Introduction
The severe acute respiratory syndrome coronavirus-2
(SARS-CoV-2) was first isolated at the end of 2019 in China
(1-5) and, as of August 3rd, 2020, almost 18
million infected patients and 686,703 deaths have been reported
globally (WHO Situation Report-196). However, the actual number of
the infected subject is under-estimated and, indeed, a recent
meta-analysis performed on 50,155 patients from 41 studies, showed
that the pooled percentage of asymptomatic infection is 15.6%
(6).
Even if SARS-CoV-2 shares similarities with the
other coronaviruses, the higher diffusion rate and the possibility
to induce fatal complications, such as severe pneumonia, acute
respiratory distress syndrome (ARDS), thrombosis, septic shock and
organ failure, make this virus a major public health threat
(7-10). Development of COVID-19
complications seems to be dependent on a dramatic release of
proinflammatory factors, such as interleukin (IL)-1β, IL-6, IL-8,
tumor necrosis factor-α (TNF-α) and CXC-chemokine ligand 10
(CXCL10) and CC-chemokine ligand 2 (CCL2) in the infected lung
tissue and other peripheral organs (2,11-13), which ultimately leads to a
reaction known as cytokine release syndrome (CRS). It is likely
that CRS promotes a self-sustaining inflammatory process that
contributes to the respiratory failure and the systemic
manifestations observed in COVID-19 patients (14). A multicenter study of 150
confirmed COVID-19 cases in Wuhan, China, identified as predictors
of mortality both elevated ferritin (15,16) and IL-6 levels, which strengthen
the hypothesis that fatality events may be due to a virus-driven
hyperinflammation (2,11).
The rapid worldwide diffusion of SARS-COV-2 has
propelled both basic science and clinical research studies for the
elucidation of the pathogenetic mechanisms underlying COVID-19. The
emerging observation that a significant percentage proportions of
individuals are asymptomatic, not only suggests that SARS-CoV-2 may
have a longer incubation period and higher transmission rate, as
compared to other coronaviruses, but also advocates potential
differences in the host immune responses to this virus. It is
therefore, of the utmost importance to characterize the immune
responses put against SARS-CoV-2 and the mechanisms of
hyperinflammation, in order to design better therapeutic strategies
for COVID-19. In the present study, we performed a transcriptomic
analysis to profile the immune signatures in lung and the
bronchoalveolar lavage fluid samples from COVID-19 patients and
controls. Our data concordantly revealed increased humoral
responses to infection. The elucidation of the host responses to
SARS-CoV-2 infection may further improve our understanding of
COVID-19 pathogenesis and suggest better therapeutic
strategies.
Materials and methods
Dataset selection
The NCBI Gene Expression Omnibus (GEO) database
(http://www.ncbi.nlm.nih.gov/geo/) was
interrogated using the terms 'SARS-CoV-2' and 'COVID-19'. The
available datasets were shortlisted if: i) they included
whole-genome transcriptomic profiling; ii) included human samples;
and iii) were not generated on cancer cell lines. Finally, the
GSE150316 and the GSE147507 (17)
datasets were selected. GSE150316 is a high throughput sequencing
dataset of five autopsy samples from patients deceased due to
SARS-CoV-2 infection (2-5 technical replicates for each sample were
averaged for the downstream analysis) and five negative control
samples.
The GSE147507 dataset was generated from three
biological replicates of primary human lung epithelium either
infected with SARS-CoV-2 (USA-WA1/2020) at a multiplicity of
infection (MOI) of 2, for 24 h, or mock infected. Total mRNA
libraries were prepared using tTruSeq Stranded mRNA LP and cDNA
libraries sequenced using an Illumina NextSeq 500 platform. Raw
reads were aligned to the human genome (hg19) using the RNA-Seq
Alignment App on Basespace (Illumina). The submitter-supplied
pre-processed and normalized gene expression matrix was used for
the analysis.
For the transcriptomic analysis of COVID-19 BALF
samples, RNA-Seq data from the Genome Sequence Archive of the
Beijing Institute of Genomics (BIG) Data Center (https://bigd.big.ac.cn/) (accession no. CRA002390),
and from the NCBI SRA database (accession nos. SRR10571724,
SRR10571730 and SRR10571732) (18) were used.
Enrichment and network analysis
Functional enrichment analysis was conducted using
the web-based utility, Metascape (19). Metascape analysis makes use of
public databanks, such as Gene Ontology, KEGG, and MSigDB, and
aggregates enriched ontology terms into non-redundant groups, by
calculating the similarity between any two terms (19). Metascape uses the hypergeometric
test and the Benjamini-Hochberg p-value correction to identify
statistically significant enriched terms. Representative terms from
the enrichment analysis are presented as a network. Each term is
represented by a node, with its size being proportional to the
number of input genes belonging to that term, and the color
representing its corresponding cluster. Terms with a similarity
score >0.3 are linked by an edge. The thicker the edge, the
higher the similarity score. The network is visualized using
Cytoscape (version 3.1.2) with 'force-directed' layout. One term
from each cluster has its description shown as a label.
Computational deconvolution of
infiltrating immune cells
In order to evaluate the relative proportions of
immune cell subsets in COVID-19 and healthy control samples, we
performed a computational deconvolution analysis. To this end, we
used the xCell software, a web computational utility that aims at
evaluating, by using gene signatures, the relative proportions in a
sample of various immune cell types, including immature dendritic
cells (iDCs), conventional DCs (cDCs), active DCs (aDCs),
plasmacytoid DCs (pDCs), B cells, CD4+ T cells, memory
cells, Th1 cells, Th2 and Treg cells and macrophages (20).
