Introduction
Cervical cancer (CC) is a type of gynecological
malignancy and a major cause of cancer-related mortality in females
worldwide (1). With ~6 million
cases per year, CC continues to be a major global health issue
(2). Recently, studies have focused
on identifying effective targets that may be useful for screening
and treating CC (3). Despite
advances in therapeutic strategies, including chemotherapy,
surgery, radiation and targeted therapy, the survival rate of
patients remains low (4,5). Thus, it is essential to investigate
the potential biological mechanism and elucidate novel targets for
improving therapeutic strategies for CC.
Long non-coding RNAs (lncRNAs), >200 nt without
protein-coding capacity are associated with various biological
functions (6). Accumulating
evidence indicates that lncRNAs play important roles in
tumorigenesis, including in CC. LncRNA XLOC_006390 promotes the
invasion and migration of CC cells and facilitates CC tumorigenesis
and metastasis (7). LncRNA small
nucleolar RNA host gene 1 knockdown inhibits cell viability and
migration in HeLa and C-33A cells (8). Recently, long intergenic non-protein
coding RNA 1123 (LINC01123) has been reported to exhibit a
carcinogenic function in certain types of cancer. For example,
LINC01123 enhances cell growth, migration, and angiogenesis in
colon cancer cells and promotes the progression of colon cancer
(9). LINC01123 promotes
proliferation and metabolic rewiring in non-small cell lung cancer
(NSCLC) cells and promotes tumor growth in vivo (10). However, to the best of our
knowledge, the function of LINC01123 in CC has not been
investigated.
MicroRNAs (miRNAs) are a type of short RNA molecule
that play an important role in gene regulation (11). Previous studies show that certain
miRNAs, such as miR-489(12),
miR-203(13) and miR-338-3p are
involved in CC proliferation (14).
Recently, several studies have shown that miR-361-3p participates
in CC progression. For example, low expression of miR-361-3p
promotes cell proliferation and invasion in CC cells (15). Moreover, miR-361-3p inhibition
enhances epithelial-mesenchymal transition, cell invasion and
growth in CC cells (16).
In the present study, the expression of LINC01123,
miR-361-3p and tetraspanin 1 (TSPAN1) was detected in CC tissue
samples and cell lines. Thereafter, the regulatory effects of
LINC01123 inhibition on cell viability, migration and invasion was
surveyed in CC cells, as well as its effect on tumor growth in
vivo. Furthermore, the mechanism by which LINC01123 knockdown
regulated CC development via miR-361-3p/TSPAN1 was evaluated. The
findings of the present study may present a novel target for CC
treatment.
Materials and methods
Patient samples
Sixty-three patients with CC (63 women; median age,
47±13 years) were recruited between April 2017 and June 2018 at
Linyi Central Hospital (Linyi, China). None of the patients with CC
underwent local or systemic treatment before surgery. CC tissue
samples and corresponding adjacent healthy cervical tissue samples
were acquired from the patients. All patients provided written
informed consent and the study was approved by the Ethics Committee
of Linyi Central Hospital (approval no. 2017013).
Cell culture and transfection
Human CC cell lines (HeLa, CaSki, SiHa and C-33A)
and the human normal cervical epithelial cell line (HCerEpiC) were
purchased from American Type Culture Collection. All cells were
cultured in Gibco Dulbecco's modified Eagle's medium (DMEM; Thermo
Fisher Scientific, Inc.) containing Gibco 10% fetal bovine serum
(FBS; Thermo Fisher Scientific, Inc.) at 37˚C in an incubator with
5% CO2 until they were employed to perform the
subsequent experiments.
Short hairpin RNA negative control (sh-NC,
5'-UUCUCCGAACGUGUCACGUTT-3'), sh-LINC01123-1
(5'-CUGAACGUCUUGCAACAGUTT-3') and sh-LINC01123-2
(5'-GCCCUAGGAAAUCCGUAAUTT-3') were acquired from Sangon Biotech
Co., Ltd. miRNA mimics NC (miR-NC, 5'-UUCUCCGAACGUGUCACGUTT-3'),
miR-361-3p mimics (5'-UCCCCCAGGUGUGAUUCUGAUUU-3'), inhibitor NC
(5'-UUCUCCGAACGUGUCACGUTT-3'), miR-361-3p inhibitor
(5'-UCCCCCAGGUGUGAUUCUGAUUU-3'), as well as pcDNA-TSPAN1 and
pcDNA-NC were purchased from GeneChem, Inc.. The above factors (50
nM) were transfected into cells with Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.). Cells were collected
48 h after transfection.
Reverse transcription-quantitative PCR
(RT-qPCR) assay
According to the manufacturer's instructions, total
RNA was extracted from tissue samples and cells using the TRIeasy
Reagent kit (Qianchen Biotechnology Co., Ltd.). A Reverse
Transcription Kit (Takara Bio, Inc.) was used according to the
manufacturer's instructions to reverse transcribe RNA into cDNA.
Thereafter, the expression levels of LINC01123, miR-361-3p and
TSPAN1 were analyzed using the SYBR Green Real-Time PCR Kit (Takara
Bio, Inc.) also according to the manufacturer's instructions. The
expression levels of LINC01123 and TSPAN1 were normalized to GAPDH
and U6 served as an endogenous control for miR-361-3p detection.
RT-qPCR primer sequences were synthesized by Sangon Biotech Co.,
Ltd. and are presented in Table I.
