-106b-5p inhibitors
(5'-ATCTGCACTGTCAGCACTTTA-3') and negative controls (miR-NC,
5'-TTCTCCGAACGTGTCACGT-3') were designed and synthesized by
Shanghai GenePharma Co., Ltd. The open reading frame of CDKN1A,
generated from RNA samples of Saos-2 cells (forward,
5'-CACCATGTCAGAACCGGCTGGGGATG-3'; reverse,
5'-TTAGGGCTTCCTCTTGGAGAAGATCAGC-3'), was inserted into the pcDNA3.1
expression vector to generate overexpressing recombinant vector
pcDNA3.1-CDKN1A (Shanghai GenePharma Co., Ltd.). Small interfering
(si)RNA for CDKN1A (si-CDKN1A) and its NC (si-NC) were synthesized
by Shanghai GenePharma Co., Ltd. Saos-2 or U2OS cells
(1x104 cells per well) were seeded into six-well plates
and transfected with 50 nM miRNA, 100 pmol siRNA and/or 4 µg
plasmid using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) for 48 h according to the manufacturer's
instructions.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Total RNA was extracted with TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. For miR-106b-5p detection, the temperature
protocol for reverse transcription of miRNA was as follows: 37˚C
for 60 min, 95˚C for 5 min and the samples were subsequently kept
at 4˚C. RT-qPCR was performed in triplicate using a miRVana™
real-time RT-PCR microRNA detection kit (Thermo Fisher Scientific,
Inc.) with U6 as an internal control. The thermocycling conditions
were as follows: Initial denaturation of 95˚C for 2 min, followed
by 40 cycles of 95˚C for 10 sec, 55˚C for 30 sec and 72˚C for 30
sec. For CDKN1A detection, cDNA was synthesized using a
PrimeScript™ RT reagent kit (Takara Bio, Inc.). The temperature
protocol for reverse transcription of RNA was as follows: 37˚C for
15 min and 85˚C for 5 sec. The expression of CDKN1A mRNA was
determined using SYBR Premix Ex Taq™ II (Takara Bio, Inc.) using
primers (forward, 5'-TCTGGGGTCTCACTTCTTGG-3' and reverse,
5'-ATGTGAGGAAGGCTCAGTGG-3') with GAPDH (forward,
5'-TGCACCACCAACTGCTTA-3' and reverse, 5'-GGATGCAGGGATGATGTTC-3') as
an internal control. The thermocycling conditions were as follows:
Initial denaturation at 95˚C for 10 min, followed by 40 cycles of
95˚C for 10 sec and 58˚C for 60 sec. Relative expression levels
were calculated using 2-ΔΔCq method (26).
Cell proliferation assay
Saos-2 or U2OS cells were seeded in 96-well plates
at a density of 4x103 cells per well and routinely
cultured (37˚C; 5% CO2) for 5 days. At the indicated
time points, 20 µl MTT reagent was added to each well
(Sigma-Aldrich; Merck KGaA) and incubated for another 2 h at 37˚C.
The blue formazan crystals in each well were subsequently dissolved
by adding 150 µl dimethylsulfoxide (Sigma-Aldrich; Merck KGaA).
Cell proliferation was evaluated by recording the absorbance value
at 595 nm using Model 680 microplate reader (Bio-Rad Laboratories,
Inc.).
Cell cycle analysis
Flow cytometry analysis was performed to determine
cell cycle distribution in OS cells. Saos-2 or U2OS cells at a
density of 4x105 cells per reaction, were washed with
PBS and fixed with pre-cooled 70% ethanol at 4˚C for 30 min.
Following centrifugation (450 x g at 4˚C and 5 min) and washing
with PBS, cells were stained with 500 µl 1% propidium iodide (PI;
Thermo Fisher Scientific, Inc.), followed by DNA content analyses
using BD FACScan™ flow cytometer equipped with CellQuest Pro 4.0.2
software (BD Biosciences).