Statistical analysis
The differential expression analysis was performed
using the DeSeq2 function. The web-based application NeworkAnalist
was used for the statistical analyses. Genes with an adjusted
P-value <0.05 were identified as differentially expressed genes
(DEGs) and selected for further analysis.
Linear regression and Spearman's correlation were
performed to compare the expression levels of genes in COVID-19
samples as compared to healthy control samples, the GSE150316 and
the GSE147507 datasets.
For the analysis of the deconvolution data,
normality was first assessed using the Shapiro-Wilk,
D'Agostino-Pearson and Kolmogorov-Smirnov tests. Based on the
results, differential analysis as performed using the
non-parametric Mann-Whitney U test.
The GraphPad Prism (version 8) software (GraphPad
Software, Inc.) and the SPSS software (SPSS, Inc.) were used for
the statistical analysis and the generation of the graphs. Unless
otherwise stated, P<0.05 was considered to indicate a
statistically significant difference.
Results
Network and enrichment analysis of
SARS-CoV-2 infection
In order to determine the transcriptomic signature
of lung tissues from COVID-19 patients, we analyzed the GSE150316
RNA-Seq dataset. A total of 55 differentially expressed genes was
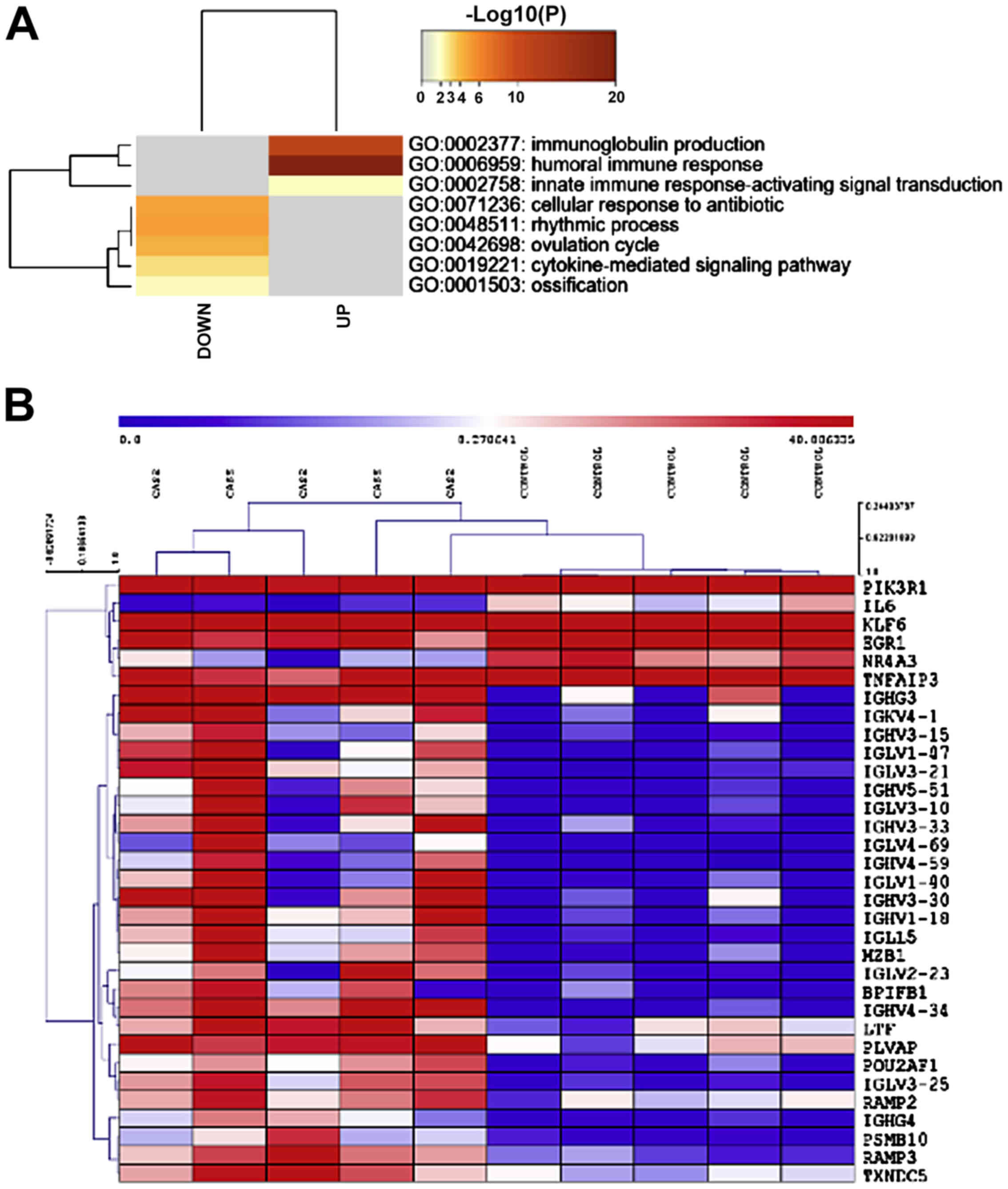
found, of which 32 were upregulated and 23 downregulated. Gene term
enrichment analysis identified GO:0002377: immunoglobulin
production, GO:0006959: humoral immune response and GO:0002758:
innate immune response-activating signal transduction, as
significantly enriched among the upregulated genes (Fig. 1A). A heatmap of the genes
belonging to the GO:0002377 (immunoglobulin production) category is
presented in Fig. 1B. Among the
downregulated genes, GO:0071236: cellular response to antibiotic,
GO:0048511: rhythmic process, GO:0042698: ovulation cycle,
GO:0019221: cytokine-mediated signaling pathway and GO:0001503:
ossification, were found to be significantly enriched (Fig. 1A).
