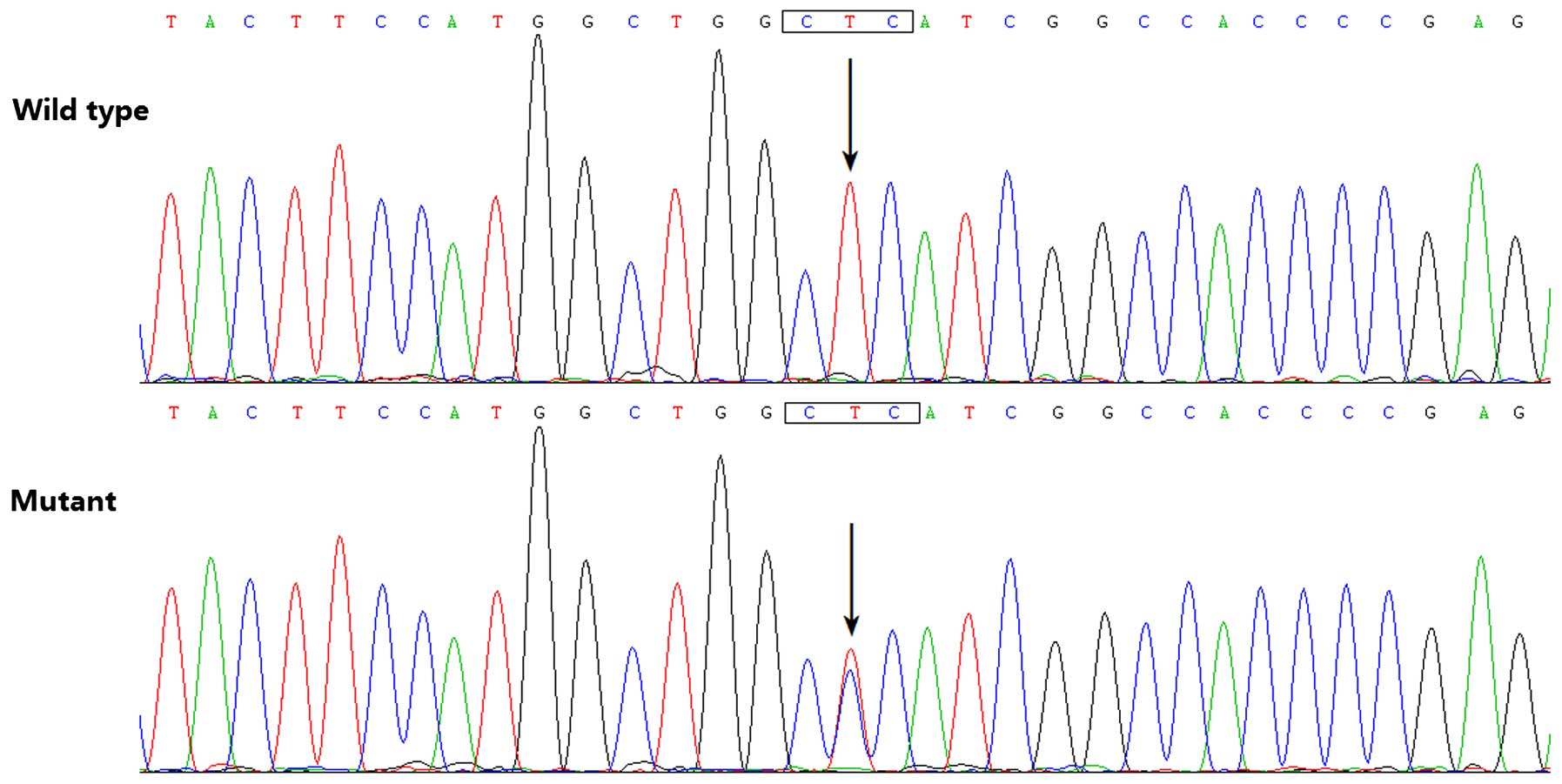
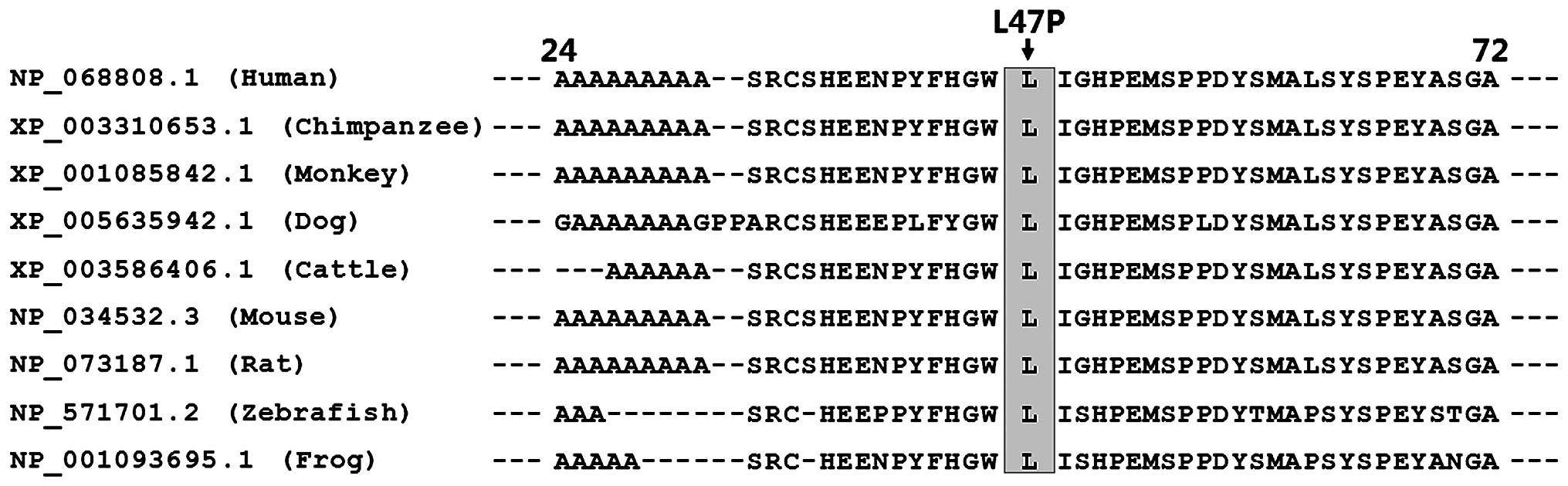
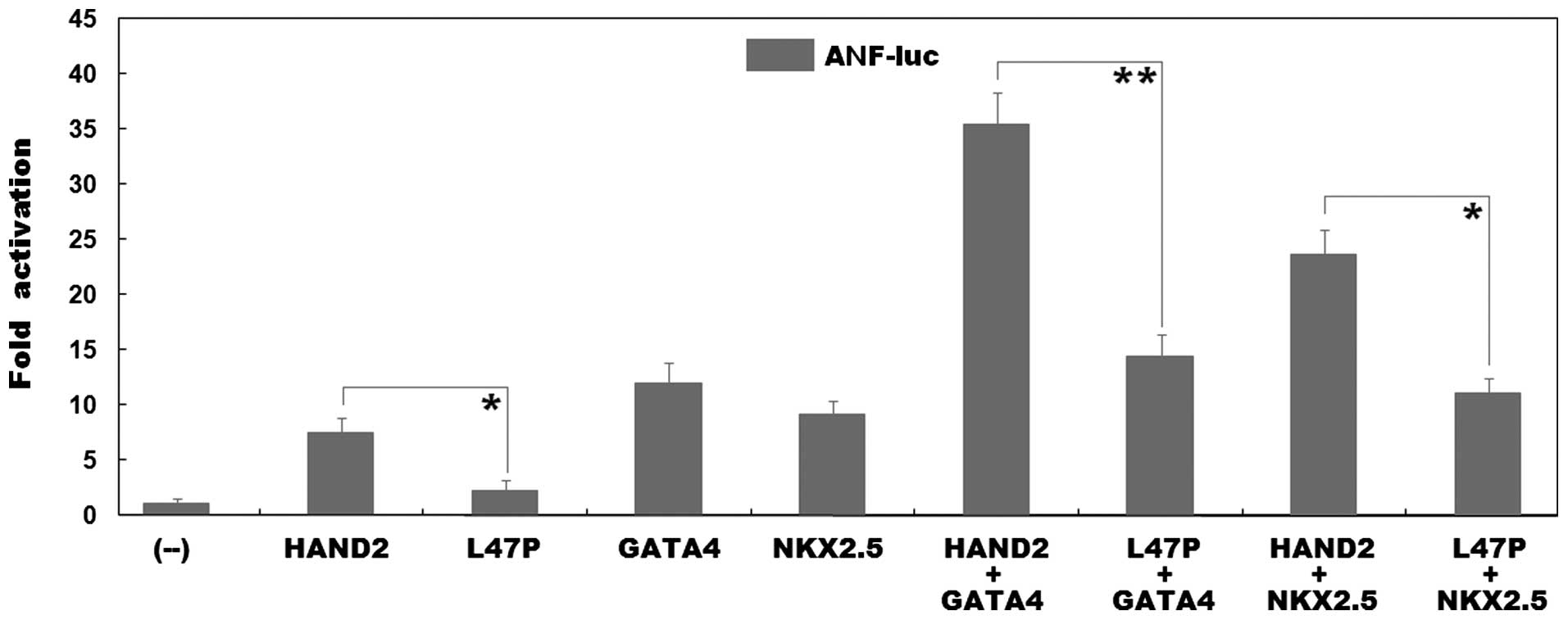
|
1
|
Fahed AC, Gelb BD, Seidman JG and Seidman
CE: Genetics of congenital heart disease: The glass half empty.
