Introduction
A previous study suggested that various biological
processes are determined by the regulatory potential of the
noncoding portions of the genome. It has been previously estimated
that ~1.5% of the genome is responsible for protein coding, whereas
a number of noncoding regulatory elements are transcribed into
noncoding RNA (ncRNA) (1). Long
noncoding (lncRNAs) (>200 nucleotides) are a novel form of
ncRNAs, which have been identified to exert their gene
transcription regulatory effect through the epigenetic regulatory
mechanism. LncRNA plasmacytoma variant translocation 1 (PVT1) has
been reported to be associated with cell proliferation, invasion
and metastasis, apoptosis and tumor prognosis (2–7).
Although, some previous studies have revealed that lncRNAs such as
MEG3 maternally expressed 3, HOX transcript antisense RNA had an
important role in angiogenesis by directly regulating vascular
endothelial growth factor (VEGF) and angiopoietin 2 (ANGPT2);
however, whether PVT1 had a similar function remains to be
elucidated (8–10). A recent study by Ma et al
(11) observed that PVT1
regulated growth, migration and angiogenesis of glioma
microvascular endothelial cells by targeting microRNA (miR)-186;
therefore, it should be considered if other pathways or downstream
targets mediate PVT1 regulation of angiogenesis in vascular
endothelial cells.
MicroRNA is a class of ~22 nucleotide noncoding
RNAs. They may regulate gene expression through recognizing and
binding to the 3′-untranslated region (3′-UTR) of target gene
mRNAs, leading to mRNA degradation or translational suppression
(12). It has been estimated that
approximately one out of three human genes are regulated by miRNAs
(13,14). Previous studies have revealed that
miRNAs have a critical role in a variety of cellular processes in
healthy and ailing individuals, including proangiogenic
therapeutics to reconstruct vasculature for patients with ischemic
heart and peripheral vascular diseases (15–18). A previous study reported that
miR-26a regulates pathological and physiological angiogenesis by
targeting its downstream bone morphogenetic protein/SMAD family
member 1 signaling (19). Another
study demonstrated that lncRNA PVT1 is a miR-26b sponge and
promoted melanoma progression, which indicated that lncRNA PVT1 may
regulate angiogenesis via interaction with miR-26b (20).
Connective tissue growth factor (CTGF) is a member
of the CCN family which consists of cysteine-rich proteins. It has
been previously demonstrated that CTGF is associated with fibrosis,
tissue remodeling and tumorigenesis (21). It is of note that CTGF has a
promoter role in the regulation of vessel growth during
development, wound healing and vascular disease, suggesting CTGF is
an angiogenetic inducer (22). In
previous studies, CTGF has been revealed to be a direct downstream
target of miR-26b, indicating that miR-26b may influence
angiogenesis via downregulation of CTGF gene expression (20).
The present study revealed that PVT1 directly
interacts with miR-26b to reduce the expression level, which
subsequently promoted CTGF and ANGPT2 expression levels, which
contributed to cell proliferation, migration and angiogenesis of
vascular endothelial cells.
Materials and methods
Cell line
Human umbilical vein endothelial cells (HUVECs) and
293T cells were purchased from America Type Culture Collection
(Manassas, VA, USA). HUVECs were cultured in vascular cell basal
medium (ScienCell Research Laboratories, Inc., San Diego, CA, USA)
supplemented with 10 ng/ml VEGF at 37°C, 5% CO2. 293T
cells were cultured in Dulbecco's modified Eagle's medium (DMEM)
(HyClone; GE Healthcare, Logan, UT, USA) supplemented with 10%
fetal bovine serum (FBS) (BI) and 1% penicillin (100
U/ml)/streptomycin (100 U/ml) (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA).
Plasmid transfection and lentivirus
package
PVT1 full-length cDNA was cloned into a pEX2 plasmid
and the short hairpin RNA (shRNA) targeting PVT1 was cloned into a
pLKO.1-TRC vector (all from Shanghai GenePharma Co., Ltd.,
Shanghai, China), where the transcription was under the control of
the U6 promoter. shPVT1 target sequence was
5′-CAGCCATCATGATGGTACT-3′. In order to generate lentiviruses, the
transducing vectors (pPAX2 and pVSVG; Shanghai GenePharma Co.,
Ltd.) were co-transfected into 293T cells with polybrene
(Sigma-Aldrich; Merck Millipore, Darmstadt, Germany). The
supernatant was harvested at 24 and 48 h after transfection,
filtered through 0.45 μm membrane and concentrated using a
centrifugal filter (EMD Millipore, Billerica, MA, USA).
Luciferase reporter assay
The CTGF mutant 3′-UTR was generated by replacing
the seed regions of the miR-26b binding sites with 5′-TTGGTT-3′ and
PVT1 mutant was generated using site-directed mutagenesis (23). Subsequently, the mutant sequence
was cloned into the firefly lucif-erase-expressing vector pGL3
(Shanghai GenePharma Co., Ltd.). As for luciferase assay, the
HUVECs were seeded in 24-well plates at 4×104 cells/well
the day before transfection and transfected with the CTGF wild type
or mutant 3′-UTR reporter vector (Shanghai GenePharma Co., Ltd.),
PVT1 or PVT1 mutant using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). The cells were
harvested and lysed 48 h after transfection and the luciferase
activity was assayed using the Dual-Luciferase Reporter system
(Promega Corporation, Madison, WI, USA). The β-lactamase gene of
the pGL3 luciferase vector was used for the normalization of the
luminescence levels. Three independent experiments were
performed.