We have previously interrogated the GSE147507
dataset, which included transcriptomic data from primary human
bronchial epithelial cells infected in vitro with the
SARS-CoV-2 virus (18). Here, we
compared this gene signature to the transcriptomic signature of
lung biopsies from COVID-19 patients. The publicly available
GSE150316 dataset was used in order to perform a correlation
analysis on the modulation of the genes perturbed upon SARS-CoV-2
infection and the corresponding genes in GSE147507. A total of 9602
genes were in common between the two datasets. As shown in Fig. 1A, a moderate but significant
correlation is found in the transcriptomic profile of in
vitro infected bronchial epithelial cells and lung samples from
patients (Fig. 2A).
Gene term enrichment analysis for the significantly
modulated genes identified pathways in common between the GSE147507
and the GSE150316 datasets. The common enriched terms for the
upregulated genes in the two datasets were: 'humoral immune
response' (GO:0006959) and 'leukocyte migration' (GO:0050900)
(Fig. 2B).
Interestingly, the top terms enriched among the
downregulated genes in the GSE150316 dataset were: 'TNF signaling
pathway' (hsa04668), 'cytokine-mediated signaling pathway'
(GO:0019221), 'myeloid leukocyte activation' (GO:0002274) and
'regulation of cytokine production' (GO:0001817) (Fig. 2B). Representative terms from the
enrichment analysis and their functional connections are presented
as a network (Fig. 2C).
Deconvolution analysis of infiltrating
immune cells in lung samples from COVID-19 patients
We next characterized the relative proportions of
infiltrating immune cells in the lungs of COVID-19 patients. A
shown in Fig. 3A, a moderate, but
not significant, increase in the immune score and microenviroment
score was detected for the COVID-19 lung samples. Also, a moderate,
non-significant increase in the percentage of infiltrating
basophils and aDCs was observed (Fig.
3B). Analysis of the lymphoid cells in the lungs of COVID-19
patients revealed a significant higher proportion of infiltrating B
cells upon SARS-CoV-2 infection, along with a moderate,
non-significant increase in NKT and Th1 cells (Fig. 3C).
Characterization of the transcriptomic
profile of BALF samples
Next, we compared the gene signature of BALF samples
from COVID-19 patients and controls. A total of 3003 genes were
found to be modulated in SARS-CoV-2 patients (adjusted P-value
<0.05 and ǀfold-changeǀ >2), with 1745 genes being
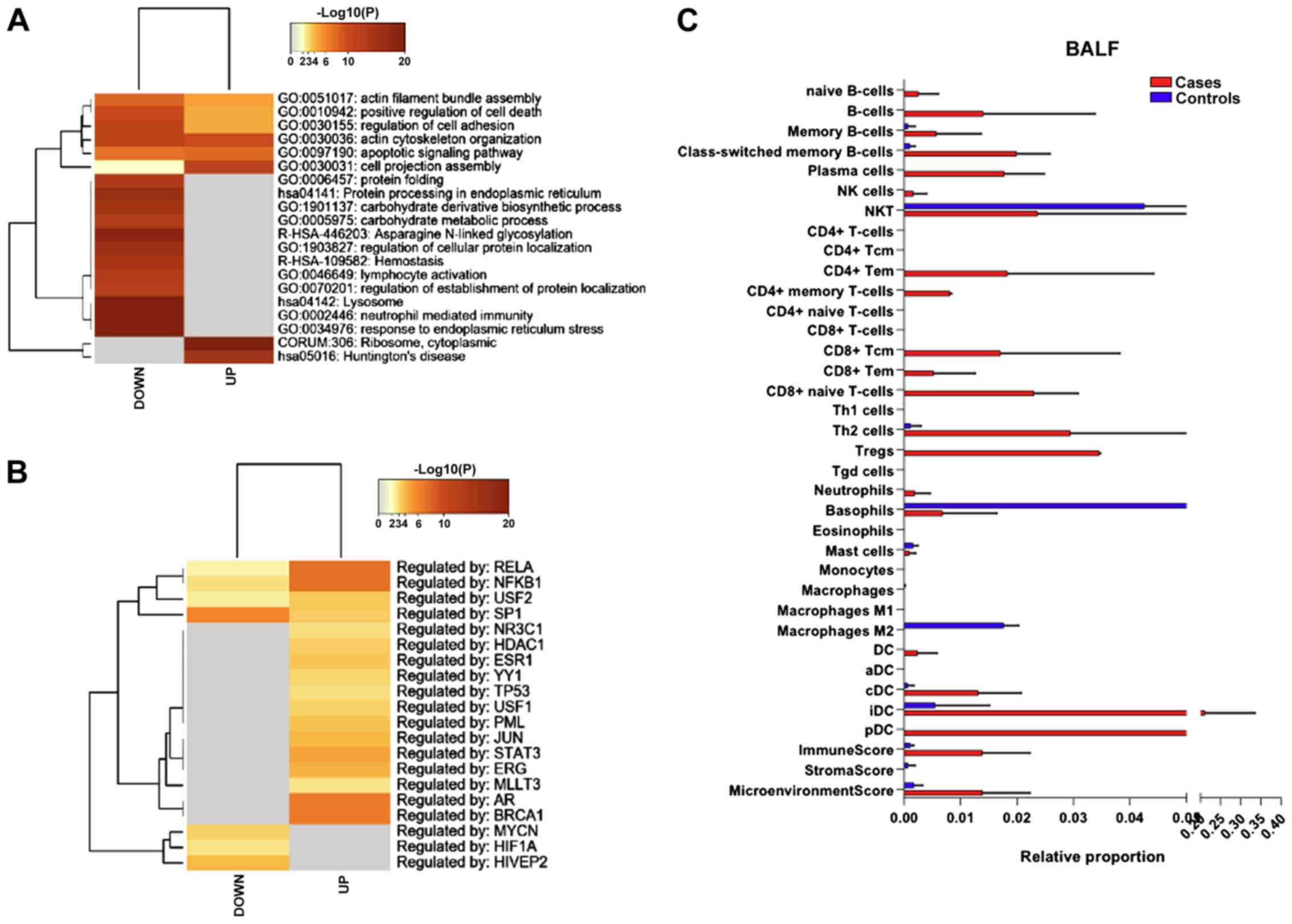
upregulated and 1258 genes downregulated. As shown in Fig. 4A, among both the upregulated and
downregulated genes, pathways related to cell morphology
(GO:0051017; GO:0030155; GO:0030036; GO:0030031) and survival
(GO:0010942; GO:0097190) were significantly enriched.
