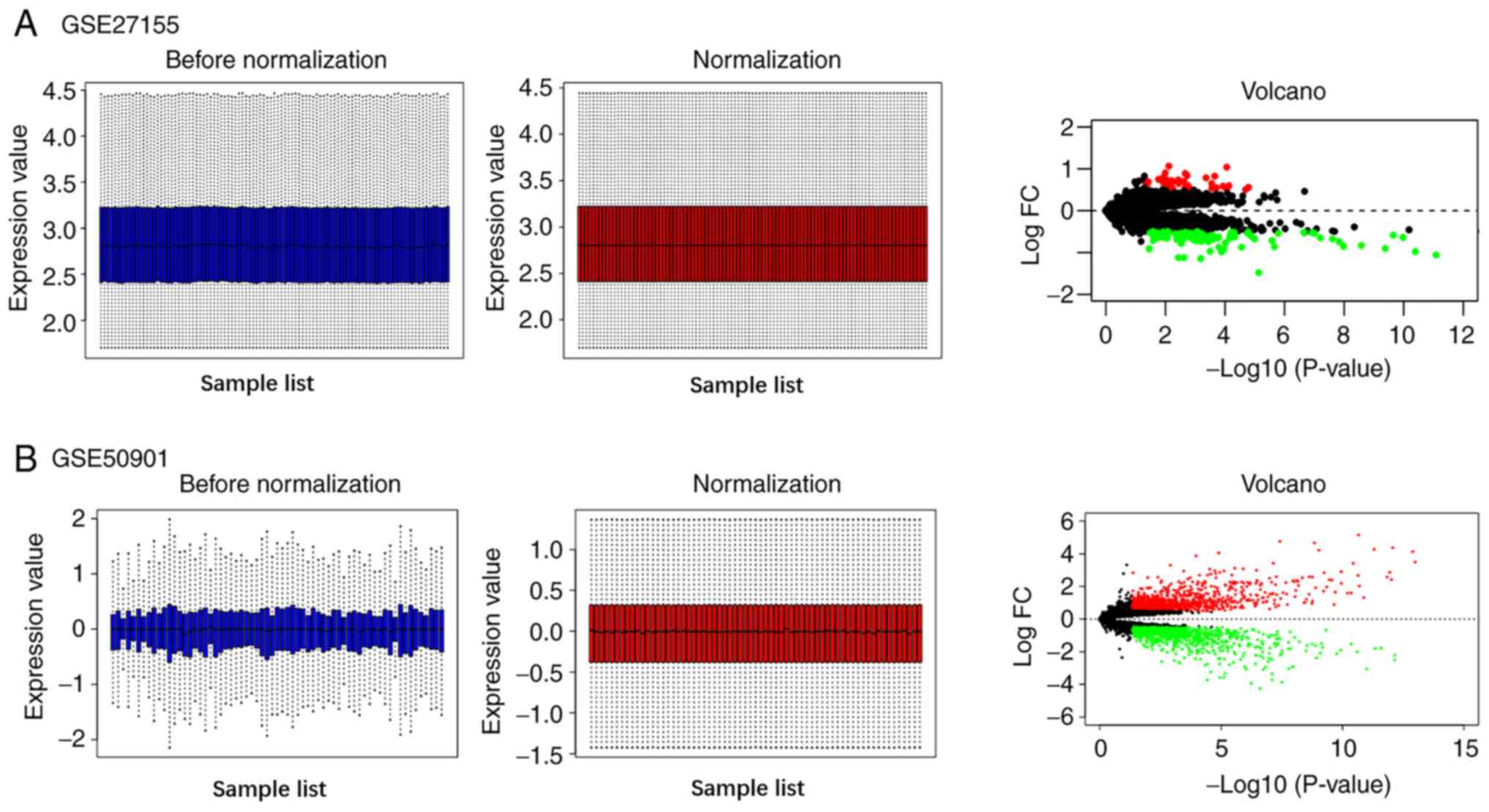
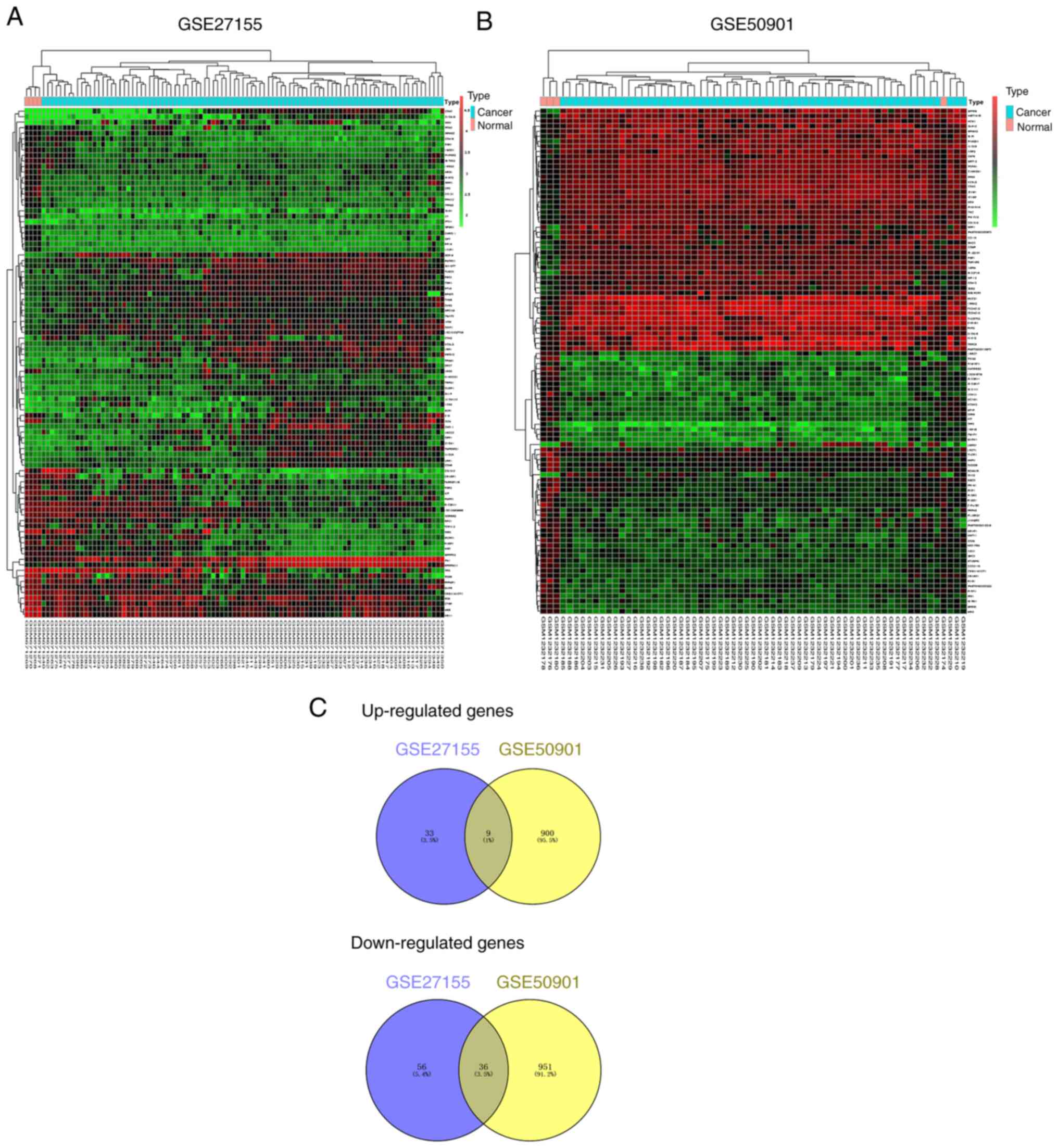
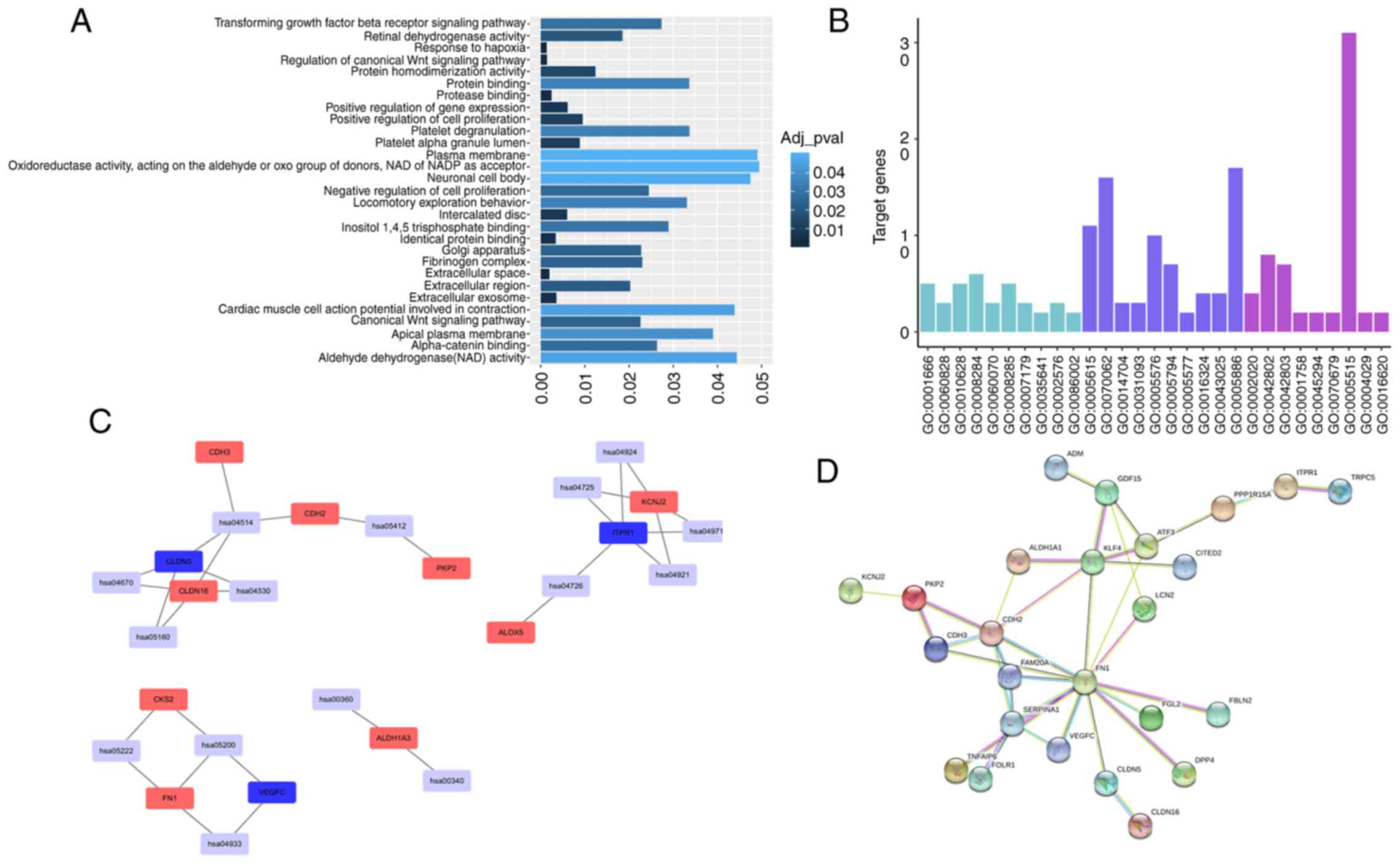
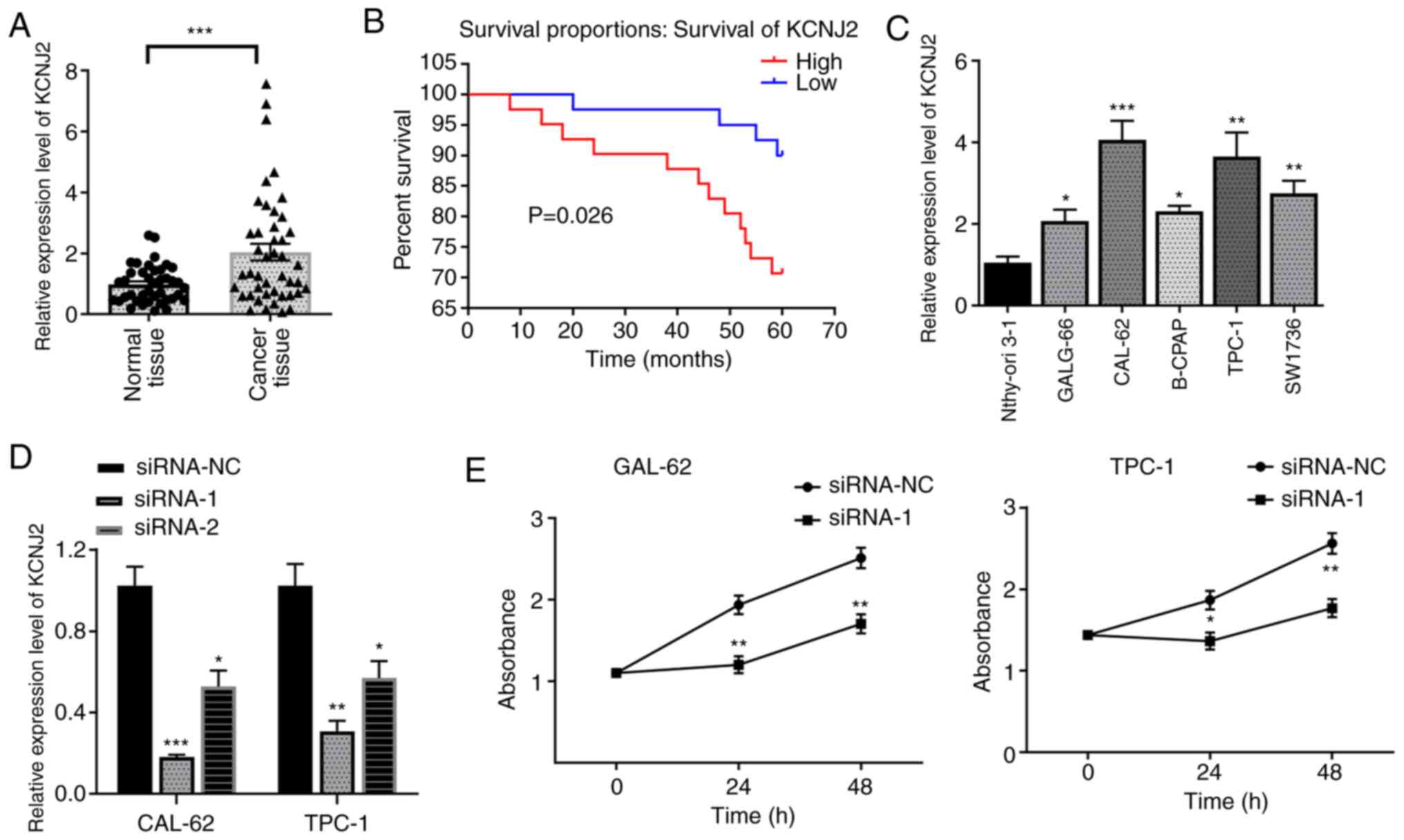
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Pacini F: Observation for newly diagnosed
micro-papillary thyroid cancer: Is now the time? J Endocrinol
Invest. 38:101–102. 2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Lodewijk L, Prins AM, Kist JW, Valk GD,
Kranenburg O, Rinkes IH and Vriens MR: The value of miRNA in
diagnosing thyroid cancer: A systematic review. Cancer Biomark.
11:229–238. 2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Yu J, Mai W, Cui Y and Kong L: Key genes
and pathways predicted in papillary thyroid carcinoma based on
bioinformatics analysis. J Endocrinol Invest. 39:1285–1293.