Luciferase reporter assay
The putative target genes of miR-106b-5p were
predicted using public available algorithms, including PicTar
(https://pictar.mdc-berlin.de),
TargetScan (http://targetscan.org) and miRDB
(http://www.mirdb.org). The predicted miR-106b-5p
binding sites in the wild-type (WT) 3'UTR of CDKN1A and the
corresponding mutant type (MUT) miR-106b-5p binding sites were
cloned into a pGL3 vector (Promega Corporation). For the luciferase
reporter assay, 293T cells (1x105 cells/well) were
seeded in 96-well plates and co-transfected with 300 ng WT-CDKN1A
or MUT-CDKN1A, and 100 nM of miR-106b-5p inhibitor or miR-NC using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) in
accordance with the manufacturer's protocol. After 48 h
transfection, the relative luciferase activities were measured
using the dual-luciferase reporter assay system (Promega
Corporation). Renilla luciferase activity was used for
normalization.
Western blot analysis
Total protein was extracted using the RIPA protein
extraction reagent (Beyotime Institute of Biotechnology) and the
protein concentration was determined using the bicinchoninic acid
Protein Assay kit (Beyotime Institute of Biotechnology).
Approximately 30 µg protein was separated by 10% SDS-PAGE and then
transferred to nitrocellulose membranes (Bio-Rad Laboratories,
Inc.). The membranes were blocked in 5% non-fat milk containing
0.1% Tween-20 (TBST) for 2 h at room temperature and then probed
with primary antibodies against p21 (1:1,000; cat. no. #2947; Cell
Signaling Technology, Inc.) and GAPDH (1:5,000; cat. no. #2118;
Cell Signaling Technology, Inc.) overnight at 4˚C. After washing
with PBS, the membranes were incubated with a horseradish
peroxidase-conjugated secondary antibody (1:20,000; cat. nos.
ab205718; Abcam) for 2 h at room temperature. Protein bands were
visualized with enhanced chemiluminescence reagents (Pierce
Biotechnology; Thermo Fisher Scientific, Inc.) following the
manufacturer's protocol.
Statistical analysis
All experiments were independently performed in
triplicate. The data were analyzed with SPSS 19.0 software (SPSS,
Inc.) and expressed as the mean ± standard deviation. Differences
between two groups were assessed using Student's t-test.
Differences amongst multiple groups were evaluated using one-way
ANOVA followed by Tukey's post-hoc test. Correlations between the
expression of miR-106b-5p and the expression of CDKN1A were
analyzed by Spearman's rank correlation. P<0.05 was considered
to indicate statistically significant difference.
Results
miR-106b-5p is highly expressed in OS
tissues and cells
RT-qPCR was used to detect the expression of
miR-106b-5p in tumor tissue and adjacent tissue derived from OS
patients. As presented in Fig. 1A,
miR-106b-5p was significantly upregulated in OS tissue compared
with adjacent tissue (P<0.001). In addition, the expression of
miR-106b-5p in several human OS cells lines including MG-63, U2OS,
SW1353 and Saos-2 was also analyzed. As presented in Fig. 1B, miR-106b-5p expression was
significantly higher in OS cell lines compared with the osteoblast
cell line hFOB1.19 (P<0.01 and P<0.001). These results
suggested that increased miR-106b-5p may serve an important role in
OS.
Downregulation of miR-106b-5p inhibits
OS cell proliferation and cell cycle progression
Saos-2 and U2OS cells demonstrated relatively higher
levels of miR-106b-5p and were therefore selected for miR-106b-5p
inhibitor transfection to knockdown miR-106b-5p. RT-qPCR was
utilized to validate transfection efficiency. The results
demonstrated that transfection with the miR-106b-5p inhibitor
significantly reduced miR-106b-5p levels in Saos-2 and U2OS cells
compared with the miR-NC group (Fig.
2A; P<0.001). The results of the MTT assay indicated that
miR-106b-5p inhibition significantly suppressed the proliferation
of Saos-2 and U2OS cells at day 3, 4 and 5 (Fig. 2B; P<0.001). As cell proliferation
may be regulated by cell cycle progression, flow cytometry analysis
was performed to examine the cell cycle distribution in Saos-2 and
U2OS cells. As presented in Fig. 2C,
downregulation of miR-106b-5p caused an increase in the cell
population in the G0/G1 phase (P<0.001). Accordingly, the cell
population in the S and G2/M phase was reduced in Saos-2 (P<0.01
and P<0.001) and U2OS cells (P<0.01 and P<0.001). These
results indicated that miR-106b-5p accelerated cell proliferation
which may be closely associated with cell cycle progression in OS
cells.