Analysis of the transcription factors identified
RELA, NFKB1, USF2 and SP1, as putatively involved in the regulation
of the differentially expressed genes (Fig. 4B).
Immune cell deconvolution analysis revealed a trend
of higher proportion in B cells (both naïve, memory and plasma
cells), along with an increase in CD4 memory T cells, CD8 T cells
and DCs (cDCs, iDCs and pDCs) (Fig.
4C).
Discussion
The characterization of the exact pathogenetic
mechanisms by which SARS-CoV-2 induces multiple organ damage are of
immediate importance. Emerging data seem to indicate that beside
lungs, other organs, including heart, kidney and the central
nervous system may also be affected in COVID-19 (21,22). Patients may show proteinuria,
hematuria and increased creatinine levels (21), and may suffer from neurological
symptoms, such as headache, epilepsy, disturbed consciousness,
anosmia and dysgeusia (22). Some
COVID-19 patients also develop thromboembolic events, with
elevation of D-dimer and other procoagulant parameters (23), which may represent a secondary
anti-phospholipid syndrome (APS) (24), as well as other autoimmune
diseases (25). Indeed,
accumulating case reports show that COVID-19 patients tested
positive for anti-CL, anti-b2-GPI autoantibodies (26-29), as well as lupus anticoagulant
(27,30,31).
The use of gene expression profiling data has been
extensively employed for the identification of novel pathogenic
pathways and therapeutic targets (32-36) for several disorders including,
autoimmune diseases (37-40) and cancer (41,42). A computational analysis was
performed in order to characterize the immune response to
SARS-CoV-2 infection. To achieve this, we exploited publicly
available RNA-seq data, generated from lung biopsies and BALF
samples from COVID-19 patients. Our data from lung and BALF samples
concordantly show that B cell responses mainly characterize
SARS-CoV-2 infection.
It has been already described that SARS-CoV-2
elicits a robust humoral cell response, with virus-specific IgM,
IgG and IgA, and neutralizing IgG antibodies following infection.
Seroconversion usually occurs in most COVID-19 patients between one
to two weeks after overt symptoms, and antibody titers last for
weeks, following virus eradication (43). It seems also that protective B
cell memory arises following infection, as a recent study of
SARS-CoV-2 infection in rhesus macaques found that animals that had
resolved the primary infection were resistant to reinfection one
month later (44). Also,
independent data show that higher virus-specific antibody titers
correlated with greater virus neutralization and are inversely
correlated with viral load (43).
However, higher titers may be associated with more severe clinical
cases (45-47), suggesting that the humoral
responses may not be sufficient to protect from severe disease. Up
to now, there is no evidence that SARS-CoV-2-induced anti-bodies
contribute to some of the pathological features observed in
COVID-19 patients. However, this possibility should be taken into
consideration in light of the above-mentioned data on secondary APS
syndrome in some COVID-19 cases. It has been proposed that
antibody-dependent enhancement (ADE), may represent, at least one,
of the causes of the CRS (48-51). When the virus infects the body,
memory B cells are activated while the activation of naive B cells
is inhibited. However, both virus-specific antibodies and
antibodies cross-reacting with other similar virus strains are
produced and secreted. These cross-reactive antibodies may elicit
the entry of viruses into macrophages in a Fc receptor-mediated
manner, and consequently, viruses undergo rapid replication and
release, resulting in immune dysregulation, and severe illness in
patients with COVID-19 (52). The
potential role of SARS-CoV-2-induced IgG antibodies in promoting
neuroinflammation in SARS-CoV-2 infection should also be mentioned,
as ADE occurrence may involve microglia cells following the binding
of Abs to Fc receptors expressed on these cells.