Transwell migration assay
The cells were transfected with miR-26b mimic using
Lipofectamine® 2000, miR-26b inhibitor (both from
Shanghai GenePharma Co., Ltd.), PVT1 or shPVT1. After 24 h, the
cells were starved in medium without serum for another 12 h and
then digested with trypsin and then 3×104 cells were
seeded in the top chamber of 24-well Transwell culture inserts
(Promega Corporation). The medium supplemented with 20% FBS used as
chemoattractant was added to bottom chamber. After 24 h incubation,
the cells were fixed for 10 min with 4% paraformalin and cells
which had not migrated were removed. The cells on the lower side of
the filter were stained with 0.005% crystal violet for 30 min in
25°C and then the number of cells was counted and photographed with
an inverted microscope at magnification of ×100 and three fields of
view.
Cell proliferation assay
The proliferation of HUVECs from various groups were
examined via MTT assay. The cells were counted and plated into
96-well plates at a density of 2×103 cells/well for 24
h, then 0.1 mg/ml MTT was added to cells at 37°C for 3 h and lysed
in DMSO at room temperature for 30 min. Finally, absorbance was
quantified at 490 nm using a microplate reader (Omega Bio-Tek,
Inc., Norcross, GA, USA).
In vitro vascular tube formation
assay
To examine the ability of endothelial tube formation
in vitro, the present study used 15-well μ-slides
(ibidi GmbH, Martinsried, Germany) coated with 10 μl
Matrigel as previously described (23). To determine the effect of PVT1 and
miR-26b on tube formation, HUVECs were pretreated with
CoCl2 and transfected with PVT1 and miR-26b mimics or
siRNAs against PVT1 or miR-26b. The cells were grown in Medium DMEM
(ScienCell Research Laboratories, Inc., San Diego, CA, USA)
supplemented 10% FBS (Thermo Fisher Scientific, Inc.) and 3.75
μg/ml endothelial cell growth supplement (BD Biosciences,
New Jersey, USA). The tube length was measured using MetaMorph
version 7.8.10 (Molecular Devices, LLC, Sunnyvale, CA, USA) and
compared with the control. The micrographs were captured and
processed with an inverted microscope.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol method. The
cells were lysed with TRIzol buffer (Thermo Fisher Scientific,
Inc.), and 200 μl chloroform was added to mixture. The
resulting solution was centrifuged at 10,000 × g for 10 min at 4°C.
The supernatant was harvested and mixed with equivalent volume of
isopropanol. The resultant was subjected to centrifugation at
10,000 × g for 10 min at 4°C. The supernatant was removed and 75%
ethanol was added to wash the pellet and centrifuged at 7,500 × g
for 5 min at 4°C. The ethanol was discarded and the pellet dried,
20-30 μl RNAse-free H2O was used to elute the RNA
pellet.
Subsequently, 1 μg total RNA underwent
reverse transcription using PrimeScript kit according to
manufacturer's protocol (Takara Biotechnology Co., Ltd., Dalian,
China). For qPCR, this experiment was performed using SYBR
(Guangzhou RiboBio Co., Ltd., Guangzhou, China) as probe dye and
detected the signal by the standard protocol. The expression of
miR-26b was detected using a Bulge-Loop™ miRNA qRT-PCR Primer set
(Guangzhou RiboBio Co., Ltd.) according to the manufacturer's
protocol. cDNA was synthe-sized from total RNA using the
PrimeScript kit at 25°C for 10 min, at 42°C for 50 min, at 95°C for
5 min. cDNA was then amplified following cycling conditions: One
initial PCR activation step at 95°C for 15 min followed by 40
cycles of denaturation at 94°C for 15 sec, annealing at 53°C for 30
sec, and elongation at 72°C for 30 sec. The U6 and GAPDH were used
as internal control. The following primers were used: PVT1 forward
(F), 5′-GGGGAATAACGCTGGTGGAA-3′ and reverse (R),
5′-CCCATGGACATCCAAGCTGT-3′; CTGFF, 5′-GAGAGTCCTTCCAGAGCAGC-3′ and
R, 5′-CATAGTTGGGTCTGGGCCAA-3′; ANGPT2 F, 5′-CCCTACGTGTCCAATGCTGT-3′
and R, 5′-CCGCTGTTTGGTTCAACAGG-3′; U6 F,
5′-CTCGCTTCGGCAGCACATATACTA-3′ and U6 R,
5′-ACGAATTTGCGTGTCATCCTTGCG-3′; GAPDHF, 5′-GAGTCAACGGATTTGGTCGT-3′
and R, 5′-TTGATTTTGGAGGGATCTCG-3′. Cq values were used for
quantification using a previously described protocol (24). cDNA was prepared for three times
and RT-qPCR was repeated in triplicate parallel experiments.
Western blotting
Cells were harvested and washed with PBS.