The PCR conditions were as follows: 95˚C for 10 min, followed by 40
cycles at 95˚C for 30 sec, 60˚C for 30 sec and 72˚C for 1 min. Data
were calculated using the 2-ΔΔCq method (17).
 | Table IPrimers for reverse
transcription-quantitative PCR. |
Table I
Primers for reverse
transcription-quantitative PCR.
| Gene | Forward
(5'-3') | Reverse
(5'-3') |
|---|
| LINC01123 |
ACAGTGGCCGCACGCATAGCTG |
CTGACGACCGAGGTGACAACGATGA |
| miR-361-3p |
ACACTCCAGCTGGGTCCCCCAG GTGTGATTC |
CTCAACTGGTGTCGTGGAGTCGGCAA
TTCAGTTGAGAAATCAGA |
| TSPAN1 |
GTGGTCTTTGCTCTTGGTTTCC |
TTTCTTGATGGCAGGCACTA |
| GAPDH |
GCGAGATCGCACTCATCATCT |
TCAGTGGTGGACCTGACC |
| U6 |
CTCGCTTCGGCAGCACA |
AACGCTTCACGAATTTGCGT |
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
Following transfection, cells were seeded in 96-well
plates (3x103 cells/well) and incubated for 0, 24, 48,
72 and 96 h at 37˚C. Thereafter, 15 µl MTT solution (0.5 mg/ml;
Millipore Sigma) was added to each well. After 4 h, 200 µl dimethyl
sulfoxide was added to each well. The optical density of each well
was read at 450 nm using a spectrophotometer (Thermo Fisher
Scientific, Inc.).
Wound healing assay
The transfected cells were seeded in 6-well plates
and grown to 100% confluence. The cells were scraped with a 10 µl
Eppendorf™ pipette tip and incubated for 24 h in a serum-free
medium (Thermo Fisher Scientific, Inc.). Thereafter, they were
washed using phosphate-buffered saline for three times to remove
cell fragments and the lengths of the scratches were subsequently
recorded using an Olympus Inverted Microscope. After 24 h, the
lengths of the scratches were recorded again. The healing rate was
analyzed using ImageTool software (version 1.46, Los Alamos
Operations). The wound closure was calculated as follows: (0-h
width-24-h width)/0-h width x 100%.
Transwell assay
To assess the invasion ability of cells, 24-well
Transwell chambers (8.0 µm; SproutStrong Biotech) were used to
conduct the Transwell assay. Transfected cells (2x105
cells/well) in serum-free medium (Thermo Fisher Scientific, Inc.)
were cultured in the upper compartment pre-coated with Matrigel
(SproutStrong Biotech). The lower compartment contained DMEM with
15% FBS. After 24 h, the cells in the lower compartment were fixed
with 75% methanol at 4˚C for 10 min and stained using 0.3% crystal
violet at 37˚C for 30 min. Images were captured using an inverted
optical microscope (Olympus).
Tumor formation assay in vivo
A total of 10 four-week-old BALB/c nude mice
(female; weight 20-25 g) were obtained from Shanghai Laboratory
Animal Centre (Shanghai, China). All mice were housed under
controlled conditions (25˚C; 50% humidity; 12 h light/dark cycle)
and were provided with free access to food and water. The mice were
randomly divided into two groups of five mice. Lentiviral vectors
(Lv) for sh-LINC01123-1 or sh-NC were constructed by Sangon Biotech
Co., Ltd.. First, HeLa cells were transfected with
Lv-sh-LINC01123-1 or Lv-sh-NC using Lipofectamine 3000. Thereafter,
mice (n=5) were injected with the HeLa cells (1x106;
s.c.) in the right flank. Tumor volume (V) was measured at 10, 15,
20, 25 and 30 days after injection using the formula: V=length x
width2 x 0.5. After 30 days, mice were anesthetized with
pentobarbital sodium (50 mg/kg) and then sacrificed by cervical
dislocation. The tumor xenograft was separated from the mice and
weighed. The present study adhered to the requirements of the
National Institutes of Health Guide for the Care and Use of
Laboratory Animals and was approved by the Ethics Committee of
Linyi Central Hospital (approval no. 2017013).
Bioinformatics analysis
The Cancer Genome Atlas (TCGA; https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga)
was used to examine LINC01123 expression in CC.
Dual-luciferase reporter (DLR)
assay
The primary binding sites of LINC01123 and
miR-361-3p, and miR-361-3p and TSPAN1 were predicted using LncBase
Predicted v.2 (https://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex-predicted)
and TargetScan (http://www.targetscan.org/vert_72/), respectively. The
wild-type (WT) or mutant (MUT) binding sites of miR-361-3p to
LINC01123 or TSPAN1 were inserted into a pGL3 vector (Promega
Corporation) to generate corresponding luciferase reporters
(LINC01123 WT, LINC01123 MUT, TSPAN1 WT and TSPAN1 MUT). Next,
Lipofectamine 3000 was used to co-transfect the luciferase
reporters and miR-361-3p mimics or miR-NC into HeLa and CaSki
cells. Then, the relative luciferase activity was analyzed using a
DLR™ Assay System (Promega Corporation) after transfection for 24
h.
Western blot analysis
HeLa and CaSki cells were lysed using
radioimmunoprecipitation assay lysis solution (Beyotime Institute
of Biotechnology). The protein concentration was detected by the
BCA Protein Assay kit (Abcam). Protein samples (20 µg per lane)
were separated equally with 10% sodium dodecyl
sulphate-polyacrylamide gel electrophoresis and subsequently
transferred to polyvinylidene difluoride membranes (EMD Millipore).