As the mTOR pathway plays a fundamental role in
B-cell development via the control of BCL6 expression in B cells
from the germinal center (53),
it is reasonable to believe that the use of inhibitors of mTOR,
i.e., rapamycin and 'rapalogs', could reduce the populations of
antigen-specific memory B cells and limit the occurrence of ADE in
SARS-CoV-2 infected patients. This further strengthen the rational
for using mTOR inhibitors in COVID-19, as previously discussed
(20). Indeed, by using an
anti-signature computational approach, our analysis showed that the
mTOR inhibitor, sirolimus, may be a candidate drug to be used in
COVID-19 patients, which is in line with data on the activation of
the phosphoinositol 3-kinase (PI3K)/AKT/mTOR pathway in response to
the infection with another coronavirus, MERS-CoV (54). Also, mTOR has been recognized as a
key factor in regulating the replication of viruses (36,54-57), and in patients with H1N1
pneumonia, early treatment with corticosteroids in combination with
rapamycin has been associated with improvement in multiple organ
dysfunction, virus clearance, and shortened time in ventilators
(58).
Hence, the use of mTOR inhibitors may have many-fold
advantages on the course od SARS-CoV-2 infection, which could
improve lung pathology, but also, the peripheral manifestations of
the disease, including the CNS.
Interestingly, our data suggest potential reasons
for the gender differences in COVID-19 susceptibility (2). Indeed, the prevalence in men is
between 55 and 68% (59) and
increased clinical severity and mortality has been reported
(60). Certainly, female-specific
hormonal factors can be involved. In this regard, it is notable
that among the upregulated genes in BALF from COVID-19 patients, 22
(Log(q) value=-4) are regulated by AR (androgen receptor), while 15
are regulated by ESR1 (estrogen receptor 1) (Log(q) value=-1.8). AR
is known to play a key role in both innate and adaptive immune
responses (61,62), and ESR1 has been recognized as a
regulator of interferon production and anti-viral responses
(63). These observations may
underly the different clinical response to SAR-CoV-2 infection in
women and men. It is important to note that selective estrogen
receptor modulators. such as toremifene, have already been proposed
as potential drugs to treat coronavirus infections (64). These observations point to
biological processes that may explain the lower female incidence
and lethality of SARS-CoV-2 infection, offering candidate
therapeutic options in patients suffering from COVID-19.
Finally, we have to acknowledge some of the
limitations of the present study. First, the differentially
expressed genes, that we have prioritized in our study, and the
deconvolution analysis have been obtained from a really small
cohort of patients, hence the data may be biased, due to the high
degree of interindividual variability that characterize SARS-CoV-2
infection. Lung-specific gene expression profiles from homo-geneous
COVID-19 patients will allow to better identify prognostic
predictors and tailored therapeutic strategies. Second, the
deconvolution analysis of the immune populations does not allow to
assess the functionality of the immune cells and their actual
involvement in COVID-19 pathology.
Funding
This study was supported by current research funds
2020 of IRCCS Centro Neurolesi 'Bonino-Pulejo', Messina, Italy.
Availability of data and materials
The datasets analyzed during the current study are
available on the NCBI Gene Expression Omnibus (GEO) database
(http://www.ncbi.nlm.nih.gov/geo/) under
the accession nos. GSE150316 and GSE147507, on the Genome Sequence
Archive of the Beijing Institute of Genomics (BIG) Data Center
(https://bigd.big.ac.cn/) (accession no.
CRA002390) and on the NCBI SRA database (accession nos.
SRR10571724, SRR10571730 and SRR10571732).
Authors' contributions
Conceptualization: DAS, YS, FN and PF; data
curation: EC, MCP and PF; formal analysis: EC, MCP, MSB and AB;
funding acquisition: PB; methodology: PF; writing original draft:
EC, AB; review and editing: PB, DAS, YS, FN and PF. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
DAS is the Editor-in-Chief for the journal, but had
no personal involvement in the reviewing process, or any influence
in terms of adjudicating on the final decision, for this article.
The other authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Zhu N, Zhang D, Wang W, Li X, Yang B, Song
J, Zhao X, Huang B, Shi W, Lu R, et al: China Novel Coronavirus
Investigating and Research Team: A Novel Coronavirus from Patients
with Pneumonia in China, 2019. N Engl J Med. 382:727–733. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xu J, Zhao S, Teng T, Abdalla AE, Zhu W,
Xie L, Wang Y and Guo X: Systematic comparison of two
animal-to-human trans-mitted human Coronaviruses: SARS-CoV-2 and
SARS-CoV. Viruses. 12:122020. View Article : Google Scholar
|
|
3
|
Cucinotta D and Vanelli M: WHO Declares
COVID-19 a Pandemic. Acta Biomed. 91:157–160. 2020.PubMed/NCBI
|
|
4
|
Chen L, Jin Q, Zhou Y, Yang J, Wang Z, Ge
K, Yang J and Wang H: Clinical characteristics of 2019 novel
coronavirus pneumonia in Zhejiang province, China. Mol Med Rep.
22:2583–2587. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kanduc D and Shoenfeld Y: On the molecular
determinants of the SARS-CoV-2 attack. Clin Immunol.
215:1084262020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
He J, Guo Y, Mao R and Zhang J: Proportion
of asymptomatic coronavirus disease 2019 (COVID-19): A systematic
review and meta-analysis. J Med Virol. jmv263262020. View Article : Google Scholar
|
|
7
|
Chen N, Zhou M, Dong X, Qu J, Gong F, Han
Y, Qiu Y, Wang J, Liu Y, Wei Y, et al: Epidemiological and clinical
characteristics of 99 cases of 2019 novel coronavirus pneumonia in
Wuhan, China: A descriptive study. Lancet. 395:507–513. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Docea AO, Tsatsakis A, Albulescu D,
Cristea O, Zlatian O, Vinceti M, Moschos SA, Tsoukalas D, Goumenou
M, Drakoulis N, et al: A new threat from an old enemy: Re-emergence
of coronavirus (Review). Int J Mol Med. 45:1631–1643.