Subsequently SDS loading buffer (Sigma-Aldrich; Merck Millipore)
was used to lyse cells. The lysates were boiled at 95°C for 10 min
and then subjected to centrifugation at 10,000 × g for 1 min at
4°C. Total protein (50 μg) was loaded onto 10% SDS-PAGE gel
and resolved at 120 V for 30 min to 1 h. Subsequently, the proteins
in the gel were transferred onto a PVDF membrane at 300 mA for 2-3
h. The membrane was blocked with 5% non-fat milk in TBST with 0.1%
Tween-20 for 1 h at room temperature, and then the membrane was
incubated with the following primary antibodies at 4°C overnight:
CTGF (cat no. ab6992; 1:1,000), ANGPT2 (cat no. ab8452; 1:1,000)
(both from Abcam, Cambridge, UK), β-actin (cat no. 60008-1-Ig;
1:5,000; ProteinTech Group, Inc., Chicago, IL, USA). The following
day, the membrane was washed with TBST 3 times and incubated with a
horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G
(cat. no. A0208; 1:5,000; Beyotime Institute of Biotechnology,
Haimen, China) at room temperature for 1 h. Finally, the membrane
was incubated with enhanced chemiluminescent reagent (7Sea Biotech,
Shanghai, China) and then exposed using Bio-Rad ChemiDoc Touch
Imaging system (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Statistical analysis
Each experiment was performed three times. All data
are presented as mean ± standard deviation. Comparisons of
parameters were performed using a two-tailed unpaired Student's
t-test using Prism version 6.0 (GraphPad Software, Inc., La Jolla,
CA, USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-26b inhibits cell proliferation,
migration and in vitro tube formation in HUVECs
In order to investigate the role of miR-26b in
angiogenesis of vascular endothelial cells, and
miR-26b-overexpressing HUVECs were constructed by transfection with
miR-26b mimic. miR-26b-depleted HUVECs were established by
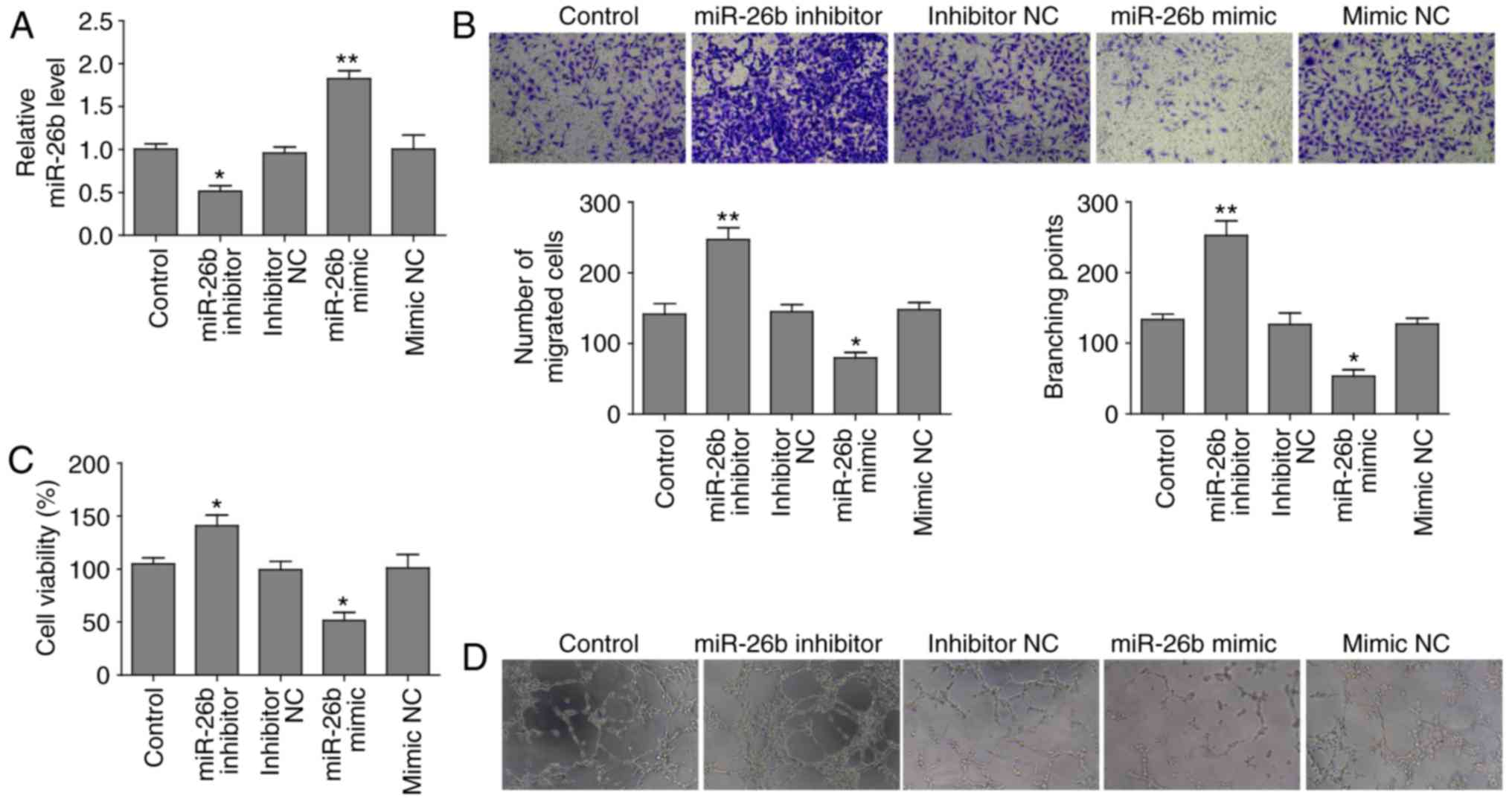
transfecting the cells with an miR-26b inhibitor. RT-qPCR revealed
that miR-26b expression was elevated in cells transfected with
miR-26b mimic compared to mimic negative control (NC), whereas
miR-26b level was attenuated following transfection with miR-26b
inhibitor relative to negative control (Fig. 1A).
The present study identified that miR-26b inhibited
the migration of HUVECs in Transwell assays and inhibition miR-26b
promoted this migration (Fig.