Circ Res. 112:707–720. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zheng JY, Tian HT, Zhu ZM, Li B, Han L,
Jiang SL, Chen Y, Li DT, He JC, Zhao Z, et al: Prevalence of
symptomatic congenital heart disease in Tibetan school children. Am
J Cardiol. 112:1468–1470. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marelli AJ, Ionescu-Ittu R, Mackie AS, Guo
L, Dendukuri N and Kaouache M: Lifetime prevalence of congenital
heart disease in the general population from 2000 to 2010.
Circulation. 130:749–756. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mozaffarian D, Benjamin EJ, Go AS, Arnett
DK, Blaha MJ, Cushman M, de Ferranti S, Després JP, Fullerton HJ,
Howard VJ, et al American Heart Association Statistics Committee
and Stroke Statistics Subcommittee: Heart disease and stroke
statistics–2015 update: A report from the American Heart
Association. Circulation. 131:e29–e322. 2015. View Article : Google Scholar
|
|
5
|
Feltez G, Coronel CC, Pellanda LC and
Lukrafka JL: Exercise capacity in children and adolescents with
corrected congenital heart disease. Pediatr Cardiol. 36:1075–1082.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Williams IA, Fifer WP and Andrews H: Fetal
growth and neuro-developmental outcome in congenital heart disease.
Pediatr Cardiol. 36:1135–1144. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Barst RJ, Ivy DD, Foreman AJ, McGoon MD
and Rosenzweig EB: Four- and seven-year outcomes of patients with
congenital heart disease-associated pulmonary arterial hypertension
(from the REVEAL Registry). Am J Cardiol. 113:147–155. 2014.
View Article : Google Scholar
|
|
8
|
Wright LK, Ehrlich A, Stauffer N, Samai C,
Kogon B and Oster ME: Relation of prenatal diagnosis with one-year
survival rate for infants with congenital heart disease. Am J
Cardiol. 113:1041–1044. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Priromprintr B, Rhodes J, Silka MJ and
Batra AS: Prevalence of arrhythmias during exercise stress testing
in patients with congenital heart disease and severe right
ventricular conduit dysfunction. Am J Cardiol. 114:468–472. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ghosh RM, Gates GJ, Walsh CA, Schiller MS,
Pass RH and Ceresnak SR: The prevalence of arrhythmias, predictors
for arrhythmias, and safety of exercise stress testing in children.
Pediatr Cardiol. 36:584–590. 2015. View Article : Google Scholar
|
|
11
|
Walsh EP: Sudden death in adult congenital
heart disease: Risk stratification in 2014. Heart Rhythm.
11:1735–1742. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Srivastava D and Olson EN: A genetic
blueprint for cardiac development. Nature. 407:221–226. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Andersen TA, Troelsen KL and Larsen LA: Of
mice and men: Molecular genetics of congenital heart disease. Cell
Mol Life Sci. 71:1327–1352. 2014. View Article : Google Scholar :
|
|
14
|
Wang X, Li P, Chen S, Xi L, Guo Y, Guo A
and Sun K: Influence of genes and the environment in familial
congenital heart defects. Mol Med Rep. 9:695–700. 2014.
|
|
15
|
Qu XK, Qiu XB, Yuan F, Wang J, Zhao CM,
Liu XY, Zhang XL, Li RG, Xu YJ, Hou XM, et al: A novel NKX2.5
loss-of-function mutation associated with congenital bicuspid
aortic valve. Am J Cardiol. 114:1891–1895. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang X, Ji W, Wang J, Zhao P, Guo Y, Xu R,
Chen S and Sun K: Identification of two novel GATA6 mutations in
patients with nonsyndromic conotruncal heart defects. Mol Med Rep.