2020.PubMed/NCBI
|
|
9
|
Kostoff RN, Briggs MB, Porter AL, Aschner
M, Spandidos DA and Tsatsakis A: [Editorial] COVID-19:
Post-lockdown guidelines. Int J Mol Med. 46:463–466. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shoenfeld Y: Corona (COVID-19) time
musings: Our involvement in COVID-19 pathogenesis, diagnosis,
treatment and vaccine planning. Autoimmun Rev. 19:1025382020.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mehta P, McAuley DF, Brown M, Sanchez E
and Tattersall RS: COVID-19: Consider cytokine storm syndromes and
immuno-suppression. Lancet. 395:1033–1034. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Stancioiu F, Papadakis GZ, Kteniadakis S,
Izotov BN, Coleman MD, Spandidos DA and Tsatsakis A: A dissection
of SARS-CoV-2 with clinical implications (Review). Int J Mol Med.
46:489–508. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kerslake R, Hall M, Randeva HS, Spandidos
DA, Chatha K, Kyrou I and Karteris E: Co-expression of peripheral
olfactory receptors with SARS-CoV-2 infection mediators: Potential
implications beyond loss of smell as a COVID-19 symptom. Int J Mol
Med. 46:949–956. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tang N, Li D, Wang X and Sun Z: Abnormal
coagulation parameters are associated with poor prognosis in
patients with novel coronavirus pneumonia. J Thromb Haemost.
18:844–847. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ruscitti P, Berardicurti O, Di Benedetto
P, Cipriani P, Iagnocco A, Shoenfeld Y and Giacomelli R: Severe
COVID-19, Another Piece in the Puzzle of the Hyperferritinemic
Syndrome. An Immunomodulatory Perspective to Alleviate the Storm.
Front Immunol. 11:11302020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Perricone C, Bartoloni E, Bursi R, Cafaro
G, Guidelli GM, Shoenfeld Y and Gerli R: COVID-19 as part of the
hyperferritinemic syndromes: The role of iron depletion therapy.
Immunol Res. 68:213–224. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Blanco-Melo D, Nilsson-Payant BE, Liu WC,
Møller R, Panis M, Sachs D and Albrecht RA: tenOever BR:SARS-CoV-2
launches a unique transcriptional signature from in vitro, ex vivo,
and in vivo systems. bioRxiv: https://doi.org/10.1101/2020.03.24.004655.
|
|
18
|
Fagone P, Ciurleo R, Lombardo SD,
Iacobello C, Palermo CI, Shoenfeld Y, Bendtzen K, Bramanti P and
Nicoletti F: Transcriptional landscape of SARS-CoV-2 infection
dismantles pathogenic pathways activated by the virus, proposes
unique sex-specific differences and predicts tailored therapeutic
strategies. Autoimmun Rev. 19:1025712020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou Y, Zhou B, Pache L, Chang M,
Khodabakhshi AH, Tanaseichuk O, Benner C and Chanda SK: Metascape
provides a biologist-oriented resource for the analysis of
systems-level datasets. Nat Commun. 10:15232019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Aran D, Hu Z and Butte AJ: xCell:
Digitally portraying the tissue cellular heterogeneity landscape.
Genome Biol. 18:2202017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cheng Y, Luo R, Wang K, Zhang M, Wang Z,
Dong L, Li J, Yao Y, Ge S and Xu G: Kidney disease is associated
with in-hospital death of patients with COVID-19. Kidney Int.
97:829–838. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu Y, Xu X, Chen Z, Duan J, Hashimoto K,
Yang L, Liu C and Yang C: Nervous system involvement after
infection with COVID-19 and other coronaviruses. Brain Behav Immun.
87:18–22. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Han H, Yang L, Liu R, Liu F, Wu KL, Li J,
Liu XH and Zhu CL: Prominent changes in blood coagulation of
patients with SARS-CoV-2 infection. Clin Chem Lab Med.
58:1116–1120. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cavalli E, Bramanti A, Ciurleo R,
Tchorbanov AI, Giordano A, Fagone P, Belizna C, Bramanti P,
Shoenfeld Y and Nicoletti F: Entangling COVID-19 associated
thrombosis into a secondary antiphospholipid antibody syndrome:
Diagnostic and therapeutic perspectives (Review). Int J Mol Med.
46:903–912. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ehrenfeld M, Tincani A, Andreoli L,
Cattalini M, Greenbaum A, Kanduc D, Alijotas-Reig J, Zinserling V,
Semenova N, Amital H, et al: Covid-19 and autoimmunity. Autoimmun
Rev. 19:1025972020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang Y, Xiao M, Zhang S, Xia P, Cao W,
Jiang W, Chen H, Ding X, Zhao H, Zhang H, et al: Coagulopathy and
antiphos-pholipid antibodies in patients with Covid-19. N Engl J
Med. 382:e382020. View Article : Google Scholar
|
|
27
|
Beyrouti R, Adams ME, Benjamin L, Cohen H,
Farmer SF, Goh YY, Humphries F, Jäger HR, Losseff NA, Perry RJ, et
al: Characteristics of ischaemic stroke associated with COVID-19. J
Neurol Neurosurg Psychiatry. 91:889–891. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Harzallah I, Debliquis A and Drenou B:
Lupus anticoagulant is frequent in patients with Covid-19. J Thromb
Haemost. 18:2064–2065. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zayet S, Klopfenstein T, Kovacs R,
Stancescu S and Hagenkötter B: Acute cerebral stroke with multiple
infarctions and COVID-19, France, 2020. Emerg Infect Dis.