1B). Subsequently, the present study investigated whether
miR-26b affected proliferation of HUVECs. The MTT assay
demonstrated that cell proliferation of cells overexpressing mir-26
was significantly reduced, conversely depletion of miR-26b
increased the cell proliferation ability of HUVECs (Fig. 1C). In vitro a vascular tube
formation assay revealed that miR-26b overexpression reduced the
length of the vascular tubes; however, miR-26b depletion led to the
formation of longer vascular tubes, which was analyzed
statistically by branching points formed by various groups of
HUVECs (Fig. 1D). These findings
indicated that miR-26b has a suppressive role in cell
proliferation, migration and tube formation in HUVECs.
miR-26b suppresses expression of PVT1 and
CTGF
In order to determine the molecular mechanism by
which miR-26b contributed to cell proliferation, migration and tube
formation in HUVECs, the present study focused on the expression
level of previously reported genes, CTGF and ANGPT2. Using RT-qPCR
it was observed that compared with the mimic NC group, miR-26b
overexpression led to an attenuated expression level of CTGF and
ANGPT2 (Fig. 2A and B) and
unexpectedly of PVT1 (Fig. 2C).
Conversely, miR-26b inhibition increased their expression levels.
Additionally, compared with the control (mimic NC), protein
expression levels of CTGF and ANGPT2 were reduced in the group
overexpressing miR-26b. In agreement with the RT-qPCR findings,
miR-26b loss led to upregulation of CTGF and ANGPT2 protein
expression levels (Fig. 2D).
Collectively, the current findings suggested that miR-26b has a
suppressive role in the angiogenesis of HUVECs primarily via
downregulation of CTGF and ANGPT2.
 | Figure 2miR-26b suppresses expression of PVT1,
CTGF and ANGPT2. (A) Reverse transcription-quantitative polymerase
chain reaction analysis of (A) CTGF, (B) ANGPT2 and (C) PVT1 mRNA
expression levels in HUVECs transfected with negative controls,
miR-26b mimics or inhibitors. GAPDH was used as internal control.
Data are presented as the mean ± standard deviation.
*P<0.05, **P<0.01 vs. respective NC
groups. (D) Western blot analysis of CTGF and ANGPT2 protein
expression levels in HUVECs transfected with negative controls,
miR-26b mimic or inhibitor. β-actin was used as internal control.
HUVEC, human umbilical vein endothelial cells; miR, microRNA; NC,
negative control; PVT1, plasmacytoma variant translocation 1; CTGF,
connective tissue growth factor; ANGPT2, angiopoietin 2. |
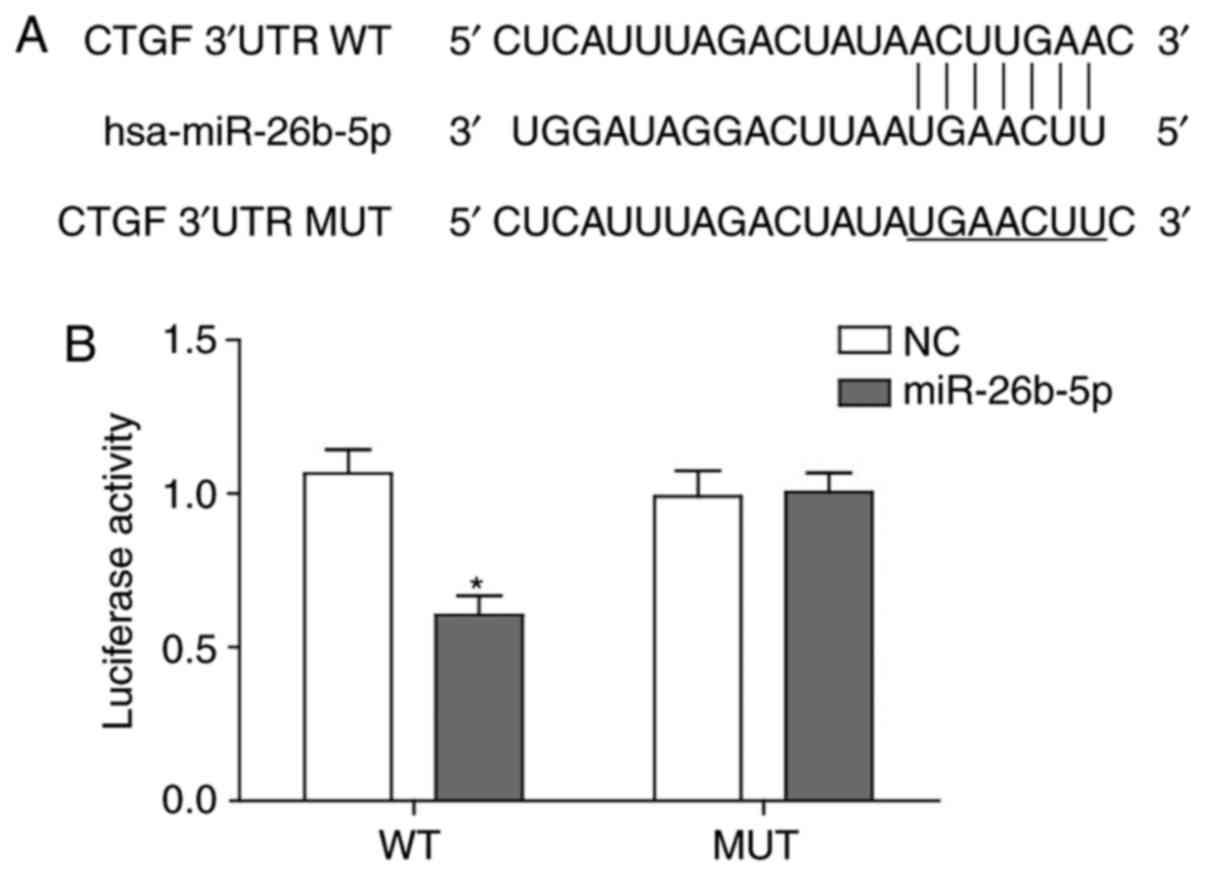
miR-26b directly interacts with 3′-UTR of
CTGF
As microRNAs exert their biological effects through
binding the cognate target sequence of mRNAs to block translation
or lead to degradation of mRNAs, the current study aimed to clarify
whether miR-26b inhibited CTGF expression by directly binding to
CTGF mRNA, which then led to accelerated degradation.