10:743–748. 2014.PubMed/NCBI
|
|
17
|
Al Turki S, Manickaraj AK, Mercer CL,
Gerety SS, Hitz MP, Lindsay S, D'Alessandro LC, Swaminathan GJ,
Bentham J, Arndt AK, et al: UK10K Consortium, Wilson DI, Mital S
and Hurles ME: Rare variants in NR2F2 cause congenital heart
defects in humans. Am J Hum Genet. 94:5745–5785. 2014.
|
|
18
|
Zhao L, Ni SH, Liu XY, Wei D, Yuan F, Xu
L, Xin-Li, Li RG, Qu XK, Xu YJ, et al: Prevalence and spectrum of
Nkx2.6 mutations in patients with congenital heart disease. Eur J
Med Genet. 57:579–586. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Werner P, Paluru P, Simpson AM, Latney B,
Iyer R, Brodeur GM and Goldmuntz E: Mutations in NTRK3 suggest a
novel signaling pathway in human congenital heart disease. Hum
Mutat. 35:1459–1468. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wei D, Gong XH, Qiu G, Wang J and Yang YQ:
Novel PITX2c loss-of-function mutations associated with complex
congenital heart disease. Int J Mol Med. 33:1201–1208.
2014.PubMed/NCBI
|
|
21
|
Cowan J, Tariq M and Ware SM: Genetic and
functional analyses of ZIC3 variants in congenital heart disease.
Hum Mutat. 35:66–75. 2014. View Article : Google Scholar :
|
|
22
|
Shi LM, Tao JW, Qiu XB, Wang J, Yuan F, Xu
L, Liu H, Li RG, Xu YJ, Wang Q, et al: GATA5 loss-of-function
mutations associated with congenital bicuspid aortic valve. Int J
Mol Med. 33:1219–1226. 2014.PubMed/NCBI
|
|
23
|
Huang RT, Xue S, Xu YJ, Zhou M and Yang
YQ: Somatic GATA5 mutations in sporadic tetralogy of Fallot. Int J
Mol Med. 33:1227–1235. 2014.PubMed/NCBI
|
|
24
|
Racedo SE, McDonald-McGinn DM, Chung JH,
Goldmuntz E, Zackai E, Emanuel BS, Zhou B, Funke B and Morrow BE:
Mouse and human CRKL is dosage sensitive for cardiac outflow tract
formation. Am J Hum Genet. 96:235–244. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pan Y, Geng R, Zhou N, Zheng GF, Zhao H,
Wang J, Zhao CM, Qiu XB, Yang YQ and Liu XY: TBX20 loss-of-function
mutation contributes to double outlet right ventricle. Int J Mol
Med. 35:1058–1066. 2015.PubMed/NCBI
|
|
26
|
McCulley DJ and Black BL: Transcription
factor pathways and congenital heart disease. Curr Top Dev Biol.
100:253–277. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vincentz JW, Barnes RM and Firulli AB:
Hand factors as regulators of cardiac morphogenesis and
implications for congenital heart defects. Birth Defects Res A Clin
Mol Teratol. 91:485–494. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Reamon-Buettner SM, Ciribilli Y, Inga A
and Borlak J: A loss-of-function mutation in the binding domain of
HAND1 predicts hypoplasia of the human hearts. Hum Mol Genet.
17:1397–1405. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Reamon-Buettner SM, Ciribilli Y, Traverso
I, Kuhls B, Inga A and Borlak J: A functional genetic study
identifies HAND1 mutations in septation defects of the human heart.
Hum Mol Genet. 18:3567–3578. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cheng Z, Lib L, Li Z, Liu M, Yan J, Wang B
and Ma X: Two novel HAND1 mutations in Chinese patients with
ventricular septal defect. Clin Chim Acta. 413:675–677. 2012.
View Article : Google Scholar
|
|
31
|
Thomas T, Yamagishi H, Overbeek PA, Olson
EN and Srivastava D: The bHLH factors, dHAND and eHAND, specify
pulmonary and systemic cardiac ventricles independent of left-right
sidedness. Dev Biol. 196:228–236. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Thattaliyath BD, Livi CB, Steinhelper ME,
Toney GM and Firulli AB: HAND1 and HAND2 are expressed in the
adult-rodent heart and are modulated during cardiac hypertrophy.
Biochem Biophys Res Commun. 297:870–875. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Srivastava D, Cserjesi P and Olson EN: A
subclass of bHLH proteins required for cardiac morphogenesis.
Science. 270:1995–1999. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Srivastava D, Thomas T, Lin Q, Kirby ML,
Brown D and Olson EN: Regulation of cardiac mesodermal and neural
crest development by the bHLH transcription factor, dHAND. Nat
Genet. 16:154–160. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
McFadden DG, Barbosa AC, Richardson JA,
Schneider MD, Srivastava D and Olson EN: The Hand1 and Hand2
transcription factors regulate expansion of the embryonic cardiac
ventricles in a gene dosage-dependent manner. Development.
