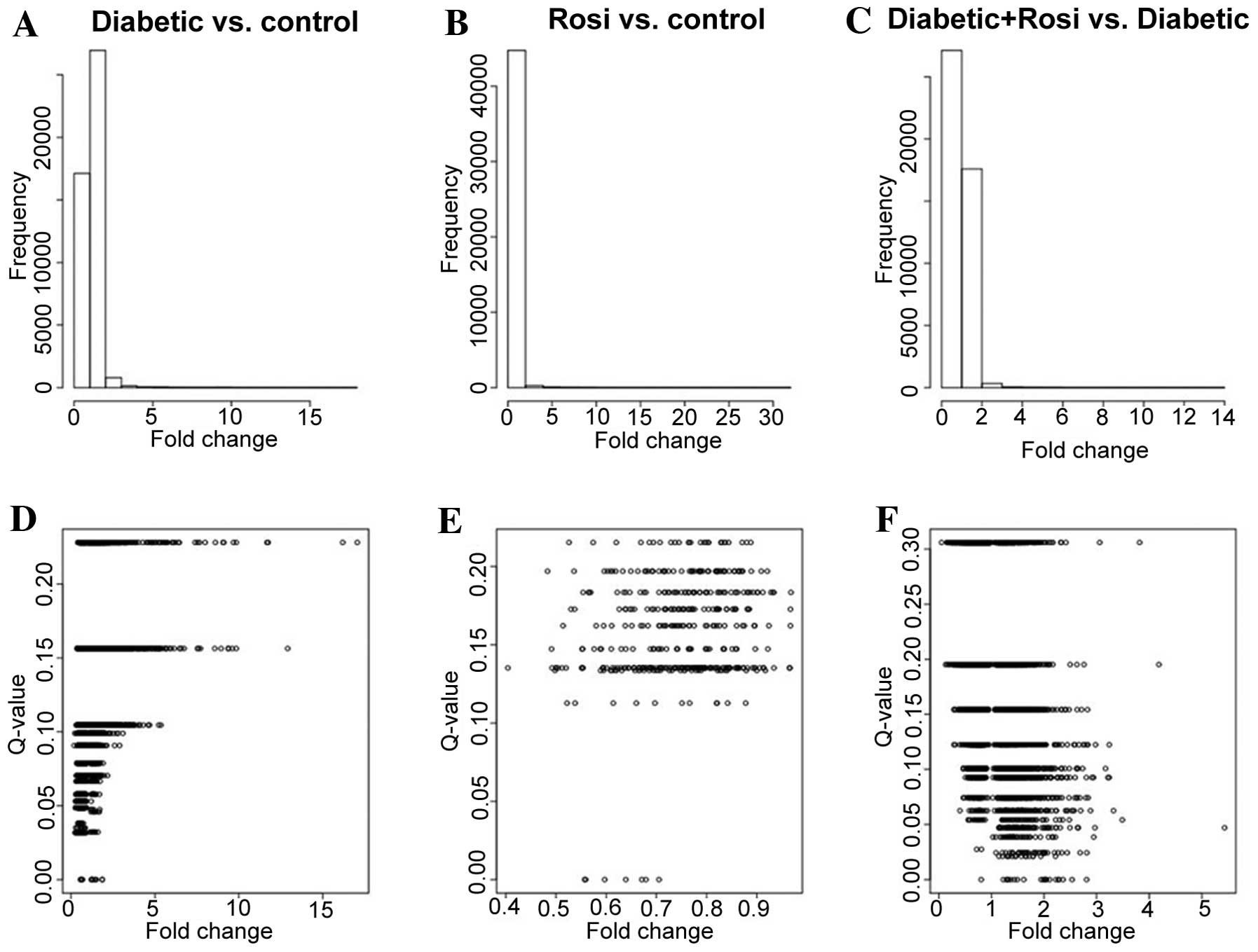
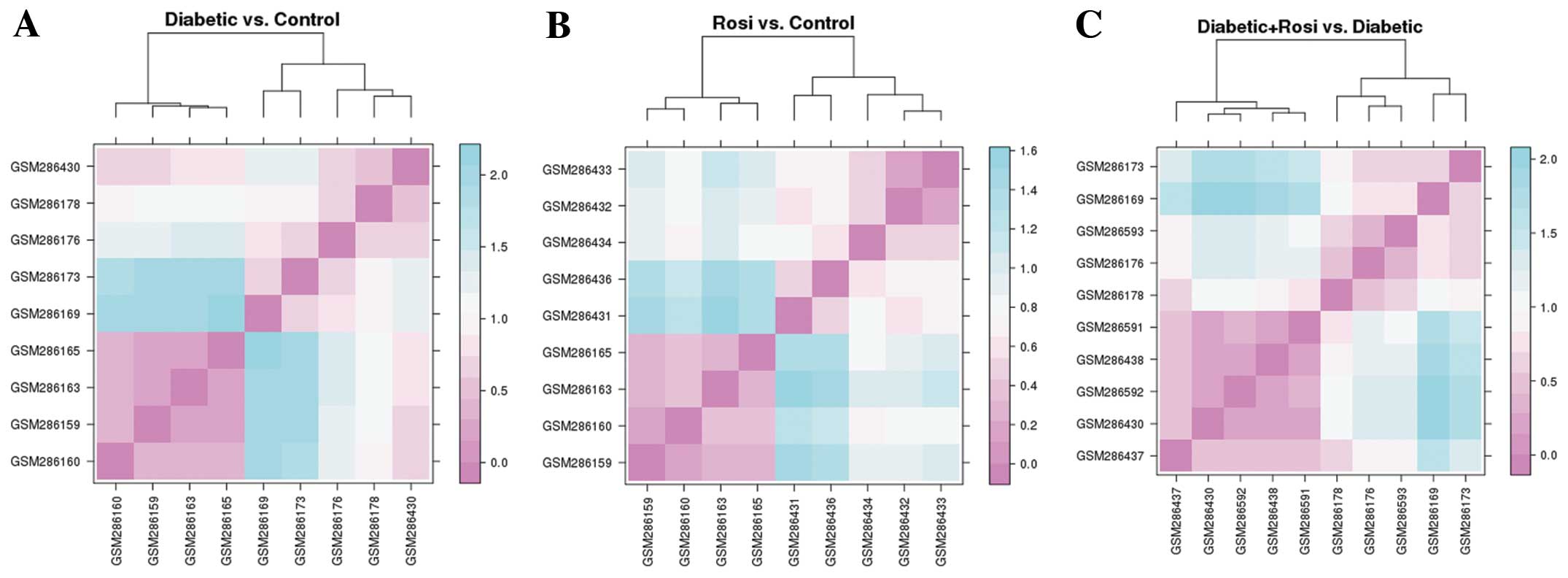
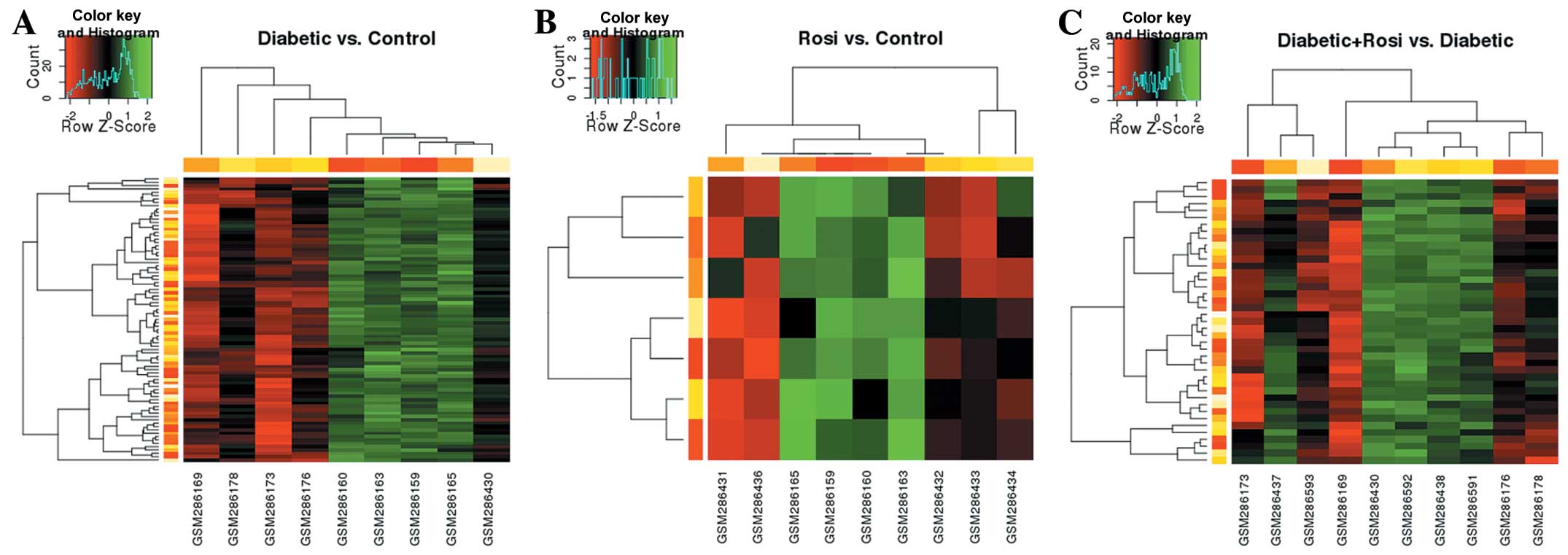
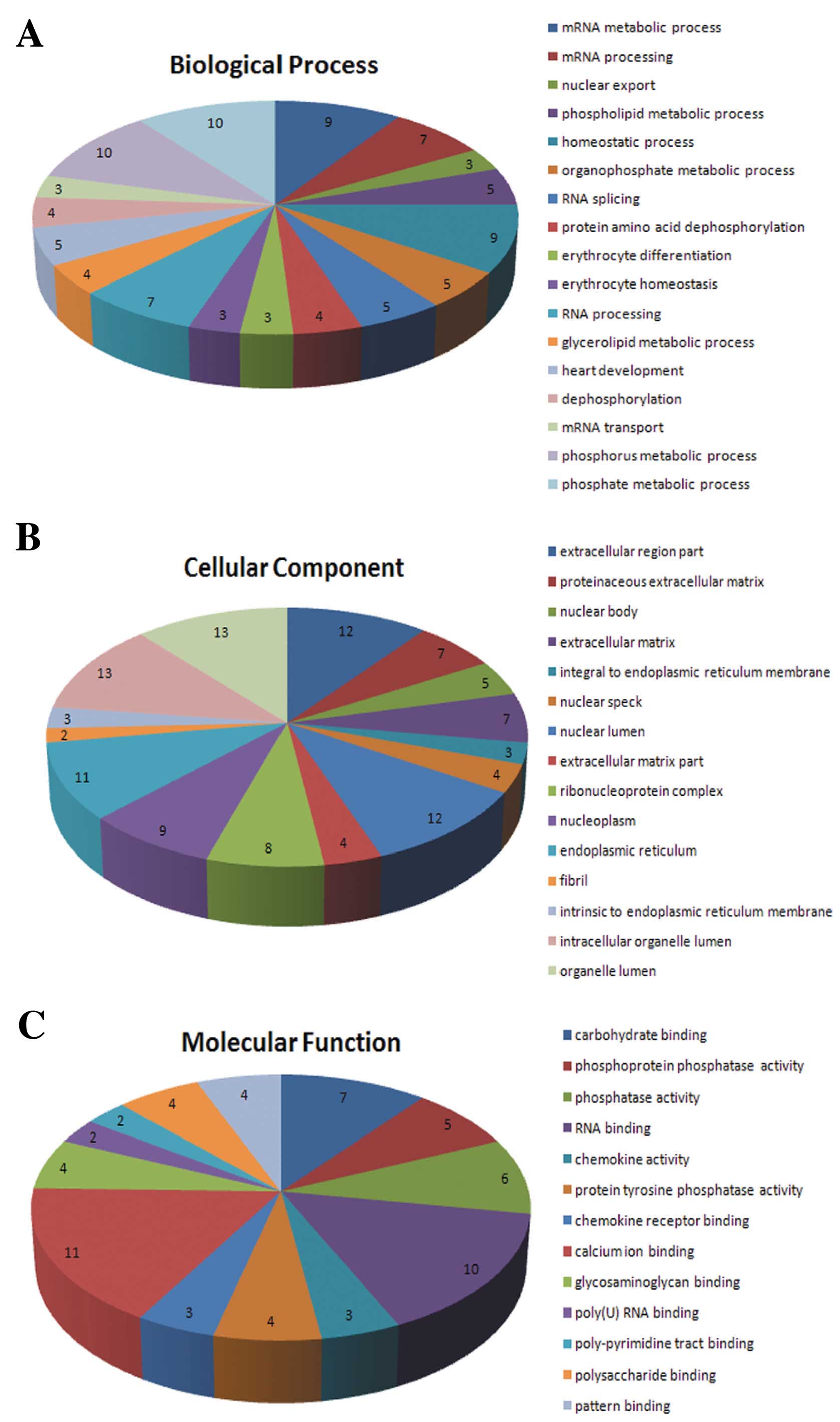
|
1
|
Workowski KA and Berman SM: Centers for
Disease Control and Prevention sexually transmitted diseases
treatment guidelines. Clin Infect Dis. 44(Suppl 3): S73–S76. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Skljarevski V and Malik RA: Clinical
diagnosis of diabetic neuropathy. Diabetic Neuropathy. Veves A and
Malik R: 2nd edition. Springer; New Jersey: pp. 275–292. 2007,
View Article : Google Scholar
|
|
3
|
Said G, Goulon-Goeau C, Slama G and
Tchobroutsky G: Severe early-onset polyneuropathy in
insulin-dependent diabetes mellitus. A clinical and pathological
study. N Engl J Med. 326:1257–1263. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Soumyanath A, Dimitrova D, Arnold G, et
al: P02.186. CAST (Centella asiatica selected triterpenes):