Bioinformatics analysis using www.microrna.org revealed that the 3′-UTR of CTGF may
be targeted by miR-26b with large extent of sequence
complementarity, which was consistent with a previous study
(23) (Fig. 3A). Based on the binding sequence,
a mutant CTCF 3′-UTR-containing luciferase reported vector and
wild-type vector were generated and verified that miR-26b was able
to recognize and bind to the wild-type CTGF 3′-UTR to impair
activity of luciferase and could not bind to the mutant form of
CTGF 3′-UTR (Fig. 3B).
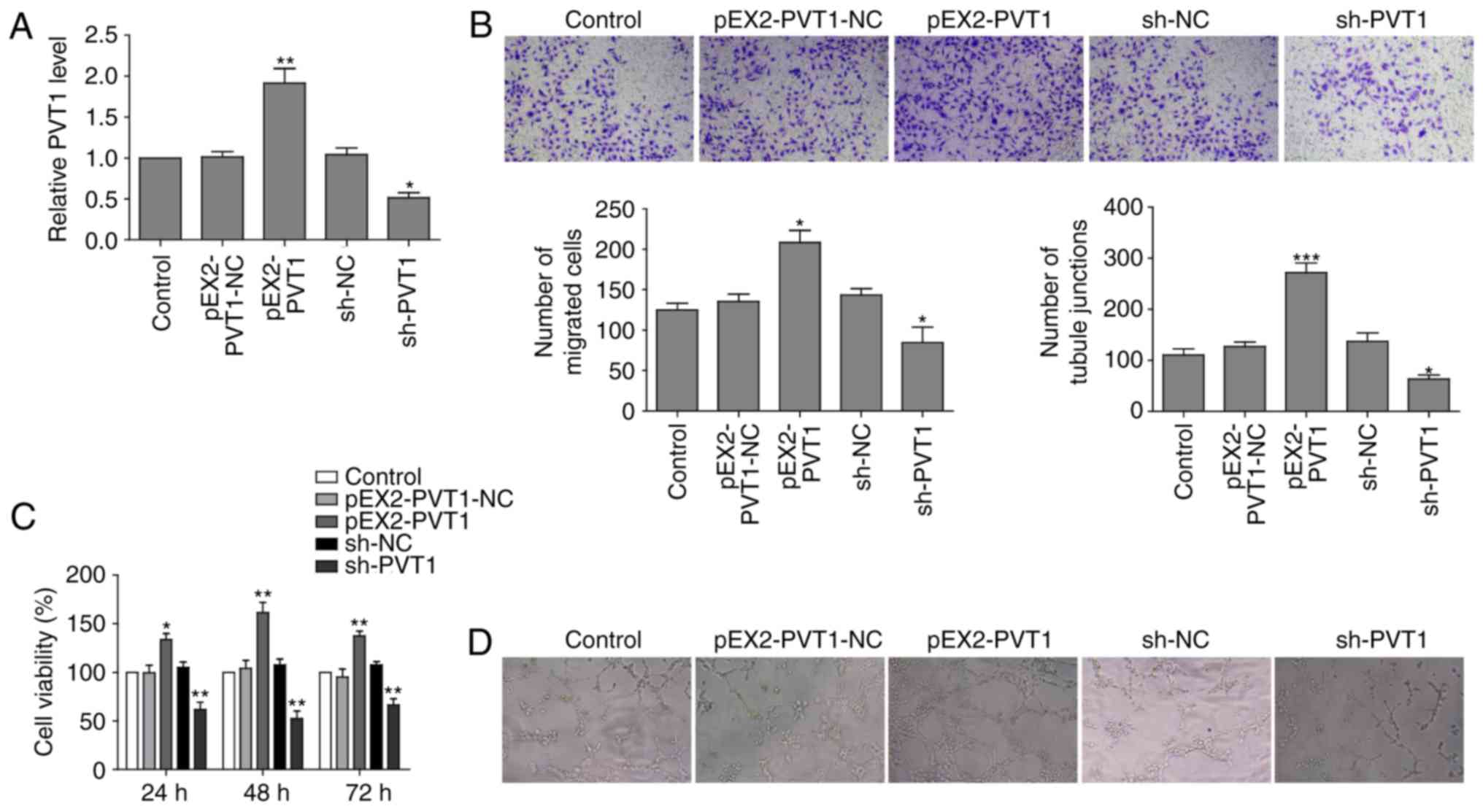
LncRNA PVT1 promotes cell proliferation,
migration and in vitro tube formation of HUVECs cell
In order to investigate the biological function of
lncRNA PVT1 in vascular endothelial cells, the present study
generated PVT1-overexpressing and PVT1-knockdown HUVECs. RT-qPCR
revealed that the relative expression of PVT1 in the PVT1
overexpressing cells (pEX2-PVT1) was higher compared with the
control cells (pEX2-PVT1-NC). The PVT1 knockdown cells (shPVT1)
exhibited substantially reduced PVT1 expression level compared with
the negative control (shNC) group cells (Fig. 4A).
Furthermore, it was also observed that PVT1 had a
stimulatory role in migration capability of HUVECs.