132:189–201. 2005. View Article : Google Scholar
|
|
36
|
Wang XH, Huang CX, Wang Q, Li RG, Xu YJ,
Liu X, Fang WY and Yang YQ: A novel GATA5 loss-of-function mutation
underlies lone atrial fibrillation. Int J Mol Med. 31:43–50.
2013.
|
|
37
|
Wei D, Bao H, Zhou N, Zheng GF, Liu XY and
Yang YQ: GATA5 loss-of-function mutation responsible for the
congenital ventriculoseptal defect. Pediatr Cardiol. 34:504–511.
2013. View Article : Google Scholar
|
|
38
|
Zhang XL, Dai N, Tang K, Chen YQ, Chen W,
Wang J, Zhao CM, Yuan F, Qiu XB, Qu XK, et al: GATA5
loss-of-function mutation in familial dilated cardiomyopathy. Int J
Mol Med. 35:763–770. 2015.
|
|
39
|
Starr JP: Tetralogy of fallot: Yesterday
and today. World J Surg. 34:658–668. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Russell MW, Kemp P, Wang L, Brody LC and
Izumo S: Molecular cloning of the human HAND2 gene. Biochim Biophys
Acta. 1443:393–399. 1998. View Article : Google Scholar
|
|
41
|
Dai YS and Cserjesi P: The basic
helix-loop-helix factor, HAND2, functions as a transcriptional
activator by binding to E-boxes as a heterodimer. J Biol Chem.
277:12604–12612. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Dai YS, Cserjesi P, Markham BE and
Molkentin JD: The transcription factors GATA4 and dHAND physically
interact to synergistically activate cardiac gene expression
through a p300-dependent mechanism. J Biol Chem. 277:24390–24398.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Thattaliyath BD, Firulli BA and Firulli
AB: The basic-helix-loop-helix transcription factor HAND2 directly
regulates transcription of the atrial naturetic peptide gene. J Mol
Cell Cardiol. 34:1335–1344. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zang MX, Li Y, Wang H, Wang JB and Jia HT:
Cooperative interaction between the basic helix-loop-helix
transcription factor dHAND and myocyte enhancer factor 2C regulates
myocardial gene expression. J Biol Chem. 279:54258–54263. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zang MX, Li Y, Xue LX, Jia HT and Jing H:
Cooperative activation of atrial naturetic peptide promoter by
dHAND and MEF2C. J Cell Biochem. 93:1255–1266. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yelon D, Ticho B, Halpern ME, Ruvinsky I,
Ho RK, Silver LM and Stainier DY: The bHLH transcription factor
hand2 plays parallel roles in zebrafish heart and pectoral fin
development. Development. 127:2573–2582. 2000.PubMed/NCBI
|
|
47
|
Garavito-Aguilar ZV, Riley HE and Yelon D:
Hand2 ensures an appropriate environment for cardiac fusion by
limiting Fibronectin function. Development. 137:3215–3220. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Morikawa Y and Cserjesi P: Cardiac neural
crest expression of Hand2 regulates outflow and second heart field
development. Circ Res. 103:1422–1429. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tsuchihashi T, Maeda J, Shin CH, Ivey KN,
Black BL, Olson EN, Yamagishi H and Srivastava D: Hand2 function in
second heart field progenitors is essential for cardiogenesis. Dev
Biol. 351:62–69. 2011. View Article : Google Scholar :
|
|
50
|
VanDusen NJ, Casanovas J, Vincentz JW,
Firulli BA, Osterwalder M, Lopez-Rios J, Zeller R, Zhou B,
Grego-Bessa J, De La Pompa JL, et al: Hand2 is an essential
regulator for two Notch-dependent functions within the embryonic
endocardium. Cell Rep. 9:2071–2083. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tamura M, Hosoya M, Fujita M, Iida T,
Amano T, Maeno A, Kataoka T, Otsuka T, Tanaka S, Tomizawa S, et al:
Overdosage of Hand2 causes limb and heart defects in the human
chromosomal disorder partial trisomy distal 4q. Hum Mol Genet.
22:2471–2481. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shen L, Li XF, Shen AD, Wang Q, Liu CX,
Guo YJ, Song ZJ and Li ZZ: Transcription factor HAND2 mutations in
sporadic Chinese patients with congenital heart disease. Chin Med J
(Engl). 123:1623–1627. 2010.
|