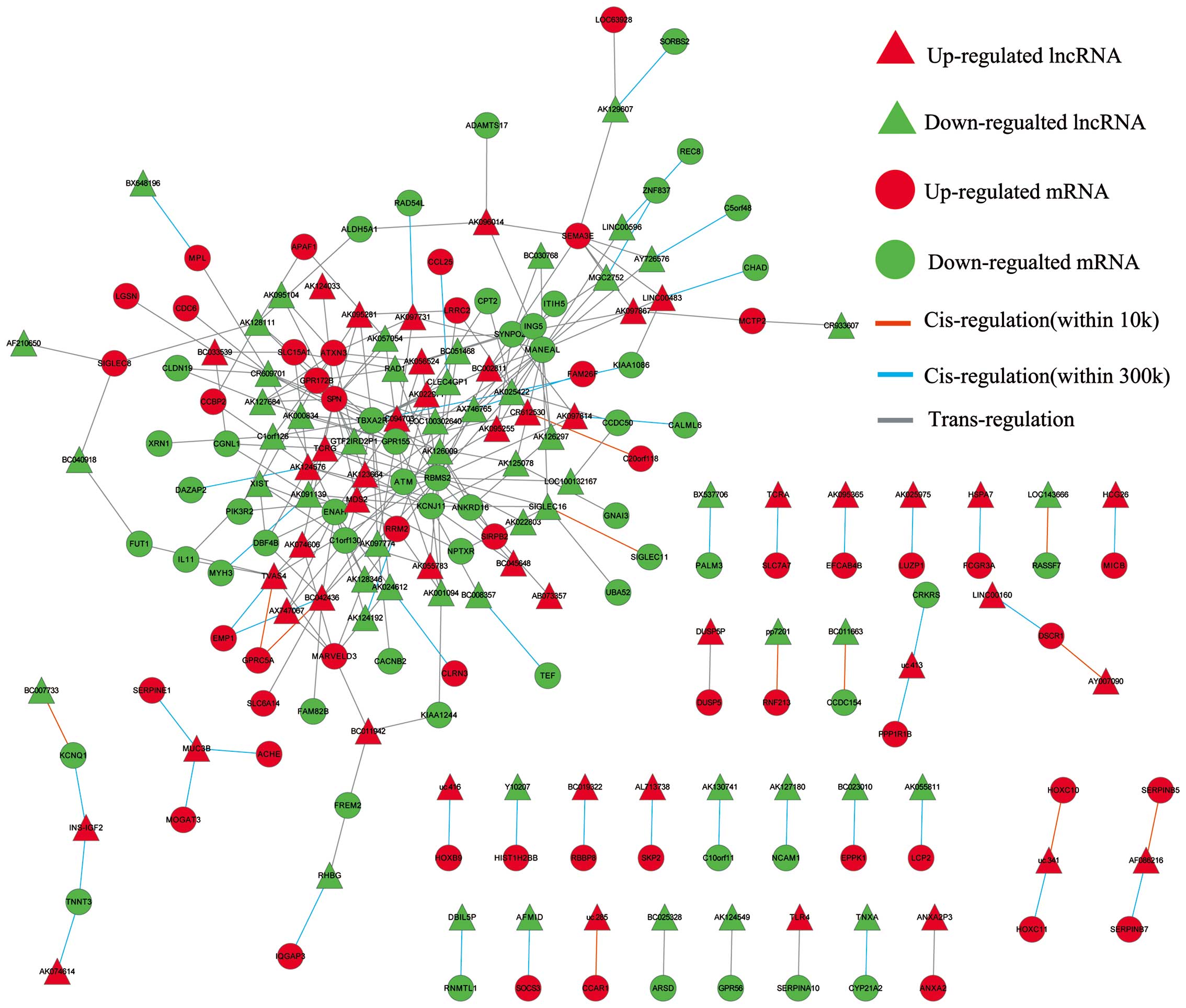
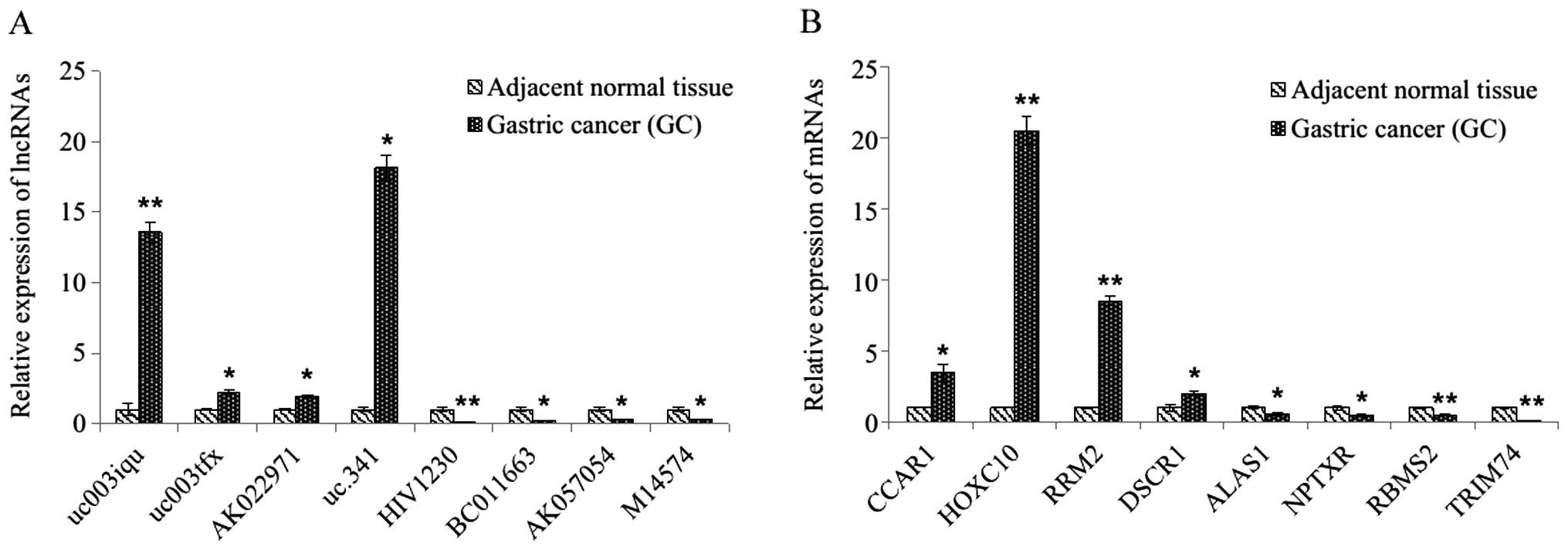
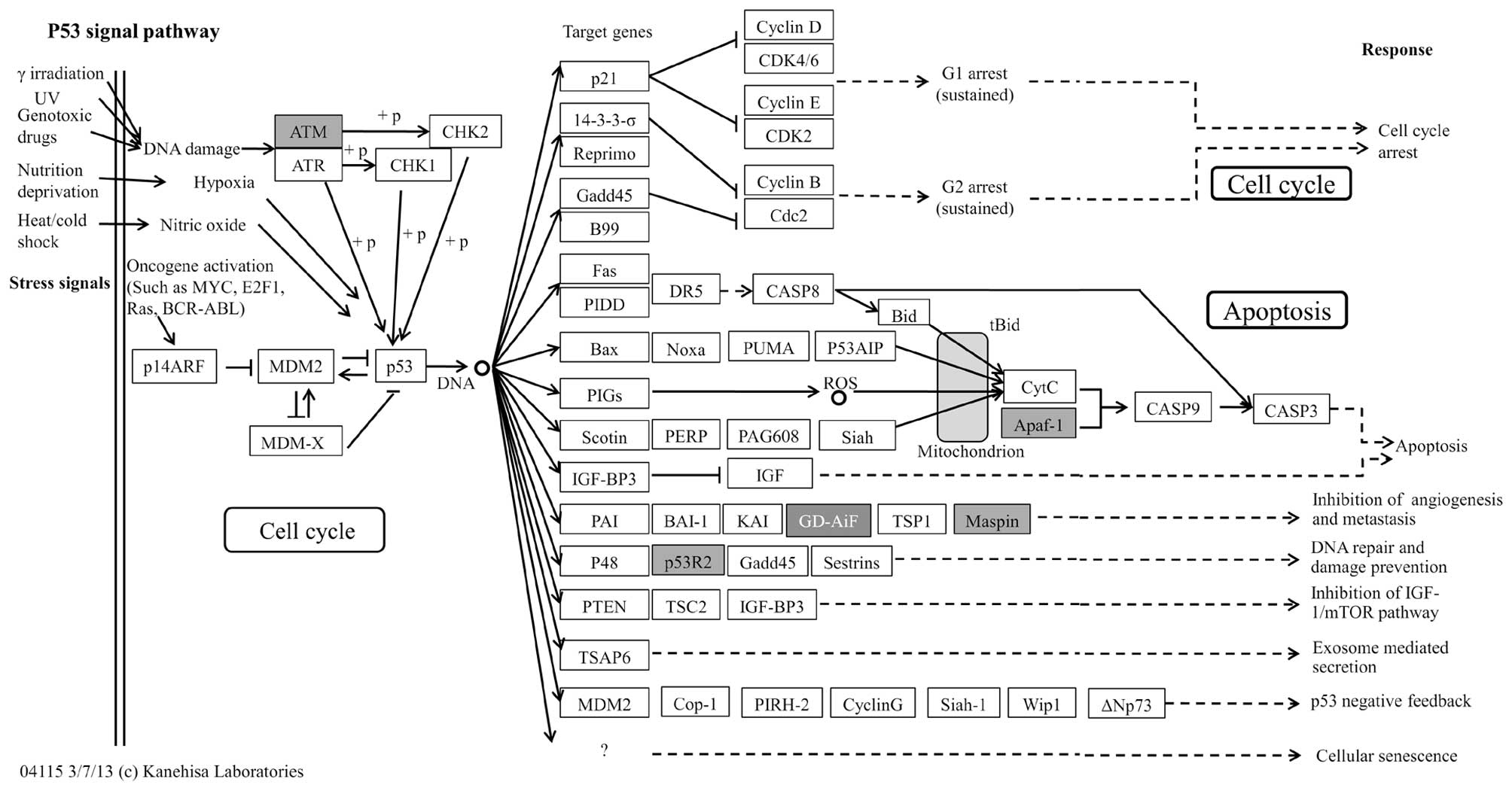
|
1
|
Thiel A and Ristimäki A: Gastric cancer:
basic aspects. Helicobacter. 1:26–29. 2012. View Article : Google Scholar
|
|
2
|
Leja M, Wex T and Malfertheiner P: Markers
for gastric cancer premalignant lesions: where do we go? Digest
Dis. 30:268–276. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guggenheim DE and Shah MA: Gastric cancer
epidemiology and risk factors. J Surg Oncol. 107:230–236. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jemal A, Bray F, Center MM, et al: Global
cancer statistics. CA Cancer J Clin. 61:69–90. 2011. View Article : Google Scholar
|
|
5
|
Wagner AD, Grothe W, Haerting J, et al:
Chemotherapy in advanced gastric cancer: a systematic review and
meta-analysis based on aggregate data. J Clin Oncol. 24:2903–2909.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang J, Wang Q, Liu H, et al: MicroRNA
expression and its implication for the diagnosis and therapeutic
strategies of gastric cancer. Cancer Lett. 297:137–143. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:38–55. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gutschner T and Diederichs S: The