miR-106b-5p negatively regulates
CDKN1A by binding to its 3'UTR
Bioinformatics analysis was performed to predict the
potential target genes of miR-106b-5p involved in cell cycle
regulation. Among the predicted targeted genes, three miR-106b-5p
targets associated with cell cycle regulation, including CDC37L1,
CDKN1A and CCNG2 were selected as potential target genes of
miR-106b-5p. Luciferase reporter assay showed that there was no
significant interaction between miR-106b-5p and the 3'UTRs of CCNG2
and CDC37L1 (Fig. S1). It was
determined that CDKN1A, a gene associated with G1-S transition, was
a putative target gene of miR-106b-5p (Fig. 3A). As expected, the luciferase
activity of the miR-106b-5p inhibitor + WT-CDKN1A was significantly
increased compared with the miR-106b-5p inhibitor + MUT-CDK1A in
293T cells (Fig. 3B). In addition,
to further confirm whether CDKN1A was regulated by miR-106b-5p, the
expression of CDKN1A in transfected Saos-2 and U2OS cells was
examined using RT-qPCR and western blotting. The results
demonstrated that miR-106b-5p downregulation significantly
upregulated the expression of CDKN1A mRNA (Fig. 3C; P<0.01) and protein levels
(Fig. 3D and E) in both Saos-2 and U2OS cells.
Furthermore, the expression of CDKN1A in OS tissues was determined.
As depicted in Fig. 3F, CDKN1A had a
significantly lower expression in OS tissues compared with adjacent
tissues (P<0.001). Through a two-tailed Pearson's correlation
analysis, it was determined that the expression of miR-106b-5p was
inversely correlated with CDKN1A expression in OS tissues (Fig. 3G; P=0.0057). Taken together, the
results suggested that CDKN1A was negatively regulated by
miR-106b-5p in OS.
Overexpressed CDKN1A imitates the
suppressive effects of miR-106b-5p inhibitor transfection on OS
cells
To illustrate whether CDKN1A acts as a downstream
effector in miR-106b-5p-mediated OS cell proliferation and cell
cycle progression, gain-of-function assays were performed in Saos-2
cells by transfecting pcDNA3.1-CDKN1A. Following transfection, the
mRNA (Fig. 4A; P<0.001) and
protein (Fig. 4B) levels of CDKN1A
were markedly increased in the pcDNA3.1-CDKN1A group compared with
the control group in Saos-2 cells. The MTT assay further indicated
that overexpression of CDKN1A suppressed the proliferation of
Saos-2 cells, which was identical to the effect exerted by
miR-106b-5p knockdown (Fig. 4C;
P<0.001). In addition, flow cytometry demonstrated that CDKN1A
upregulation induced cell cycle arrest at the G0/G1 stage in Saos-2
cells, similar to the effect of miR-106b-5p knockdown (Fig. 4D; P<0.001). Collectively, these
results suggested that CDKN1A might be a direct effector involved
in the miR-106b-5p-mediated cell proliferation in OS.