PVT1-overexpressing cells had a higher migration ability compared
with the control, whereas shPVT1 cells had decreased migration
ability (Fig. 4B). It is of note
that PVT1 overexpression markedly promoted cell proliferation of
HUVECs, whereas PVT1 depletion impaired cell growth rate of HUVECs
(Fig. 4C). Additionally, PVT1
overexpression promoted in vitro vascular tube formation as
tube length was increased in pEX2-PVT1 cells compared with shPVT1
cells, which formed fewer tubule junctions (Fig. 4D).
PVT1 suppresses miR-26b and CTGF
expression levels
In order to investigate PVT-promoted HUVECs
angiogenesis, the present study determined that whether PVT1
promoted angiogenesis through the miR-26b/CTGF axis. Therefore, the
present study aimed to determine whether PVT1 affected the
expression of miR-26b and its downstream angiogenesis-associated
targets. RT-qPCR analysis revealed that PVT1 overexpression led to
reduced expression level of miR-26b and increased CTGF and ANGPT2
mRNA expression level. The cells lacking PVT1 following an
infection with a PVT1 shRNA-containing virus exhibited elevated
level of miR-26b and downregulated mRNA levels of CTGF and ANGPT2
(Fig. 5A–C). In addition, similar
changes were observed in terms of the CTGF and ANGPT2 protein
expression levels in PVT1-overexpressed and PVT1-knockdown HUVECs
(Fig. 5D). These findings
suggested that PVT1 was able to bind to and subsequently
downregulate miR-26b expression to promote the expression of CTGF
and ANGPT2, facilitating angiogenesis of vascular endothelial
cells.
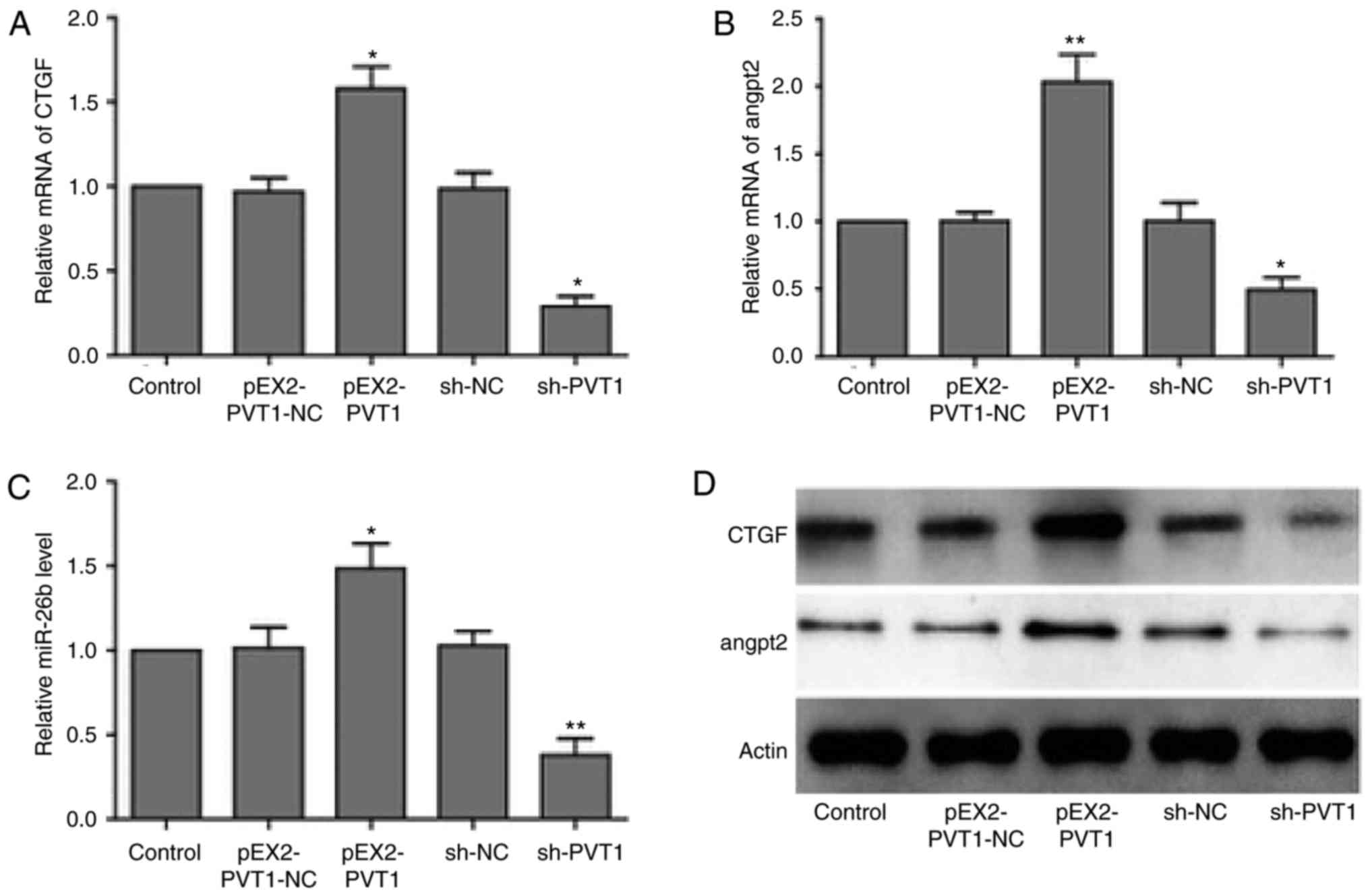 | Figure 5PVT1 suppresses expression of
miR-26b, CTGF and ANGPT2. RT-qPCR analysis of (A) CTGF, (B) ANGPT2
and (C) miR-26b mRNA expression levels in HUVECs transfected with
negative controls, PVT1 or shRNA against PVT1. GAPDH was used as
internal control. *P<0.05, **P<0.01 vs.
respective NC groups. (D) Western blot analysis of CTGF and ANGPT2
protein level in HUVECs cell transfected with negative controls,
PVT1 or shRNA against PVT1. GAPDH was used as internal control.