The membranes were blocked with 5% non-fat milk for 1 h at 25˚C and
incubated overnight at 4˚C with primary antibodies against TSPAN1
(1:1,000; cat. no. ab254730) and α-tubulin (1:1,000; cat. no.
ab176560) purchased from Abcam. After the membranes were washed
with TBST three times, the membranes were incubated at 37˚C for 2 h
with a horseradish peroxidase-conjugated secondary antibody
(1:5,000; cat. no. ab205718) purchased from Abcam. The signals were
determined using ECL Luminous Liquid (Pierce; Thermo Fisher
Scientific, Inc.) and the immunoblots were analyzed using Image
Lab™ Software (Bio-Rad Laboratories, Inc.).
Statistical analysis
In vitro experiments were performed in
triplicate and each experiment was repeated at least three times.
In vivo experiments were performed using five mice in each
of the two groups. Data analysis was performed using SPSS software
version 20.0 (IBM Corp.) and data are presented as means ± standard
deviation (SD). Differences between two groups or among multiple
groups were determined using Student's t-test or one-way ANOVA
followed by Tukey's post-hoc test, respectively. Pearson's
correlation analysis was also implemented to evaluate variate
correlation. P<0.05 was considered to indicate a statistically
significant difference.
Results
LINC01123 expression is increased in
CC tissue samples and cell lines
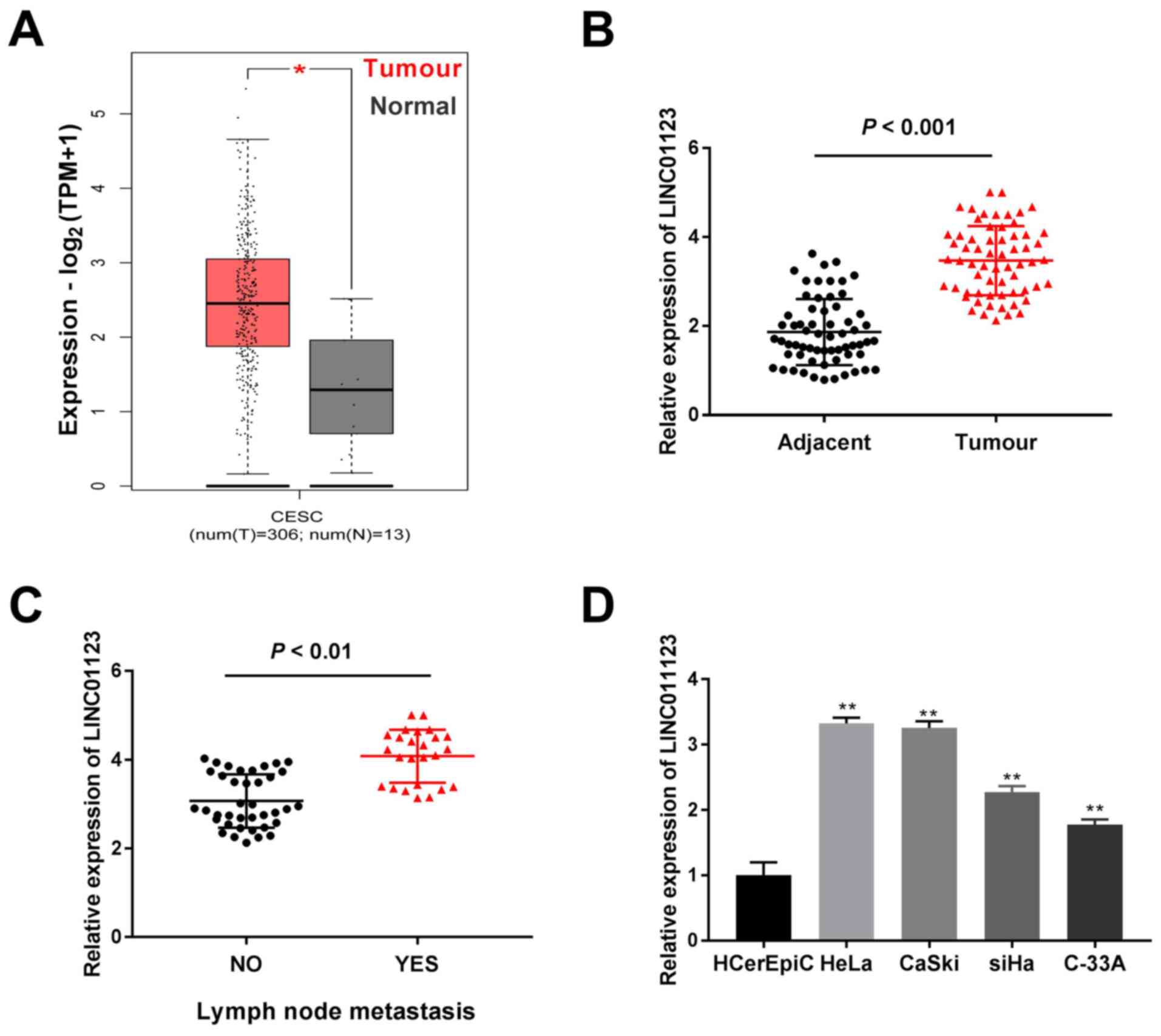
TCGA analysis demonstrated that LINC01123 expression
was significantly increased in cervical squamous cell carcinoma and
endocervical adenocarcinoma (CESC) tumors when compared to that in
healthy tissue samples (P<0.05; Fig.