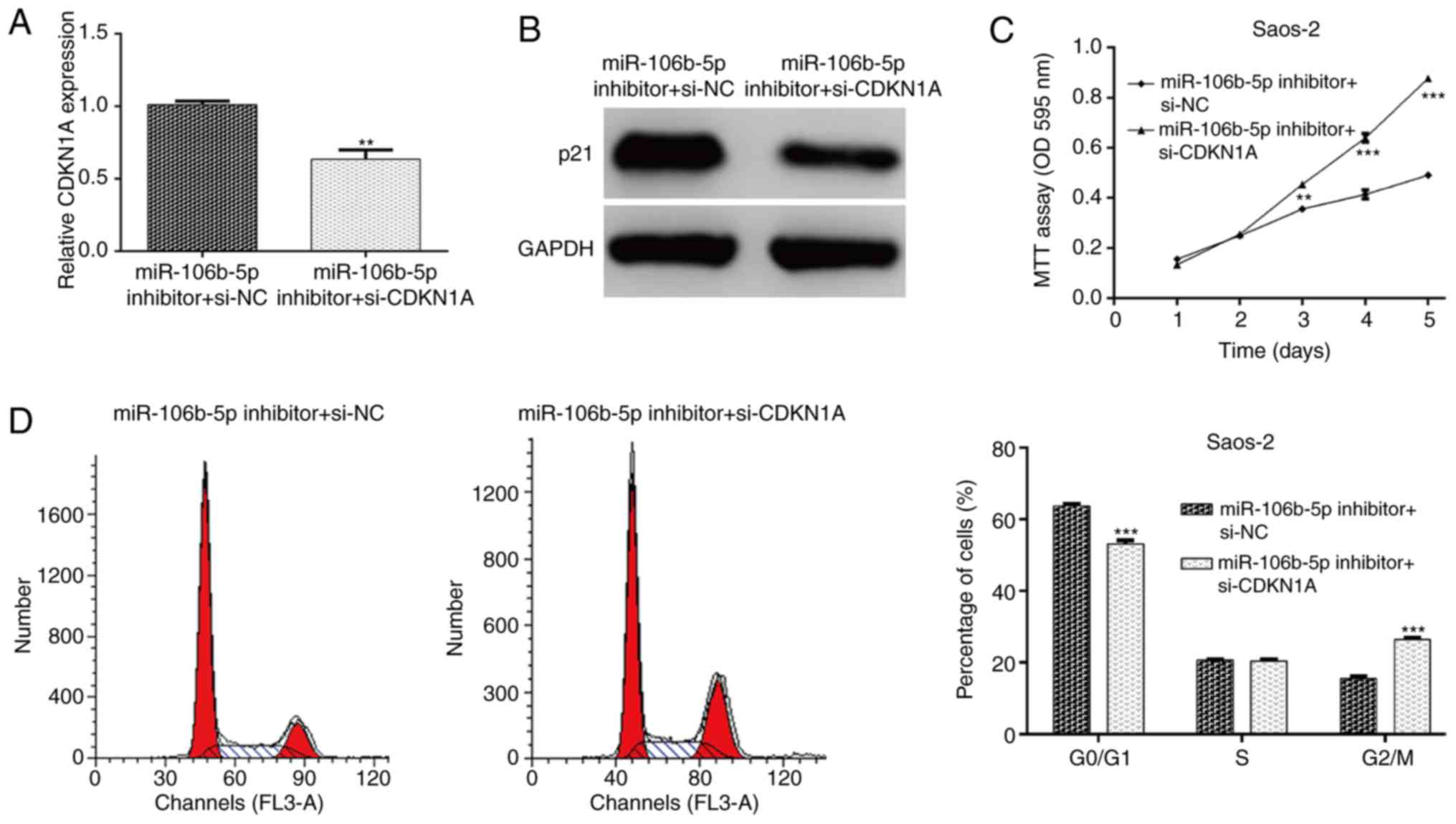
CDKN1A knockdown abolishes the
suppressive effects of miR-106b-5p inhibitor transfection on OS
cells
To further confirm whether miR-106b-5p-mediated cell
proliferation and cell cycle progression was dependent on its
capacity to modulate CDKN1A expression, rescue experiments were
performed by transfecting si-CDKN1A plasmid into Saos-2 cells
treated with miR-106b-5p inhibitor. The expression of CDKN1A in
Saos-2 cells was detected and it was determined that si-CDKN1A
transfection markedly attenuated the increased CDKN1A mRNA
(P<0.01; Fig. 5A) and protein
(Fig. 5B) expression. As expected,
the decreased cell proliferation and induced cell cycle G0/G1 phase
arrest by miR-106b-5p inhibitor were partially abolished by CDKN1A
knockdown in Saos-2 cells, as determined by an MTT assay (P<0.01
and P<0.001; Fig. 5C) and flow
cytometry analysis (P<0.001; Fig.
5D). These results further demonstrated that CDKN1A might be a
key regulator of miR-106b-5p-mediated cell proliferation and cell
cycle progression in OS cells.