26:262020. View Article : Google Scholar
|
|
30
|
Helms J, Tacquard C, Severac F,
Leonard-Lorant I, Ohana M, Delabranche X, Merdji H, Clere-Jehl R,
Schenck M, Fagot Gandet F, et al: CRICS TRIGGERSEP Group (Clinical
Research in Intensive Care and Sepsis Trial Group for Global
Evaluation and Research in Sepsis): High risk of thrombosis in
patients with severe SARS-CoV-2 infection: A multicenter
prospective cohort study. Intensive Care Med. 46:1089–1098. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sieiro Santos C, Nogal Arias C, Moriano
Morales C, Ballesteros Pomar M, Diez Alvarez E and Perez Sandoval
T: Antiphospholipid antibodies in patient with acute lower member
ischemia and pulmonary thromboembolism as a result of infection by
SARS-CoV2. Clin Rheumatol. 39:2105–2106. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fagone P, Mangano K, Quattrocchi C,
Motterlini R, Di Marco R, Magro G, Penacho N, Romao CC and
Nicoletti F: Prevention of clinical and histological signs of
proteolipid protein (PLP)-induced experimental allergic
encephalomyelitis (EAE) in mice by the water-soluble carbon
monoxide-releasing molecule (CORM)-A1. Clin Exp Immunol.
163:368–374. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fagone P, Mangano K, Coco M, Perciavalle
V, Garotta G, Romao CC and Nicoletti F: Therapeutic potential of
carbon monoxide in multiple sclerosis. Clin Exp Immunol.
167:179–187. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cavalli E, Mazzon E, Basile MS, Mangano K,
Di Marco R, Bramanti P, Nicoletti F, Fagone P and Petralia MC:
Upregulated expression of macrophage migration inhibitory factor,
its analogue D-Dopachrome Tautomerase, and the CD44 receptor in
peripheral CD4 T cells from clinically isolated syndrome patients
with rapid conversion to clinical defined multiple sclerosis.
Medicina (Kaunas). 55:6672019. View Article : Google Scholar
|
|
35
|
Rothweiler F, Michaelis M, Brauer P, Otte
J, Weber K, Fehse B, Doerr HW, Wiese M, Kreuter J, Al-Abed Y, et
al: Anticancer effects of the nitric oxide-modified saquinavir
derivative saquinavir-NO against multidrug-resistant cancer cells.
Neoplasia. 12:1023–1030. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nicoletti F, Fagone P, Meroni P, McCubrey
J and Bendtzen K: mTOR as a multifunctional therapeutic target in
HIV infection. Drug Discov Today. 16:715–721. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lombardo SD, Mazzon E, Basile MS, Cavalli
E, Bramanti P, Nania R, Fagone P, Nicoletti F and Petralia MC:
Upregulation of IL-1 receptor antagonist in a mouse model of
migraine. Brain Sci. 9:1722019. View Article : Google Scholar :
|
|
38
|
Petralia MC, Mazzon E, Fagone P, Falzone
L, Bramanti P, Nicoletti F and Basile MS: Retrospective follow-up
analysis of the transcriptomic patterns of cytokines, cytokine
receptors and chemokines at preconception and during pregnancy, in
women with post-partum depression. Exp Ther Med. 18:2055–2062.
2019.PubMed/NCBI
|
|
39
|
Lombardo SD, Mazzon E, Mangano K, Basile
MS, Cavalli E, Mammana S, Fagone P, Nicoletti F and Petralia MC:
Transcriptomic analysis reveals involvement of the macrophage
migration inhibitory factor gene network in Duchenne Muscular
Dystrophy. Genes (Basel). 10. pp. 9392019, View Article : Google Scholar
|
|
40
|
Kosiewicz MM, Auci DL, Fagone P, Mangano
K, Caponnetto S, Tucker CF, Azeem N, White SK, Frincke JM, Reading
CL, et al: HE3286, an orally bioavailable synthetic analogue of an
active DHEA metabolite suppresses spontaneous autoimmune diabetes
in the non-obese diabetic (NOD) mouse. Eur J Pharmacol.
658:257–262. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lombardo SD, Presti M, Mangano K, Petralia
MC, Basile MS, Libra M, Candido S, Fagone P, Mazzon E, Nicoletti F,
et al: Prediction of PD-L1 expression in neuroblastoma via
computational modeling. Brain Sci. 9:2212019. View Article : Google Scholar :
|
|
42
|
Basile MS, Mazzon E, Russo A, Mammana S,
Longo A, Bonfiglio V, Fallico M, Caltabiano R, Fagone P, Nicoletti
F, et al: Differential modulation and prognostic values of
immuneescape genes in uveal melanoma. PLoS One. 14:e02102762019.