1A). Furthermore, LINC01123 expression was highly enhanced in
CC tissue samples when compared to that in adjacent healthy
cervical tissue samples (P<0.001; Fig. 1B). Meanwhile, LINC01123 expression
was significantly enhanced in CC tissue samples with lymph node
metastasis compared to that in tissue samples without metastasis
(P<0.01; Fig. 1C). Furthermore,
LINC01123 expression in CC cell lines (HeLa, CaSki, SiHa and C-33A)
and the healthy cervical epithelial cell line (HCerEpiC) was
examined. It was confirmed that LINC01123 expression was
significantly enhanced in CC cell lines, particularly in CaSki and
HeLa cells, when compared to that in HCerEpiC cells (P<0.01;
Fig. 1D). Thus, these two cells
were selected for subsequent experiments. As presented in Table II, LINC01123 expression was
significantly correlated with lymph node metastasis and
International Federation of Gynecology and Obstetrics stage
(18) (P<0.05). Thus, these data
revealed that LINC01123 may contribute to CC tumorigenesis.
 | Table IICorrection of LINC01123 expression
with clinicopathologic features in cervical cancer. |
Table II
Correction of LINC01123 expression
with clinicopathologic features in cervical cancer.
| | LINC01123
expression | |
|---|
| Characteristic | n | Low | High | P-value |
|---|
| Age (years) | | | | 0.693 |
|
<45 | 28 | 13 | 15 | |
|
≥45 | 35 | 18 | 17 | |
| Menopause | | | | 0.537 |
|
No | 37 | 17 | 20 | |
|
Yes | 26 | 14 | 12 | |
| Tumor size
(cm) | | | | 0.382 |
|
<4 | 25 | 14 | 11 | |
|
≥4 cm | 38 | 17 | 21 | |
| Depth of cervical
invasion | | | | 0.098 |
|
<2/3 | 24 | 15 | 9 | |
|
≥2/3 | 39 | 16 | 23 | |
| Histology | | | | 0.154 |
|
Squamous | 42 | 18 | 24 | |
|
Adenocarcinoma | 21 | 13 | 8 | |
| Lymph node
metastasis | | | | 0.027a |
|
No | 38 | 23 | 15 | |
|
Yes | 25 | 8 | 17 | |
| FIGO stage | | | | 0.006a |
|
I/II | 25 | 7 | 18 | |
|
III/IV | 38 | 24 | 14 | |
Knockdown of LINC01123 inhibits CC
progression in vitro and in vivo
Based on the current findings, the biological
function of LINC01123 in CC was investigated. Following
transfection with sh-LINC01123-1 and sh-LINC01123-2, LINC01123
expression in HeLa and CaSki cells was significantly reduced
compared to that in the sh-NC group, and significantly greater
inhibition of LINC01123 expression was observed in the
sh-LINC1123-1 group than in the sh-LINC01123-2 group (P<0.01;
Fig. 2A). Thus, sh-LINC01123-1 was
selected for subsequent experiments. Thereafter, the knockdown of
LINC01123 was observed to significantly inhibit cell viability in
CaSki and HeLa cells (P<0.01; Fig.
2B). Furthermore, silencing LINC01123 notably suppressed cell
invasion and migration in CaSki and HeLa cells (P<0.01; Fig. 2C and D). Meanwhile, the influence of LINC01123
knockdown on CC tumor growth in vivo was investigated by
establishing mouse xenograft models. The volume and weight of CC
tumors were found to be significantly decreased in the
Lv-sh-LINC01123-1 group when compared to those in the Lv-sh-NC
group (P<0.01; Fig. 2E).
Collectively, the results indicate that silencing LINC01123
inhibits CC cell viability, invasion and migration in vitro,
and inhibits CC tumor growth in vivo.
 | Figure 2Silencing of LINC01123 inhibited CC
progression in vitro and in vivo. (A) LINC01123
expression was detected by RT-qPCR in HeLa and CaSki cells
transfected with sh-NC, sh-LINC01123-1 or sh-LINC01123-2.
**P<0.01 vs. sh-NC. (B) Cell viability was detected
by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
assay in HeLa and CaSki cells transfected with sh-LINC01123-1 or
sh-NC. *P<0.05 and **P<0.01 vs. sh-NC.
(C) Cell migration was analyzed by wound healing assay in HeLa and
CaSki cells transfected with sh-LINC01123-1 or sh-NC.
**P< 0.01 vs. sh-NC. Magnification, x400. (D) Cell
invasion was detected by Transwell assay in HeLa and CaSki cells
transfected with sh-LINC01123-1 or sh-NC. **P<0.01
vs. sh-NC. The in vitro experiments were performed in
triplicate and repeated at least three times. Magnification, x400.
(E) Images, volume and weight change of tumors detected in the
mouse xenograft models. **P<0.01 vs. Lv-sh-NC. The
in vivo experiments were performed on five mice in each
group. LINC01123, long intergenic non-protein coding RNA 1123; CC,
cervical cancer; RT-qPCR, reverse transcription-quantitative PCR;
sh-NC, short hairpin RNA negative control; Lv, lentiviral
vector. |
miR-361-3p is a direct target of
LINC01123
The primary target site between LINC01123 and
miR-361-3p was predicted using LncBase Predicted v.2 (Fig. 3A). Knockdown of LINC01123 increased
miR-361-3p expression in HeLa and CaSki cells (P<0.01; Fig. 3B). Furthermore, the DLR assay
demonstrated that miR-361-3p mimics significantly decreased the
luciferase activity of LINC01123 WT in HeLa and CaSki cells
(P<0.01; Fig. 3C). Furthermore,
miR-361-3p expression in CC tissue samples was lower than that in
adjacent healthy cervical tissue samples (P<0.001; Fig. 3D). Additionally, miR-361-3p
expression was negatively correlated with LINC01123 expression in
CC tissue samples (r=-0.5158; P<0.0001) as shown in Fig. 3E. Thereafter, miR-361-3p expression
in vitro was investigated. The results indicated that
miR-361-3p expression in the CC cell lines was significantly
reduced when compared to that in HCerEpiC cells (P<0.01;
Fig. 3F). Collectively, these
results indicated that LINC01123 directly targeted miR-361-3p and
negatively modulated miR-361-3p expression in CC.