hallmarks of cancer: a long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Griffiths JS, Saini HK, van Dongen S, et
al: miRBase: tools for microRNA genomics. Nucleic Acids Res.
36:D154–D158. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Storz G, Opdyke JA and Zhang A:
Controlling mRNA stability and translation with small, noncoding
RNAs. Curr Opin Microbiol. 7:140–144. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Morey C and Avner P: Employment
opportunities for non-coding RNAs. FEBS Lett. 567:27–34. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liao Q, Liu C, Yuan X, et al: Large-scale
prediction of long non-coding RNA functions in a coding-non-coding
gene co-expression network. Nucleic Acids Res. 39:3864–3878. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Au PC, Zhu QH, Dennis ES, et al: Long
non-coding RNA-mediated mechanisms independent of the RNAi pathway
in animals and plants. RNA Biol. 8:404–414. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ma H, Hao Y, Dong X, et al: Molecular
mechanisms and function prediction of long noncoding RNA. Sci World
J. 2012:5417862012.PubMed/NCBI
|
|
15
|
Sui W, Yan Q, Li H, et al: Genome-wide
analysis of long noncoding RNA expression in peripheral blood
mononuclear cells of uremia patients. J Nephrol. 26:731–738. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yin Z, Guan D, Fan Q, et al: lncRNA
expression signatures in response to enterovirus 71 infection.
Biochem Biophys Res Commun. 430:629–633. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu G, Yao W, Wang J, et al: LncRNAs
expression signatures of renal clear cell carcinoma revealed by
microarray. PLoS One. 7:e423772012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen G, Wang Z, Wang D, et al:
LncRNADisease: a database for long-non-coding RNA-associated
diseases. Nucleic Acids Res. 41:D983–D986. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Harries LW: Long non-coding RNAs and human
disease. Biochem Soc. 40:902–906. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cao WJ, Wu HL, He BS, et al: Analysis of
long non-coding RNA expression profiles in gastric cancer. World J
Gastroenterol. 19:3658–3664. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pfaffl MW: A new mathematical model for
relative quantification in real-time RT-PCR. Nucleic Acids Res.