stability, safety, and effect on diabetic neuropathy (DN). BMC
Complement Altern Med. 12(Suppl 1): P2422012. View Article : Google Scholar :
|
|
5
|
Diep QN, El Mabrouk M, Cohn JS, et al:
Structure, endothelial function, cell growth, and inflammation in
blood vessels of angiotensin II-infused rats role of peroxisome
proliferator-activated receptor-gamma. Circulation. 105:2296–2302.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lygate CA, Hulbert K, Monfared M, Cole MA,
Clarke K and Neubauer S: The PPARγ-activator rosiglitazone does not
alter remodeling but increases mortality in rats post-myocardial
infarction. Cardiovasc Res. 58:632–637. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Szkudelski T: The mechanism of alloxan and
streptozotocin action in B cells of the rat pancreas. Physiol Res.
50:537–546. 2001.
|
|
8
|
Yamamoto H, Shimoshige Y, Yamaji T, Murai
N, Aoki T and Matsuoka N: Pharmacological characterization of
standard analgesics on mechanical allodynia in
streptozotocin-induced diabetic rats. Neuropharmacology.
57:403–408. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Edgar R, Domrachev M and Lash AE: Gene
Expression Omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar :
|
|
10
|
Tusher VG, Tibshirani R and Chu G:
Significance analysis of microarrays applied to the ionizing
radiation response. Proc Natl Acad Sci USA. 98:5116–5121. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kanehisa M: The KEGG database. Novartis
Found Symp. 247:91–103. 119–128. 244–252. 2002. View Article : Google Scholar
|
|
12
|
Dennis G Jr, Sherman BT, Hosack DA, et al:
DAVID: database for annotation, visualization, and integrated
discovery. Genome Biol. 4:P32003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nishimura D: BioCarta. Biotech Software
and Internet Report. 2:117–120. 2001. View Article : Google Scholar
|
|
14
|
Franceschini A, Szklarczyk D, Frankild S,
et al: STRING v9.1: protein-protein interaction networks, with
increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar :
|
|
15
|
Berman HM, Westbrook J, Feng Z, et al: The
Protein Data Bank. Nucleic Acids Res. 28:235–242. 2000. View Article : Google Scholar
|
|
16
|
Bairoch A, Apweiler R, Wu CH, et al: The
universal protein resource (UniProt). Nucleic Acids Res.
33:D154–D159. 2005. View Article : Google Scholar :
|
|
17
|
Altschul SF, Madden TL, Schäffer AA, et
al: Gapped BLAST and PSI-BLAST: a new generation of protein
database search programs. Nucleic Acids Res. 25:3389–3402. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yue TI TL, Chen J, Bao W, et al: In vivo
myocardial protection from ischemia/reperfusion injury by the
peroxisome proliferator-activated receptor-γ agonist rosiglitazone.