miR-361-3p overexpression inhibits CC
cell viability, migration and invasion
To elucidate the potential mechanism of miR-361-3p
in CC progression, miR-361-3p mimics or miR-361-3p inhibitor were
transfected into CaSki and HeLa cells. The results indicated that
miR-361-3p expression was significantly increased in the miR-361-3p
mimics group when compared to that in the miR-NC group, whereas
miR-361-3p expression was decreased in the miR-361-3p inhibitor
group when compared with the inhibitor NC group (P<0.01;
Fig. 4A). Subsequently, miR-361-3p
overexpression reduced cell viability in CaSki and HeLa cells
(P<0.01; Fig. 4B). Similarly,
cell migration and invasion in the miR-361-3p mimics group were
significantly inhibited when compared to that in the miR-NC group
(P<0.01; Fig. 4C and D). Collectively, these results confirmed
that miR-361-3p could inhibit cell viability, invasion and
migration in CC cells.
TSPAN1 is a target of miR-361-3p
The primary target site between TSPAN1 and
miR-361-3p was predicted using TargetScan (Fig. 5A). The results of the DLR assay
indicated that miR-361-3p mimics significantly impaired luciferase
activities of TSPAN1 WT in HeLa and CaSki cells (P<0.01;
Fig. 5B). Furthermore, the protein
expression of TSPAN1 in HeLa and CaSki cells was suppressed by
transfection of miR-361-3p mimics and sh-LINC01123-1 (P<0.01;
Fig. 5C and D). TCGA analysis showed that TSPAN1
expression was significantly upregulated in CESC tumors compared to
that in healthy tissues (P<0.05; Fig. 5E). Additionally, the results from
RT-qPCR confirmed that TSPAN1 expression was significantly enhanced
in CC tissue samples when compared to that in adjacent healthy
cervical tissue samples (P<0.001; Fig. 5F). Furthermore, in CC tissue
samples, TSPAN1 expression was positively correlated with LINC01123
expression (r=0.4455; P=0.0003) as shown in Fig. 5G and was negatively correlated with
miR-361-3p expression (r=-0.4273; P=0.0005) as shown in Fig. 5H. Collectively, these outcomes
indicated that miR-361-3p targeted TSPAN1 and negatively regulated
TSPAN1 expression in CC.
 | Figure 5TSPAN1 was the direct target gene of
miR-361-3p. (A) The putative binding site of miR-361-3p was
predicted by TargetScan. (B) Dual-luciferase reporter assay
confirmed the association between miR-361-3p and TSPAN1 in HeLa and
CaSki cells transfected with miR-NC, miR-361-3p mimics, TSPAN1 WT
or TSPAN1 MUT. **P<0.01 vs. miR-NC. (C) The
expression of TSPAN1 protein was detected by western blotting in
HeLa and CaSki cells transfected with miR-NC and miR-361-3p mimics.
**P<0.01 vs. miR-NC. (D) The expression of TSPAN1
protein was detected by western blotting in HeLa and CaSki cells
transfected with sh-NC and sh-LINC01123-1. **P<0.01
vs. sh-NC. (E) Relative expression of TSPAN1 was evaluated by The
Cancer Genome Atlas database. *P<0.05. (F) TSPAN1
expression was detected by reverse transcription-quantitative PCR
in CC and adjacent healthy cervical tissue samples. P<0.001. The
correlation between (G) TSPAN1 and LINC01123 (P=0.0003) and (H)
TSPAN1 and miR-361-3p (P=0.0005) was evaluated by Pearson's
correlation analysis. The experiments were performed in triplicate
and repeated at least three times. TSPAN1, tetraspanin 1;
miR-361-3p, microRNA-361-3p; CC, cervical cancer; NC, negative
control; WT, wild-type; MUT, mutant; sh, short hairpin RNA;
LINC01123, long intergenic non-protein coding RNA 1123. |
miR-361-3p interacts with TSPAN1 to
mediate CC progression
To investigate the possible function of the
miR-361-3p/TSPAN1 axis on CC progression in vitro,
pcDNA-TSPAN1/NC was primarily transfected into HeLa cells to
evaluate the protein expression of TSPAN1. As shown in Fig. 6A, the TSPAN1 expression level was
elevated by pcDNA-TSPAN1 transfection (P<0.01). Using MTT, wound
healing and Transwell assays, the inhibitory effects of miR-361-3p
mimics on viability, migration and invasion of HeLa cells were
observed to be ameliorated by pcDNA-TSPAN1 (P<0.01; Fig. 6B-D).
 | Figure 6miR-361-3p interacted with TSPAN1 to
mediate CC progression. (A) The expression of TSPAN1 protein was
detected by western blotting in HeLa cells transfected with
pcDNA-NC or pcDNA-TSPAN1. **P<0.01 vs. pcDNA-NC. (B)
Cell viability was detected by
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay
in HeLa cells transfected with miR-NC, miR-361-3p mimics or
miR-361-3p mimics + pcDNA-TSPAN1. **P<0.01 vs.
miR-NC. ##P<0.01 vs. miR-361-3p mimics. (C) Cell
migration was analyzed by wound healing assay in HeLa cells
transfected with miR-NC, miR-361-3p mimics or miR-361-3p mimics +
pcDNA-TSPAN1. **P<0.01 vs. miR-NC.
##P<0.01 vs. miR-361-3p mimics. Magnification, x400.