2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Smallridge RC, Marlow LA and Copland JA:
Anaplastic thyroid cancer: Molecular pathogenesis and emerging
therapies. Endocr Relat Cancer. 16:17–44. 2009.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Geraldo MV and Kimura ET: Integrated
analysis of thyroid cancer public datasets reveals role of
post-transcriptional regulation on tumor progression by targeting
of immune system mediators. PLoS One. 10(e0141726)2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hu S, Liao Y and Chen L: Identification of
key pathways and genes in anaplastic thyroid carcinoma via
integrated bioinformatics analysis. Med Sci Monit. 24:6438–6448.
2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Fluge Ø, Bruland O, Akslen LA, Lillehaug
JR and Varhaug JE: Gene expression in poorly differentiated
papillary thyroid carcinomas. Thyroid. 16:161–175. 2006.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Garg M, Kanojia D, Okamoto R, Jain S,
Madan V, Chien W, Sampath Ding LW, Xuan M, Said JW, et al:
Laminin-5γ-2 (LAMC2) is highly expressed in anaplastic thyroid
carcinoma and is associated with tumor progression, migration, and
invasion by modulating signaling of EGFR. J Clin Endocrinol Metab.
99:E62–E72. 2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kulasingam V and Diamandis EP: Strategies
for discovering novel cancer biomarkers through utilization of
emerging technologies. Nat Clin Pract Oncol. 5:588–599.
2008.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Liu YJ, Zhang S, Hou K, Li YT, Liu Z, Ren
HL, Luo D and Li SH: Analysis of key genes and pathways associated
with colorectal cancer with microarray technology. Asian Pac J
Cancer Prev. 14:1819–1823. 2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang J, Wei B, Cao S, Xu F, Chen W, Lin H,
Du C and Sun Z: Identification by microarray technology of key
genes involved in the progression of carotid atherosclerotic
plaque. Genes Genet Syst. 89:253–258. 2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wang Y and Zheng T: Screening of hub genes
and pathways in colorectal cancer with microarray technology.
Pathol Oncol Res. 20:611–618. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Goujon M, McWilliam H, Li W, Valentin F,
Squizzato S, Paern J and Lopez R: A new bioinformatics analysis
tools framework at EMBL-EBI. Nucleic Acids Res. 38:W695–9.
2010.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Giordano TJ, Kuick R, Thomas DG, Misek DE,
Vinco M, Sanders D, Zhu Z, Ciampi R, Roh M, Shedden K, et al:
Molecular classification of papillary thyroid carcinoma: Distinct
BRAF, RAS, and RET/PTC mutation-specific gene expression profiles
discovered by DNA microarray analysis. Oncogene. 24:6646–6656.
2005.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Barros-Filho MC, Marchi FA, Pinto CA,
Rogatto SR and Kowalski LP: High diagnostic accuracy based on
CLDN10, HMGA2, and LAMB3 transcripts in papillary thyroid
carcinoma. J Clin Endocrinol Metab. 100:E890–E899. 2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kenneth L and Thomas S: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2005.
|
|
20
|
Kubo Y, Baldwin TJ, Jan YN and Jan LY:
Primary structure and functional expression of a mouse inward
rectifier potassium channel. Nature. 362:127–133. 1993.PubMed/NCBI View
Article : Google Scholar
|
|
21
|
Liu H, Huang J, Peng J, Wu X, Zhang Y, Zhu
W and Guo L: Upregulation of the inwardly rectifying potassium
channel Kir2.1 (KCNJ2) modulates multidrug resistance of small-cell
lung cancer under the regulation of miR-7 and the Ras/MAPK pathway.
Mol Cancer. 14(59)2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Li M, Kanda Y, Ashihara T, Sasano T, Nakai
Y, Kodama M, Hayashi E, Sekino Y, Furukawa T and Kurokawa J:
Overexpression of KCNJ2 in induced pluripotent stem cell-derived
cardiomyocytes for the assessment of QT-prolonging drugs. J
Pharmacol Sci. 134:75–85. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Giovannardi S, Forlani G, Balestrini M,
Bossi E, Tonini R, Sturani E, Peres A and Zippel R: Modulation of
the inward rectifier potassium channel IRK1 by the Ras signaling
pathway. J Biol Chem. 277:12158–12163. 2002.PubMed/NCBI View Article : Google Scholar
|