Discussion
Changes in miR-106b-5p levels are correlated with
and facilitate cancer tumorigenesis. Some examples include the
amplification of miR-106b-5p triggering stem cell-like properties
of hepatocellular carcinoma cells (12), miR-106b-5p conferring proliferative
advantage and inhibiting apoptosis in non-small cell lung cancer
and overexpression of miR-106b-5p attenuating invasion and
metastasis in colorectal cancer (27). By contrast, miR-106b-5p is reported
to inhibit metastasis in colorectal cancer (27) and is associated with better survival
in bladder cancer (28). However,
whether miR-106-5p functions as an oncogene or tumor suppressor in
OS still remains unclear. The present study assessed the
differential expression of miR-106b-5p in OS clinical tissue and
adjacent tissue, as well as in OS cell lines and a normal
osteoblast cell line (hFOB1.19). The present results demonstrated
that miR-106b-5p was overexpressed in OS tissues and cell lines
using RT-qPCR. Furthermore, miR-106b-5p inhibition resulted in
reduction of cell proliferation, suggesting that miR-106a-5p could
act as oncomiR in OS cell growth.
Uncontrolled cell proliferation is a pathological
manifestation of cancer (29).
Beneath the complexity of cancer cell proliferation lies a critical
event involving defects in cell cycle progression, that have
propelled the tumor cells and its progeny into uncontrolled mitotic
division (29). The present study
investigated the cell-cycle profile to better understand the
modulation of OS cell proliferation by miR-106b-5p. Knockdown of
miR-106b-5p caused a significant increase in the percentage of
cells in G0/G1 phase and a decrease in the number of cells in S and
G2/M phases, suggesting that depletion of miR-106b-5p prevented
cell-cycle progression by arresting cells at the G0/G1 phase.
A number of miR-106b-5p target genes have been
identified, including SET domain containing 2, histone lysine
methyltransferase (11), cathepsin A
(27), PTEN (14) and BTG anti-proliferation factor
3(15), which have pivotal roles in
anti-oncogenic processes. Since miRNAs depress the expression of
their target mRNAs, the present study hypothesized that miR-106b-5p
may target certain genes that function as tumor suppressors. Using
bioinformatic prediction and a luciferase reporter assay, it was
identified and validated that a cell cycle related gene, CDKN1A,
was a direct target gene of miR-106b-5p. A negative correlation
between miR-106b-5p and CDKN1A was confirmed by RT-qPCR and western
blot analysis. In mammalians, cell cycle progression is partly
controlled by a catalytic subunit CDK and its essential activation
partner, cyclin (30).
Cyclin-dependent kinase inhibitors are known to exert effects by
binding to CDK monomers or CDK/cyclin complexes (31). As a universal inhibitor (especially
the CDK4/6-cyclin D complexes in G1 phase), CDKN1A serves a
significant role in a p53-dependent and independent manner, leading
to G0/G1 extension and suppression of further cell proliferation
(32).
Transfection experiments were performed to validate
that whether CDKN1A was implicated in the miR-106b-5p
knockdown-induced reduction of OS cell proliferation. Upregulation
of CDKN1A expression mimicked, while CDKN1A knockdown reversed the
suppressive effects of miR-106b-5p inhibitor transfection on OS
cell proliferation and cell cycle progression. Similarly,
miR-106-5p targeting CDKN1A has been reported in gastric cancer
(33) and renal cell carcinoma
(34). The present study
hypothesized that knockdown of miR-106b-3p inhibited proliferation
via G0/G1 cell cycle arrest by negatively regulating CDKN1A.
However, the present study had a number of limitations including i)
lack of immunohistochemical staining of CDKN1A in OS clinical
specimens and no investigation into its relationship with
miR-106b-5p; ii) the association between miR-106-5p and the
clinicopathological features in The Cancer Genome Atlas was not
investigated and iii) the effects of miR-106b-5p on cell apoptosis,
migration and invasion still need to be explored.
In conclusion, the present study provided strong
evidence that miR-106b-5p acts as an oncogene to attenuate the
tumor suppressor CDKN1A. Characterization of the miR-106b-5p/CDKN1A
functional axis correlation will deepen our understanding of OS
etiology and data also indicate the potential of miR-106-5p/CDKN1A
axis as a therapeutic target in the treatment of OS.