(D) Cell invasion was detected by Transwell assay in HeLa cells
transfected with miR-NC, miR-361-3p mimics or miR-361-3p mimics +
pcDNA-TSPAN1. **P<0.01 vs. miR-NC.
##P<0.01 vs. miR-361-3p mimics. Magnification, x400.
The experiments were performed in triplicate and repeated at least
three times. TSPAN1, tetraspanin 1; miR-361-3p, microRNA-361-3p;
CC, cervical cancer; NC, negative control. magnification x400. |
LINC01123 regulates CC tumorigenesis
by targeting miR-361-3p/TSPAN1
Whether the tumor-suppressive effect of LINC01123
knockdown was dependent on miR-361-3p and TSPAN1 in CC progression
was subsequently investigated. As demonstrated in Fig. 7A, LINC01123 knockdown decreased HeLa
cell viability, while miR-361-3p inhibition or TSPAN1
overexpression ameliorated this trend (P<0.01). Furthermore,
silencing LINC01123 inhibited the migration and invasion of HeLa
cells, while miR-361-3p inhibition or TSPAN1 overexpression was
found to ameliorate the inhibitory effect (P<0.01; Fig. 7B and C). These data demonstrated that the
silencing of LINC01123 inhibited CC tumorigenesis via
miR-361-3p/TSPAN1 interaction.
 | Figure 7LINC01123 regulated tumorigenesis of
CC by targeting miR-361-3p/TSPAN1. (A) Cell viability was detected
by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
assay in HeLa cells transfected with sh-NC, sh-LINC01123-1,
sh-LINC01123-1 + pcDNA-TSPAN1 or sh-LINC01123-1 + miR-361-3p.
**P<0.01 vs. sh-NC. ##P<0.01 vs.
sh-LINC01123-1. (B) Cell migration was analyzed by wound healing
assay in HeLa cells transfected with sh-NC, sh-LINC01123-1,
sh-LINC01123-1 + pcDNA-TSPAN1 or sh-LINC01123-1 + miR-361-3p.
**P<0.01 vs. sh-NC. ##P<0.01 vs.
sh-LINC01123-1. Magnification, x400. (C) Cell invasion was detected
by Transwell assay in HeLa cells transfected with sh-NC,
sh-LINC01123-1, sh-LINC01123-1 + pcDNA-TSPAN1 or sh-LINC01123-1 +
miR-361-3p. **P<0.01 vs. sh-NC.
##P<0.01 vs. sh-LINC01123-1. Magnification, x400. The
experiments were performed in triplicate and repeated at least
three times. LINC01123, long intergenic non-protein coding RNA
1123; CC, cervical cancer; miR-361-3p, microRNA-361-3p; TSPAN1,
tetraspanin 1; NC, negative control; sh, short hairpin RNA. |
Discussion
CC is a gynecological malignancy, which poses an
enormous threat to females (19).
Thus, identifying available targets to improve prognosis and
effectively treat CC is considered critical. Numerous lncRNAs have
been reported to play vital roles in CC tumorigenesis (20,21).
In the current study, LINC01123 expression was enhanced in CC
tissue samples and cell lines. Silencing of LINC01123 inhibited
cell viability, migration and invasion in vitro and
suppressed tumor growth in vivo. Furthermore, LINC01123
negatively regulated miR-361-3p and miR-361-3p negatively regulated
TSPAN1. Further experiments demonstrated that LINC01123 knockdown
regulated the progression of CC by targeting miR-361-3p/TSPAN1;
thus, it could likely serve as a primary target for the treatment
of CC.
LINC01123 is a lncRNA activated by c-Myc, which
exerts oncogenic effects in certain types of cancer. Previous
studies show that LINC01123 is upregulated in colon cancer
(9), NSCLC (10) and triple-negative breast cancer
(22). In the present study,
LINC01123 expression was also increased in CC tissue samples and
cell lines, suggesting its association with CC development.
Numerous studies show that LINC01123 promotes cell growth, invasion
and migration in several cancer cell lines, such as colon (9) and endometrial (23) cancer cells. Similarly, the present
results demonstrated that silencing LINC01123 inhibited cell
viability, migration and invasion in HeLa and CaSki cells,
indicating that LINC01123 knockdown has a tumor-suppressive effect
in vitro. To further verify the inhibitory function of
LINC01123 knockdown on CC tumor growth in vivo, mouse
xenograft models were established. Results showed that the
knockdown of LINC01123 suppressed the volume and weight of CC
tumors in vivo. These results indicated that silencing of
LINC01123 plays a vital role in inhibiting CC development.
Previous studies have reported that miR-361-3p
participates in the progression of several types of cancer,
including colorectal cancer (CRC) (24), thyroid cancer (25) and CC (16) and its expression is downregulated in
these cancer types. Similar to the above results (16,24,25),
the present study showed that miR-361-3p expression was also
downregulated in CC tissue samples and cell lines, suggesting that
miR-361-3p may be a tumor-suppressor gene in CC. LncRNAs can act as
competing endogenous RNAs to exert their functions by targeting
miRNAs (26). LINC01123 facilitates
cell proliferation and tumor growth in NSCLC by targeting
miR-199a-5p (10). LINC01123
facilitates the proliferation and invasion of CRC cells via
miR-625-5p sponging (27). Notably,
both LINC00460 and BBOX1 antisense RNA 1 interact with miR-361-3p
to affect CC progression (24,28).
In the current study, LINC01123 directly targeted miR-361-3p and
negatively modulated miR-361-3p expression in CC tissue samples. In
addition, overexpression of miR-361-3p inhibits cell growth and
invasion in NSCLC (29) and HeLa
(16) cells.