Circulation. 104:2588–2594. 2001. View Article : Google Scholar
|
|
19
|
Hussein Z, Wentworth JM, Nankervis AJ, et
al: Effectiveness and side effects of thiazolidinediones for type 2
diabetes: real-life experience from a tertiary hospital. Med J
Aust. 181:536–539. 2004.PubMed/NCBI
|
|
20
|
Vanopdenbosch N, Cramer R and Giarrusso
FF: Sybyl, the Integrated Molecular Modeling System. J Mol Graph.
3:110–111. 1985.
|
|
21
|
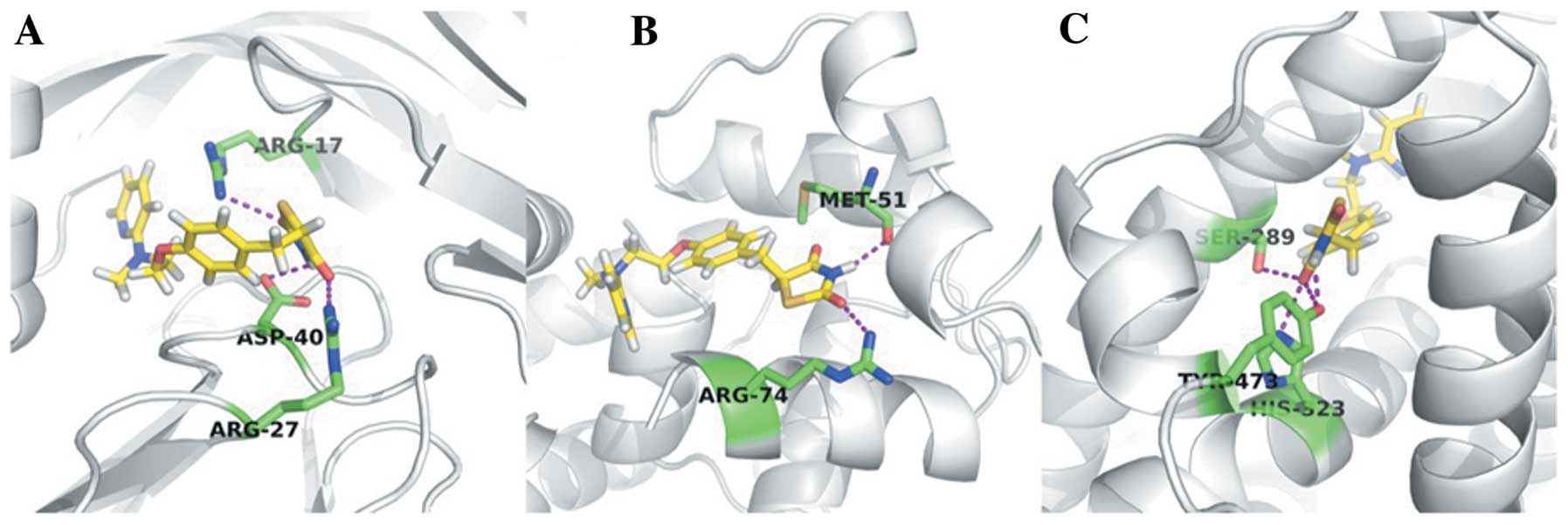
Morris GM, Huey R, Lindstrom W, et al:
AutoDock4 and AutoDockTools4: Automated docking with selective
receptor flexibility. J Comput Chem. 30:2785–2791. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
De Groot M, Anderson R, Freedland KE,
Clouse RE and Lustman PJ: Association of depression and diabetes
complications: a meta-analysis. Psychosom Med. 63:619–630. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Price SA, Zeef LaH, Wardleworth L, Hayes A
and Tomlinson DR: Identification of changes in gene expression in
dorsal root ganglia in diabetic neuropathy: correlation with
functional deficits. J Neuropathol Exp Neur. 65:722–732. 2006.
View Article : Google Scholar
|
|
24
|
Guarguaglini G, Duncan PI, Stierhof YD,
Holmström T, Duensing S and Nigg EA: The forkhead-associated domain
protein Cep170 interacts with Polo-like kinase 1 and serves as a
marker for mature centrioles. Mol Biol Cell. 16:1095–1107. 2005.