View Article : Google Scholar
|
|
43
|
Vabret N, Britton GJ, Gruber C, Hegde S,
Kim J, Kuksin M, Levantovsky R, Malle L, Moreira A, Park MD, et al:
Sinai Immunology Review Project: Immunology of COVID-19: Current
State of the Science. Immunity. 52:910–941. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bao L, Deng W, Gao H, Xiao C, Liu J, Xue
J, Lv Q, Liu J, Yu P, Xu Y, et al: Reinfection could not occur in
SARS-CoV-2 infected rhesus macaques. bioRxiv: https://doi.org/10.1101/2020.03.13990226.
|
|
45
|
Okba NMA, Müller MA, Li W, Wang C, Geurts
van Kessel CH, Corman VM, Lamers MM, Sikkema RS, de Bruin E,
Chandler FD, et al: Severe acute respiratory syndrome coronavirus
2-specific antibody responses in coronavirus disease patients.
Emerg Infect Dis. 26:1478–1488. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su
Y, Wang X, Yuan J, Li T, Li J, et al: Antibody responses to
SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin
Infect Dis. March 28–2020.Epub ahead of print. View Article : Google Scholar
|
|
47
|
Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z,
Xiang J, Wang Y, Song B, Gu X, et al: Clinical course and risk
factors for mortality of adult inpatients with COVID-19 in Wuhan,
China: A retro-spective cohort study. Lancet. 395:1054–1062. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tetro JA: Is COVID-19 receiving ADE from
other coronaviruses? Microbes Infect. 22:72–73. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Cao X: COVID-19: Immunopathology and its
implications for therapy. Nat Rev Immunol. 20:269–270. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Iwasaki A and Yang Y: The potential danger
of suboptimal antibody responses in COVID-19. Nat Rev Immunol.
20:339–341. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Huang A, Garcia-Carreras B, Hitchings M,
Yang B, Katzelnick LC, Rattigan SM, Borgert BA, Moreno CA, Solomon
BD, Rodriguez-Barraquer I, et al: A systematic review of antibody
mediated immunity to coronaviruses: antibody kinetics, correlates
of protection, and association of antibody responses with severity
of disease. medRxiv. View Article : Google Scholar
|
|
52
|
Zheng Y, Li R and Liu S: Immunoregulation
with mTOR inhibitors to prevent COVID-19 severity: A novel
intervention strategy beyond vaccines and specific antiviral
medicines. J Med Virol. May 22–2020.Epub ahead of print.
|
|
53
|
Raybuck AL, Cho SH, Li J, Rogers MC, Lee
K, Williams CL, Shlomchik M, Thomas JW, Chen J, Williams JV, et al:
B cell-intrinsic mTORC1 promotes germinal center-defining
transcription factor gene expression, somatic hypermutation, and
memory B cell generation in humoral immunity. J Immunol.
200:2627–2639. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kindrachuk J, Ork B, Hart BJ, Mazur S,
Holbrook MR, Frieman MB, Traynor D, Johnson RF, Dyall J, Kuhn JH,
et al: Antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling
modulation for Middle East respiratory syndrome coronavirus
infection as identified by temporal kinome analysis. Antimicrob
Agents Chemother. 59:1088–1099. 2015. View Article : Google Scholar :
|
|
55
|
Kuss-Duerkop SK, Wang J, Mena I, White K,
Metreveli G, Sakthivel R, Mata MA, Muñoz-Moreno R, Chen X, Krammer
F, et al: Influenza virus differentially activates mTORC1 and
mTORC2 signaling to maximize late stage replication. PLoS Pathog.
13:e10066352017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Nicoletti F, Lapenta C, Donati S, Spada M,
Ranazzi A, Cacopardo B, Mangano K, Belardelli F, Perno C and Aquaro
S: Inhibition of human immunodeficiency virus (HIV-1) infection in
human peripheral blood leucocytes - SCID reconstituted mice by
rapamycin. Clin Exp Immunol. 155:28–34. 2009. View Article : Google Scholar :
|
|
57
|
Donia M, McCubrey JA, Bendtzen K and
Nicoletti F: Potential use of rapamycin in HIV infection. Br J Clin
Pharmacol. 70:784–793. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang CH, Chung FT, Lin SM, Huang SY, Chou
CL, Lee KY, Lin TY and Kuo HP: Adjuvant treatment with a mammalian
target of rapamycin inhibitor, sirolimus, and steroids improves
outcomes in patients with severe H1N1 pneumonia and acute
respiratory failure. Crit Care Med. 42:313–321. 2014. View Article : Google Scholar
|
|
59
|
Cai H: Sex difference and smoking
predisposition in patients with COVID-19. Lancet Respir Med.
8:e202020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li L, Huang T and Wang Y, Wang ZP, Liang
Y, Huang TB, Zhang HY, Sun W and Wang Y: COVID-19 patients'
clinical characteristics, discharge rate, and fatality rate of
meta-analysis. J Med Virol. 92:577–583. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lai JJ, Lai KP, Zeng W, Chuang KH,
Altuwaijri S and Chang C: Androgen receptor influences on body
defense system via modulation of innate and adaptive immune
systems: Lessons from conditional AR knockout mice. Am J Pathol.
181:1504–1512. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Gubbels Bupp MR and Jorgensen TN:
Androgen-induced immunosuppression. Front Immunol. 9:7942018.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kovats S: Estrogen receptors regulate
innate immune cells and signaling pathways. Cell Immunol.
294:63–69. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhou Y, Hou Y, Shen J, Huang Y, Martin W
and Cheng F: Network-based drug repurposing for novel coronavirus
2019-nCoV/SARS-CoV-2. Cell Discov. 6:142020. View Article : Google Scholar : PubMed/NCBI
|