In the present study, overexpression of miR-361-3p
was observed to inhibit cell viability, invasion and migration in
HeLa and CaSki cells. The present study further hypothesized that
LINC01123 silencing may ameliorate the malignant behavior of CC by
regulating miR-361-3p. TSPAN1, a transmembrane protein, is involved
in cancer progression (30).
Previous studies have confirmed that TSPAN1 is targeted by
miR-491-3p in osteosarcoma (31)
and miR-216a in pancreatic cancer (32). In the present study, TSPAN1 was
found to be targeted by miR-361-3p and its expression was
negatively modulated by miR-361-3p in CC. It has been reported that
TSPAN1 expression is upregulated in gastric carcinoma (33), ovarian carcinoma (34), pancreatic cancer (35) and CC (36). Similarly, the present study found
that TSPAN1 expression was also upregulated in CC tissue samples,
indicating that TSPAN1 may be an oncogene in CC.
Previous studies have shown that TSPAN1 increases
cell growth and invasion in certain types of cancer. For example,
the overexpression of TSPAN1 enhances cell invasion in CC cell
lines (36). In addition, miR-573
overexpression inhibits cell viability and invasion by decreasing
TSPAN1 expression in gastric cancer (37). Thus, it was hypothesized that
miR-361-3p might regulate cell viability, migration and invasion by
regulating TSPAN1 in CC cells. As LINC01123 directly targeted
miR-361-3p, it was hypothesized that LINC01123 might facilitate CC
tumorigenesis by regulating the miR-361-3p/TSPAN1 axis. Further
rescue studies confirmed that miR-361-3p inhibition and TSPAN1
overexpression reversed LINC01123 knockdown-induced inhibition of
cell viability, migration and invasion in HeLa cells, indicating
that LINC01123 knockdown inhibited CC tumorigenesis by targeting
miR-361-3p/TSPAN1.
Thus, the present study revealed that LINC01123
expression was increased in CC tissue samples and cell lines.
Silencing of LINC01123 inhibited cell viability, invasion and
migration in vitro and suppressed tumor growth in
vivo. miR-361-3p was targeted by LINC01123, whereas TSPAN1 was
targeted by miR-361-3p. Furthermore, knockdown of LINC01123
inhibited CC tumorigenesis by regulating the miR-361-3p/TSPAN1
axis. However, there may be some upstream factors or related
signaling pathways interaction with the LINC01123/miR-361-3p/TSPAN1
axis in the progression of CC. This is a direction for future
studies. However, the findings of the present study may also
provide valuable information in the therapy of CC.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
CL and CH made substantial contributions to the
conception and design of the study. CL, YL, YZ and HY made
substantial contributions to the acquisition, analysis and
interpretation of data, as well as the drafting and revision of the
manuscript. CL, YL, YZ HY and CH confirm the authenticity of all
the raw data and approved the final manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
All patients provided signed informed consent and
the Ethics Committee of Linyi Central Hospital approved the study.
(approval no. 2017013). Animal experiments were performed according
to the National Institutes of Health Guide for the Care and Use of
Laboratory Animals (approval no. 2017013).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Morris E and Roett MA: Genital cancers in
Women: Cervical cancer. FP Essent. 438:18–23. 2015.PubMed/NCBI
|
|
2
|
Arbyn M, Weiderpass E, Bruni L, de Sanjosé
S, Saraiya M, Ferlay J and Bray F: Estimates of incidence and
mortality of cervical cancer in 2018: A worldwide analysis. Lancet
Glob Health. 8:e191–e203. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Kori M and Yalcin Arga K: Potential
biomarkers and therapeutic targets in cervical cancer: Insights
from the meta-analysis of transcriptomics data within network
biomedicine perspective. PLoS One. 13(e0200717)2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Li H, Wu X and Cheng X: Advances in
diagnosis and treatment of metastatic cervical cancer. J Gynecol
Oncol. 27(e43)2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Yee GP, de Souza P and Khachigian LM:
Current and potential treatments for cervical cancer. Curr Cancer
Drug Targets. 13:205–220. 2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Sanchez Calle A, Kawamura Y, Yamamoto Y,
Takeshita F and Ochiya T: Emerging roles of long non-coding RNA in
cancer. Cancer Sci. 109:2093–2100. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Luan X and Wang Y: LncRNA XLOC_006390
facilitates cervical cancer tumorigenesis and metastasis as a ceRNA
against miR-331-3p and miR-338-3p. J Gynecol Oncol.
29(e95)2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Liu Y, Yang Y, Li L, Liu Y, Geng P, Li G
and Song H: LncRNA SNHG1 enhances cell proliferation, migration,
and invasion in cervical cancer. Biochem Cell Biol. 96:38–43.
2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ye S, Sun B, Wu W, Yu C, Tian T, Lian Z,
Liang Q and Zhou Y: LINC01123 facilitates proliferation, invasion
and chemoresistance of colon cancer cells. Biosci Rep.
40(BSR20194062)2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Hua Q, Jin M, Mi B, Xu F, Li T, Zhao L,
Liu J and Huang G: LINC01123, a c-Myc-activated long non-coding
RNA, promotes proliferation and aerobic glycolysis of non-small
cell lung cancer through miR-199a-5p/c-Myc axis. J Hematol Oncol.
12(91)2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Juan C, Hua Q, Ruping Z and Tingting W:
miRNA-489 as a biomarker in diagnosis and treatment of cervical
cancer. Bratisl Lek Listy. 119:278–283. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Yin XZ, Zhao DM, Zhang GX and Liu L:
Effect of miRNA-203 on cervical cancer cells and its underlying
mechanism. Genet Mol Res. 15:2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Hua FF, Liu SS, Zhu LH, Wang YH, Liang X,
Ma N and Shi HR: MiRNA-338-3p regulates cervical cancer cells
proliferation by targeting MACC1 through MAPK signaling pathway.