View Article : Google Scholar :
|
|
25
|
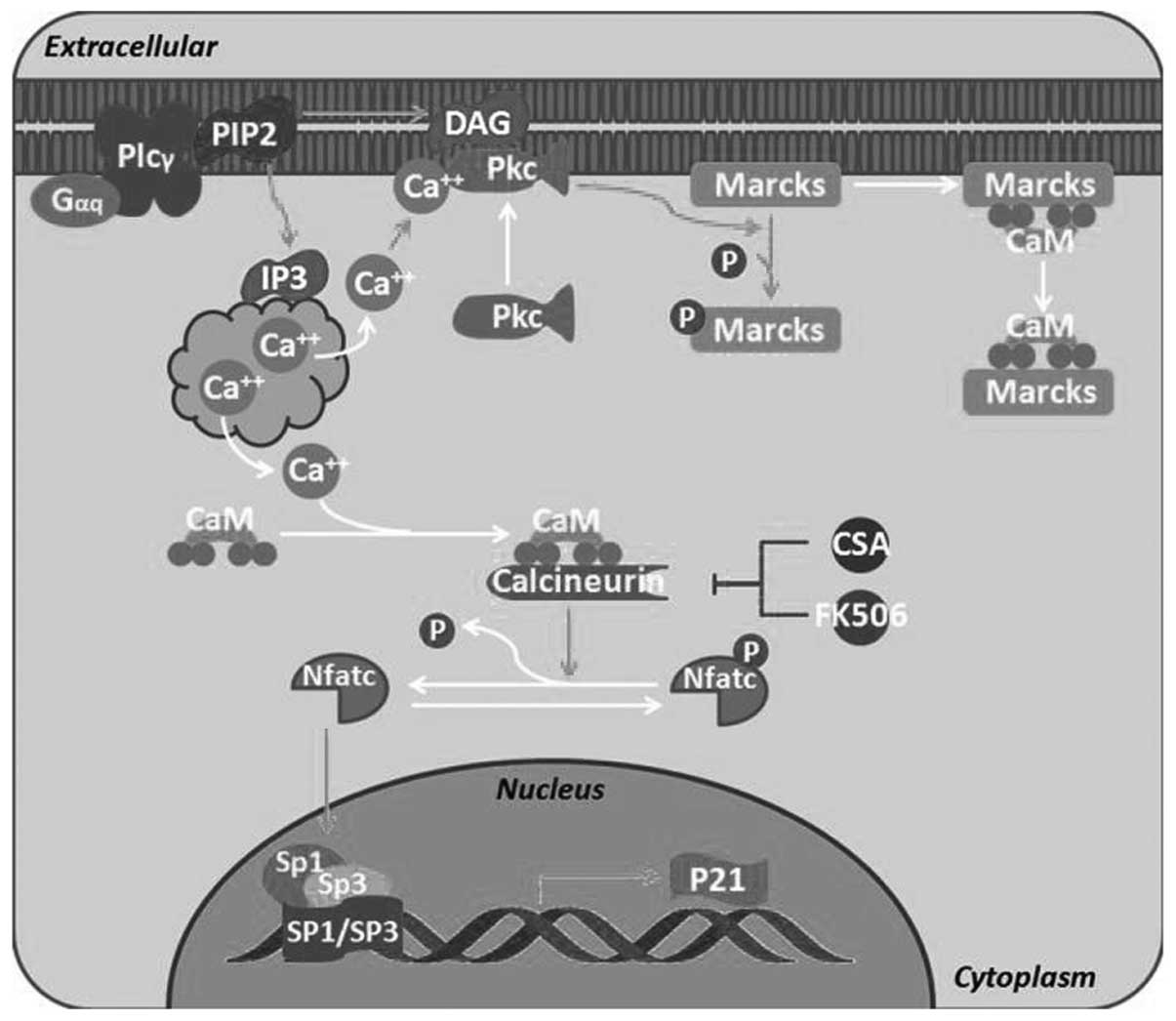
Laux T, Fukami K, Thelen M, Golub T, Frey
D and Caroni P: GAP43, MARCKS, and CAP23 modulate PI(4,5)P(2) at
plasmalemmal rafts, and regulate cell cortex actin dynamics through
a common mechanism. J Cell Biol. 149:1455–1472. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hooi LS, Wong HS and Morad Z: Prevention
of renal failure: The Malaysian experience. Kidney Int Suppl.
67:S70–S74. 2005. View Article : Google Scholar
|
|
27
|
Lokman FE, Seman NA, Al-Safi Ismail A, et
al: Gene expression profiling in ethnic Malays with type 2 diabetes
mellitus, with and without diabetic nephropathy. J Nephrol.