RT-qPCR, reverse transcription-quantitative polymerase chain
reaction; shRNA, short hairpin RNA; CTGF, connective tissue growth
factor; ANGPT2, angiopoietin 2; PVT1, plasmacytoma variant
translocation 1; miR, microRNA; sh, short hairpin RNA; NC, negative
control. |
PVT1 directly interacts with miR-26b
According to a previous study PVT1 has been
identified to negatively regulate miR-26b expression, a previously
reported regulator of angiogenesis (20). To confirm whether PVT1 regulated
miR-26b through an interaction the present study used
bioinformatics to predict the potential physical interaction
between PVT1 and miR-26b (Fig.
6A). Subsequently, the present study conducted a dual
luciferase reporter assay to confirm that PVT1 was able to bind
directly and degrade miR-26b. It was demonstrated that only
wild-type PVT1 significantly lowered the luciferase activity of
miR-26b and the mutant form of PVT1 did not alter the luciferase
activity (Fig. 6B).
Discussion
The present study demonstrated that PVT1 enhanced
angiogenesis of vascular endothelial cells through binding miR-26b
and subsequently increasing the expression of CTGF and ANGPT2.
Bioinformatics and biochemical analyses demonstrated that PVT1 was
able to directly bind to miR-26b and thus reduce the level of
miR-26b. Subsequently, miR-26b regulation of two
angiogenesis-promoting genes was investigated using RT-qPCR and
western blot analysis. In addition, bioinformatics prediction and
dual reporter luciferase assay revealed that miR-26b directly
targeted the CTGF 3′-UTR. An MTT, Transwell assay and in
vitro tube formation assays revealed that PVT1 promoted cell
proliferation, migration and tube formation of HUVECs,
respectively, whereas miR-26b exerted the opposite effects.
Considered together, PVT1 had a stimulatory role in the
angiogenesis of vascular endothelial cells by reducing the miR-26b
expression level to increase CTGF and ANGPT2 expression.
Previous studies revealed that microRNAs regulate a
variety of physiological and pathological processes, such as
cardiovascular diseases. It has been reported that miR-143/145 are
associated with vascular injury and hypertension and miR-499 is
involved in myocardial ischemia (25–27). Additionally, miR-26a was
identified to target the SMAD1-Id1-p21WAF/CIP1 signaling
axis and inhibit angiogenesis in endothelial cells (18). However, the association between
miR-26b and angiogenesis of vascular endothelial cells remains to
be fully elucidated. To the best of our knowledge, the present
study is the first to determine that miR-26b has an inhibitory role
in angiogenesis of human endothelial cell through targeting CTGF
and regulating its expression, which is consistent with previous
studies that state that miR-26b inhibits tumorigenesis and
metastasis of cancer cells partially by regulating CTGF (28,29).
LncRNAs function to regulate gene expression by an
epigenetic or post-transcriptional mechanism, including mRNA
processing and degradation by interacting with a splicing factor
and the 3′-UTR of the mRNA. Previous studies have reported that
lncRNAs may act as sponges for microRNAs, which were coined as
competing endogenous RNAs (ceRNAs) (30–32). These ceRNAs share sequences
recognized by miRNAs termed microRNA recognition elements (MREs).
Paci et al (33) developed
a computational model to identify putative ceRNAs among lncRNAs in
breast cancer. They determined that PVT1 had binding preference
towards the miR-200 family, regulating the expression of hundreds
of mRNAs. The present study verified that PVT1 was able to bind
miR-26b via a complementary sequence. Therefore, PVT1 was important
in angiogenesis of vascular endothelial cell as ceRNA to bind and
degrade miR-26b, which expanded the current understanding of PVT1
as ceRNA to regulate biological processes.
In conclusion, the present study highlighted the
importance of lncRNA PVT1 in promoting angiogenesis of vascular
endothelial cells, which broadened the knowledge of how PVT1 has an
effect in this process. It is of note that the findings of the
present study provide the rationale for targeting PVT1 to treat
angiogenesis abnormality-associated diseases, including cancer
metastasis.
Acknowledgments
Not applicable.
References
|
1
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Colombo T, Farina L, Macino G and Paci P:
PVT1: A rising star among oncogenic long noncoding RNAs. BioMed Res
Int. 2015:3042082015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang YR, Zang SZ, Zhong CL, Li YX, Zhao SS
and Feng XJ: Increased expression of the lncRNA PVT1 promotes
tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol.
7:6929–6935. 2014.PubMed/NCBI
|
|
4
|
Ding C, Yang Z, Lv Z, Du C, Xiao H, Peng
C, Cheng S, Xie H, Zhou L, Wu J and Zheng S: Long non-coding RNA
PVT1 is associated with tumor progression and predicts recurrence
in hepatocellular carcinoma patients. Oncol Lett. 9:955–963. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ding J, Li D, Gong M, Wang J, Huang X, Wu
T and Wang C: Expression and clinical significance of the long
non-coding RNA PVT1 in human gastric cancer. Onco Targets Ther.
7:1625–1630. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zheng X, Hu H and Li S: High expression of
lncRNA PVT1 promotes invasion by inducing epithelial-to-mesenchymal
transition in esophageal cancer. Oncol Lett. 12:2357–2362. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shen CJ, Cheng YM and Wang CL: LncRNA PVT1
epigenetically silences miR-195 and modulates EMT and
chemoresistance in cervical cancer cells. J Drug Target. 25:1–8.