Supplementary Material
Luciferase reporter assays were
performed on 293T cells following transfection. (A) CDC37L1 and (B)
CCNG2 3'-UTR vectors and miR-106b-5p inhibitor or miR-NC were
transfected. WT, wild-type; MUT, mutant; UTR, untranslated region;
miR, microRNA; NC, negative control.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
QP designed the study. CH and HC performed reverse
transcription-quantitative PCR analysis, MTT assay, luciferase
reporter assay and western blotting. YL and XL conducted the other
functional experiments. CZ collected and analyzed the data. QQ
performed the statistical analysis, researched the literature and
contributed the manuscript editing. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
A signed written informed consent was obtained from
each patient and the experimental procedures were all in accordance
with the guideline of the Ethics Committee of Jingzhou Traditional
Chinese Medicine Hospital. The present study was approved by the
Ethics Committee of Jingzhou Traditional Chinese Medicine Hospital
(grant no. ZA3029C; Jingzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare they have no competing
interests.
References
|
1
|
Verrecchia F and Rédini F: Transforming
growth factor-β signaling plays a pivotal role in the interplay
between osteosarcoma cells and their microenvironment. Front Oncol.
8(133)2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Luetke A, Meyers PA, Lewis I and Juergens
H: Osteosarcoma treatment-Where do we stand? A state of the art
review. Cancer Treat Rev. 40:523–532. 2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Piletic K and Kunej T: MicroRNA epigenetic
signatures in human disease. Arch Toxicol. 90:2405–2419.
2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Apprey V, Wang S, Tang W, Kittles RA,
Southerland WM, Ittmann M and Kwabi-Addo B: Association of Genetic
ancestry with DNA methylation changes in prostate cancer disparity.
Anticancer Res. 39:5861–5866. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Wang J, Liu S, Shi J, Li J, Wang S, Liu H,
Zhao S, Duan K, Pan X and Yi Z: The Role of miRNA in the diagnosis,
prognosis, and treatment of osteosarcoma. Cancer Biother
Radiopharm. 34:605–613. 2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ma X, Li D, Gao Y and Liu C: miR-451a
inhibits the growth and invasion of osteosarcoma via targeting
TRIM66. Technol Cancer Res Treat.
18(1533033819870209)2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Marx V: Meet some code-breakers of
noncoding RNAs. Nat Methods. 15:103–106. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Yamamoto N, Nishikawa R, Chiyomaru T, Goto
Y, Fukumoto I, Usui H, Mitsuhashi A, Enokida H, Nakagawa M, Shozu M
and Seki N: The tumor-suppressive microRNA-1/133a cluster targets
PDE7A and inhibits cancer cell migration and invasion in
endometrial cancer. Int J Oncol. 47:325–334. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43:D146–D152. 2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Xiang W, He J, Huang C, Chen L, Tao D, Wu
X, Wang M, Luo G, Xiao X, Zeng F and Jiang G: miR-106b-5p targets
tumor suppressor gene SETD2 to inactive its function in clear cell
renal cell carcinoma. Oncotarget. 6:4066–4079. 2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Shi DM, Bian XY, Qin CD and Wu WZ:
miR-106b-5p promotes stem cell-like properties of hepatocellular
carcinoma cells by targeting PTEN via PI3K/Akt pathway. Onco
Targets Ther. 11:571–585. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Liu F, Gong J, Huang W, Wang Z, Wang M,
Yang J, Wu C, Wu Z and Han B: MicroRNA-106b-5p boosts glioma
tumorigensis by targeting multiple tumor suppressor genes.
Oncogene. 33:4813–4822. 2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Chen X, Chen P, Chen SS, Ma T, Shi G, Zhou
Y, Li J and Sheng L: miR106b5p promotes cell cycle progression of
malignant melanoma by targeting PTEN. Oncol Rep. 39:331–337.
2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wei K, Pan C, Yao G, Liu B, Ma T, Xia Y,
Jiang W, Chen L and Chen Y: MiR-106b-5p promotes proliferation and
inhibits apoptosis by regulating BTG3 in Non-small cell lung
cancer. Cell Physiol Biochem. 44:1545–1558. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Malumbres M: Cyclin-dependent kinases.
Genome Biol. 15(122)2014.PubMed/NCBI View
Article : Google Scholar
|
|
17
|
Kreis NN, Louwen F and Yuan J: Less
understood issues: p21(Cip1) in mitosis and its therapeutic
potential. Oncogene. 34:1758–1767. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Abbas T and Dutta A: p21 in cancer:
Intricate networks and multiple activities. Nat Rev Cancer.