29:452001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sui W, Lin H, Peng W, et al: Molecular
dysfunctions in acute rejection after renal transplantation
revealed by integrated analysis of transcription factor, microRNA
and long noncoding RNA. Genomics. 102:310–322. 2013. View Article : Google Scholar
|
|
23
|
Szafranski P, Dharmadhikari AV, Brosens E,
et al: Small noncoding differentially methylated copy-number
variants, including lncRNA genes, cause a lethal lung developmental
disorder. Genome Res. 23:23–33. 2013. View Article : Google Scholar
|
|
24
|
Jia H, Osak M, Bogu GK, et al: Genome-wide
computational identification and manual annotation of human long
noncoding RNA genes. RNA. 16:1478–1487. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Han L, Zhang K, Shi Z, et al: LncRNA
profile of glioblastoma reveals the potential role of lncRNAs in
contributing to glioblastoma pathogenesis. Int J Oncol.
40:2004–2012. 2012.PubMed/NCBI
|
|
26
|
Bu Q, Hu Z, Chen F, et al: Transcriptome
analysis of long non-coding RNAs of the nucleus accumbens in
cocaine-conditioned mice. J Neurochem. 123:790–799. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tafer H and Hofacker IL: RNAplex: a fast
tool for RNA-RNA interaction search. Bioinformatics. 24:2657–2663.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tafer H, Amman F, Eggenhofer F, et al:
Fast accessibility-based prediction of RNA-RNA interactions.
Bioinformatics. 27:1934–1940. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ryu DS, Lee HS, Lee GS, et al: Effects of
the ethylacetate extract of Orostachys japonicus on induction of
apoptosis through the p53-mediated signaling pathway in human
gastric cancer cells. Biol Pharm Bull. 35:660–665. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu J, Xie YS, Wang FL, et al:
Cytotoxicity of 5-Aza-2′-deoxycytidine against gastric cancer
involves DNA damage in an ATM-P53 dependent signaling pathway and
demethylation of P16(INK4A). Biomed Pharmacother. 67:78–87.
2013.
|
|
31
|
Ma GF, Chen SY, Sun ZR, et al: FoxP3
inhibits proliferation and induces apoptosis of gastric cancer
cells by activating the apoptotic signaling pathway. Biochem
Biophys Res Commun. 430:804–809. 2013. View Article : Google Scholar
|
|
32
|
Yu RX, Hu XM, Xu SQ, et al: Effects of
fucoxanthin on proliferation and apoptosis in human gastric
adenocarcinoma MGC-803 cells via JAK/STAT signal pathway. Eur J
Pharmacol. 657:10–19. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
To KF, Chan MW, Leung WK, et al:
Constitutional activation of IL-6-mediated JAK/STAT pathway through
hypermethylation of SOCS-1 in human gastric cancer cell line. Br J
Cancer. 91:1335–1341. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim JW, Im SA, Kim MA, et al:
Ataxia-telangiectasia mutated (ATM) protein expression with
microsatellite instability in gastric cancer as prognostic marker.
Int J Cancer. 134:72–80. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Morikawa T, Hino R, Uozaki H, et al:
Expression of ribonucleotide reductase M2 subunit in gastric cancer
and effects of RRM2 inhibition in vitro. Hum Pathol. 41:1742–1748.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Terashima M, Maesawa C, Oyama K, et al:
Gene expression profiles in human gastric cancer: expression of
maspin correlates with lymph node metastasis. Br J Cancer.
92:1130–1136. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hudler P: Genetic aspects of gastric
cancer instability. Sci World J. 2012:7619092012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Qiu MT, Hu JW, Yin R, et al: Long
noncoding RNA: an emerging paradigm of cancer research. Tumour
Biol. 34:613–620. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Qi P and Du X: The long non-coding RNAs, a
new cancer diagnostic and therapeutic gold mine. Mod Pathol.
26:155–165. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang F, Bi J, Xue X, et al: Up-regulated
long non-coding RNA H19 contributes to proliferation of gastric
cancer cells. FEBS J. 279:3159–3165. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kim ED and Sung S: Long noncoding RNA:
unveiling hidden layer of gene regulatory networks. Trends Plant
Sci. 17:16–21. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
De LF and Dean C: Long non-coding RNAs and
chromatin regulation. Curr Opin Plant Biol. 14:168–173. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Qu Z and Adelson DL: Identification and
comparative analysis of ncRNAs in human, mouse and zebrafish
indicate a conserved role in regulation of genes expressed in
brain. PLoS One. 7:e522752012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ponjavic J, Oliver PL, Lunter G, et al:
Genomic and transcriptional co-localization of protein-coding and
long non-coding RNA pairs in the developing brain. PLoS Genet.
5:e10006172009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|