2017. View Article : Google Scholar
|
|
8
|
Chunharojrith P, Nakayama Y, Jiang X, Kery
RE, Ma J, De La Hoz Ulloa CS, Zhang X, Zhou Y and Klibanski A:
Tumor suppression by MEG3 lncRNA in a human pituitary tumor derived
cell line. Mol Cell Endocrinol. 416:27–35. 2015. View Article : Google Scholar :
|
|
9
|
Gordon FE, Nutt CL, Cheunsuchon P,
Nakayama Y, Provencher KA, Rice KA, Zhou Y, Zhang X and Klibanski
A: Increased expression of angiogenic genes in the brains of mouse
meg3-null embryos. Endocrinology. 151:2443–2452. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fu WM, Lu YF, Hu BG, Liang WC, Zhu X, Yang
HD, Li G and Zhang JF: Long noncoding RNA Hotair mediated
angiogenesis in nasopharyngeal carcinoma by direct and indirect
signaling pathways. Oncotarget. 7:4712–4723. 2016. View Article : Google Scholar :
|
|
11
|
Ma Y, Wang P, Xue Y, Qu C, Zheng J, Liu X,
Ma J and Liu Y: PVT1 affects growth of glioma microvascular
endothelial cells by negatively regulating miR-186. Tumour Biol.
39:1010428317694326. 2017. View Article : Google Scholar
|
|
12
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu J, Wang F, Yang GH, Wang FL, Ma YN, Du
ZW and Zhang JW: Human microRNA clusters: Genomic organization and
expression profile in leukemia cell lines. Biochem Biophys Res
Commun. 349:59–68. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bentwich I, Avniel A, Karov Y, Aharonov R,
Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al:
Identification of hundreds of conserved and nonconserved human
microRNAs. Nature Genet. 37:766–770. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Brennecke J, Hipfner DR, Stark A, Russell
RB and Cohen SM: Bantam encodes a developmentally regulated
microRNA that controls cell proliferation and regulates the
proapoptotic gene hid in Drosophila. Cell. 113:25–36. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Calame K: MicroRNA-155 function in B
Cells. Immunity. 27:825–827. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jopling CL, Yi M, Lancaster AM, Lemon SM
and Sarnow P: Modulation of hepatitis C virus RNA abundance by a
liver-specific MicroRNA. Science. 309:1577–1581. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Icli B, Dorbala P and Feinberg MW: An
emerging role for the miR-26 family in cardiovascular disease.
Trends Cardiovasc Med. 24:241–248. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Icli B, Wara AK, Moslehi J, Sun X, Plovie
E, Cahill M, Marchini JF, Schissler A, Padera RF, Shi J, et al:
MicroRNA-26a regulates pathological and physiological angiogenesis
by targeting BMP/SMAD1 signaling. Circ Res. 113:1231–1241. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu SC, Chuang SM, Hsu CJ, Tsai CH, Wang
SW and Tang CH: CTGF increases vascular endothelial growth
factor-dependent angiogenesis in human synovial fibroblasts by
increasing miR-210 expression. Cell Death Dis. 5:e14852014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Babic AM, Chen CC and Lau LF: Fisp12/mouse
connective tissue growth factor mediates endothelial cell adhesion
and migration through integrin alphavbeta3, promotes endothelial
cell survival, and induces angiogenesis in vivo. Mol Cell Biol.
19:2958–2966. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang BJ, Ding HW and Ma GA: Long noncoding
RNA PVT1 promotes melanoma progression via endogenous sponging
miR-26b. Oncol Res. 2017. View Article : Google Scholar
|
|
23
|
Wang R, Ding X, Zhou S, Li M, Sun L, Xu X
and Fei G: Microrna-26b attenuates monocrotaline-induced pulmonary
vascular remodeling via targeting connective tissue growth factor
(CTGF) and cyclin D1 (CCND1). Oncotarget. 7:72746–72757. 2016.
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Kuo CH, Chen PK, Chang BI, Sung MC, Shi
CS, Lee JS, Chang CF, Shi GY and Wu HL: The recombinant lectin-like
domain of thrombomodulin inhibits angiogenesis through interaction
with Lewis Y antigen. Blood. 119:1302–1313. 2012. View Article : Google Scholar
|
|
26
|
Boettger T, Beetz N, Kostin S, Schneider
J, Kruger M, Hein L and Braun T: Acquisition of the contractile
phenotype by murine arterial smooth muscle cells depends on the
Mir143/145 gene cluster. J Clin Invest. 119:2634–2647. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xin M, Small EM, Sutherland LB, Qi X,
McAnally J, Plato CF, Richardson JA, Bassel-Duby R and Olson EN:
MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and
responsiveness of smooth muscle cells to injury. Genes Dev.
23:2166–2178. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dorn GW 2nd, Matkovich SJ, Eschenbacher WH
and Zhang Y: A human 3′ miR-499 mutation alters cardiac mRNA
targeting and function. Circ Res. 110:958–967. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Duan G, Ren C, Zhang Y and Feng S:
MicroRNA-26b inhibits metastasis of osteosarcoma via targeting CTGF
and Smad1. Tumour Biol. 36:6201–6209. 2015. View Article : Google Scholar
|
|
30
|
Guttman M, Amit I, Ga rber M, French C,
Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huarte M, Guttman M, Feldser D, Garber M,
Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M,
et al: A large intergenic noncoding RNA induced by p53 mediates
global gene repression in the p53 response. Cell. 142:409–419.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Paci P, Colombo T and Farina L:
Computational analysis identifies a sponge interaction network
between long non-coding RNAs and messenger RNAs in human breast
cancer. BMC Syst Biol. 8:832014. View Article : Google Scholar : PubMed/NCBI
|