9:400–414. 2009.PubMed/NCBI View
Article : Google Scholar
|
|
19
|
Fitzgerald AL, Osman AA, Xie TX, Patel A,
Skinner H, Sandulache V and Myers JN: Reactive oxygen species and
p21Waf1/Cip1 are both essential for p53-mediated senescence of head
and neck cancer cells. Cell Death Dis. 6(e1678)2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wu Z, Liu K, Wang Y, Xu Z, Meng J and Gu
S: Upregulation of microRNA-96 and its oncogenic functions by
targeting CDKN1A in bladder cancer. Cancer Cell Int.
15(107)2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Ohta K, Hoshino H, Wang J, Ono S, Iida Y,
Hata K, Huang SK, Colquhoun S and Hoon DS: MicroRNA-93 activates
c-Met/PI3K/Akt pathway activity in hepatocellular carcinoma by
directly inhibiting PTEN and CDKN1A. Oncotarget. 6:3211–3224.
2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Brock M, Haider TJ, Vogel J, Gassmann M,
Speich R, Trenkmann M, Ulrich S, Kohler M and Huber LC: The
hypoxia-induced microRNA-130a controls pulmonary smooth muscle cell
proliferation by directly targeting CDKN1A. Int J Biochem Cell
Biol. 61:129–137. 2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Fornari F, Milazzo M, Chieco P, Negrini M,
Marasco E, Capranico G, Mantovani V, Marinello J, Sabbioni S,
Callegari E, et al: In hepatocellular carcinoma miR-519d is
up-regulated by p53 and DNA hypomethylation and targets CDKN1A/p21,
PTEN, AKT3 and TIMP2. J Pathol. 227:275–285. 2012.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Shao M, Geng Y, Lu P, Xi Y, Wei S, Wang L,
Fan Q and Ma W: miR-4295 promotes cell proliferation and invasion
in anaplastic thyroid carcinoma via CDKN1A. Biochem Biophys Res
Commun. 464:1309–1313. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhao X, Yang Y, Xu J, Luo Y, Xin Y and
Wang Y: Downregulation of microRNA-95-3p suppresses cell growth of
osteosarcoma via CDKN1A/p21 expression. Oncol Rep. 39:289–297.
2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Livak K and Schmittgen T: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2000.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ni S, Weng W, Xu M, Wang Q, Tan C, Sun H,
Wang L, Huang D, Du X and Sheng W: miR-106b-5p inhibits the
invasion and metastasis of colorectal cancer by targeting CTSA.
Onco Targets Ther. 11:3835–3845. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Lee E, Collazo-Lorduy A, Castillo-Martin
M, Gong Y, Wang L, Oh WK, Galsky MD, Cordon-Cardo C and Zhu J:
Identification of microR-106b as a prognostic biomarker of p53-like
bladder cancers by ActMiR. Oncogene. 37:5858–5872. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Evan GI and Vousden KH: Proliferation,
cell cycle and apoptosis in cancer. Nature. 411:342–348.
2001.PubMed/NCBI View
Article : Google Scholar
|
|
30
|
Wood DJ and Endicott JA: Structural
insights into the functional diversity of the CDK-cyclin family.
Open Biol. 8(180112)2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Sánchez-Martínez C, Gelbert LM, Lallena MJ
and de Dios A: Cyclin dependent kinase (CDK) inhibitors as
anticancer drugs. Bioorg Med Chem Lett. 25:3420–3435.
2015.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Abbadie C, Pluquet O and Pourtier A:
Epithelial cell senescence: An adaptive response to
pre-carcinogenic stresses? Cell Mol Life Sci. 74:4471–4509.
2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Dong X, Hu X, Chen J, Hu D and Chen LF:
BRD4 regulates cellular senescence in gastric cancer cells via
E2F/miR-106b/p21 axis. Cell Death Dis. 9(203)2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Sun K, Jia Z, Duan R, Yan Z, Jin Z, Yan L,
Li Q and Yang J: Long non-coding RNA XIST regulates miR-106b-5p/P21
axis to suppress tumor progression in renal cell carcinoma. Biochem
Biophys Res Commun. 510:416–420. 2019.PubMed/NCBI View Article : Google Scholar
|