Eur Rev Med Pharmacol Sci. 21:5342–5352. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Liu S, Song L, Yao H, Zhang L, Xu D, Li Q
and Li Y: Preserved miR-361-3p Expression is an independent
prognostic indicator of favorable survival in cervical cancer. Dis
Markers. 2018(8949606)2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wang J, Li H and Liang Z: circ-MYBL2
serves as a sponge for miR-361-3p promoting cervical cancer cells
proliferation and invasion. Onco Targets Ther. 12:9957–9964.
2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Matsuo K, Machida H, Mandelbaum RS,
Konishi I and Mikami M: Validation of the 2018 FIGO cervical cancer
staging system. Gynecol Oncol. 152:97–93. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Tewari KS, Sill MW, Long HJ III, Penson
RT, Huang H, Ramondetta LM, Landrum LM, Oaknin A, Reid TJ, Leitao
MM, et al: Improved survival with bevacizumab in advanced cervical
cancer. N Engl J Med. 370:734–743. 2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Hsu W, Liu L, Chen X, Zhang Y and Zhu W:
LncRNA CASC11 promotes the cervical cancer progression by
activating Wnt/beta-catenin signaling pathway. Biol Res.
52(33)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang R, Li Y, Du P, Zhang X, Li X and
Cheng G: Hypomethylation of the lncRNA SOX21-AS1 has clinical
prognostic value in cervical cancer. Life Sci.
233(116708)2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhang P, Long Q, Zeng S, Wen M and Lu Q:
FOXC1-induced LINC01123 acts as a mediator in triple negative
breast cancer. Cancer Cell Int. 20(199)2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Yang Y, Wu J, Zhou H, Liu W, Wang J and
Zhang Q: STAT1-induced upregulation of lncRNA LINC01123 predicts
poor prognosis and promotes the progression of endometrial cancer
through miR-516b/KIF4A. Cell Cycle. 19:1502–1516. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Liu J, Zhu J, Xiao Z, Wang X and Luo J:
BBOX1-AS1 contributes to colorectal cancer progression by sponging
hsa-miR-361-3p and targeting SH2B1. FEBS Open Bio, Jan 27, 2020
(Epub ahead of print).
|
|
25
|
Xia F, Chen Y, Jiang B, Bai N and Li X:
Hsa_circ_0011385 accelerates the progression of thyroid cancer by
targeting miR-361-3p. Cancer Cell Int. 20(49)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Shang T, Zhou X and Chen W: LINC01123
promotes progression of colorectal cancer via miR-625-5p/LASP1
axis. Cancer Biother Radiopharm, May 18, 2020 (Epub ahead of
print).
|
|
28
|
Li F, Zhu W and Wang Z: Long noncoding RNA
LINC00460 promotes the progression of cervical cancer via
regulation of the miR-361-3p/Gli1 axis. Hum Cell. 34:229–237.
2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Chen W, Wang J, Liu S, Wang S, Cheng Y,
Zhou W, Duan C and Zhang C: MicroRNA-361-3p suppresses tumor cell
proliferation and metastasis by directly targeting SH2B1 in NSCLC.
J Exp Clin Cancer Res. 35(76)2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Huang S, Yuan S, Dong M, Su J, Yu C, Shen
Y, Xie X, Yu Y, Yu X, Chen S, et al: The phylogenetic analysis of
tetraspanins projects the evolution of cell-cell interactions from
unicellular to multicellular organisms. Genomics. 86:674–684.
2005.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Duan J, Liu J, Liu Y, Huang B and Rao L:
miR-491-3p suppresses the growth and invasion of osteosarcoma cells
by targeting TSPAN1. Mol Med Rep. 16:5568–5574. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Wang S, Liu X, Khan AA, Li H, Tahir M, Yan
X, Wang J and Huang H: miR-216a-mediated upregulation of TSPAN1
contributes to pancreatic cancer progression via transcriptional
regulation of ITGA2. Am J Cancer Res. 10:1115–1129. 2020.PubMed/NCBI
|
|
33
|
Chen L, Li X, Wang GL, Wang Y, Zhu YY and
Zhu J: Clinicopathological significance of overexpression of
TSPAN1, Ki67 and CD34 in gastric carcinoma. Tumori. 94:531–538.
2008.PubMed/NCBI
|
|
34
|
Scholz CJ, Kurzeder C, Koretz K, Windisch
J, Kreienberg R, Sauer G and Deissler H: Tspan-1 is a tetraspanin
preferentially expressed by mucinous and endometrioid subtypes of
human ovarian carcinomas. Cancer Lett. 275:198–203. 2009.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Tian J, Zhang R, Piao H, Li X, Sheng W,
Zhou J, Dong M, Zhang X, Yan X, Shang W, et al: Silencing Tspan1
inhibits migration and invasion, and induces the apoptosis of human
pancreatic cancer cells. Mol Med Rep. 18:3280–3288. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Hölters S, Anacker J, Jansen L,
Beer-Grondke K, Dürst M and Rubio I: Tetraspanin 1 promotes
invasiveness of cervical cancer cells. Int J Oncol. 43:503–512.
2013.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Lu Z, Luo T, Nie M, Pang T, Zhang X, Shen
X, Ma L, Bi J, Wei G, Fang G and Xue X: TSPAN1 functions as an
oncogene in gastric cancer and is downregulated by miR-573. FEBS
Lett. 589:1988–1994. 2015.PubMed/NCBI View Article : Google Scholar
|