24:778–789. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
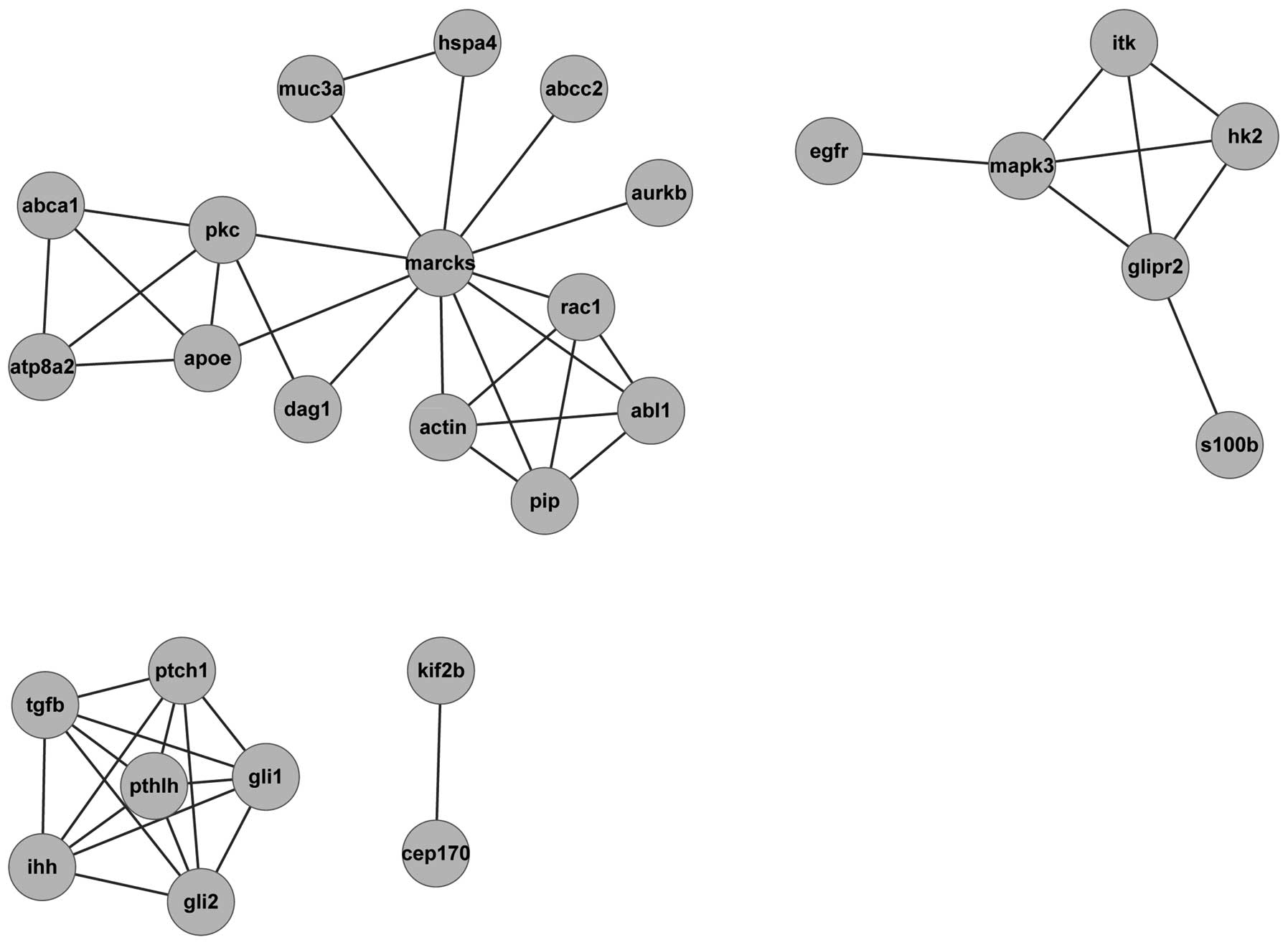
Huang S, Liu F, Niu Q, et al: GLIPR-2
overexpression in HK-2 cells promotes cell EMT and migration
through ERK1/2 activation. PLoS One. 8:e585742013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Krentz A: Thiazolidinediones: effects on
the development and progression of type 2 diabetes and associated
vascular complications. Diabetes Metab Res Rev. 25:112–126. 2009.
View Article : Google Scholar : PubMed/NCBI
|