Introduction
Although salivary gland adenoid cystic carcinoma
(ACC) is a rare disease with roughly 500 cases annually in the USA
(1), the prognosis is grave due to
the clinical behavior of ACC, such as indolent growth, but frequent
perineural invasion and distant metastasis (2,3).
ACCs are currently managed by surgery, with postoperative
radiotherapy utilized for the more aggressive cases. The
effectiveness of adjuvant chemotherapy or targeted therapy to date
has been limited. No molecular biomarkers are yet available that
can either be used to predict the chemotherapeutic response or
prognosis of ACC, or serve as chemotherapeutic targets, and is due
to the fact that the molecular pathogenesis remains poorly
understood (2,4).
Recent whole-genome and exome sequencing studies
(5,6) have confirmed some known mutations and
uncovered a handful of new genomic alterations in the ACC
mutational landscape, such as somatic driver mutations in PIK3CA,
FGFR2, NOTCH1/2 and the negative NOTCH signaling regulator, SPEN
(6). Ho et al (5) found somatic mutations in genes
belonging to the DNA damage response and protein kinase A signaling
pathways. Both Ho et al (5)
and Stephens et al (6)
identified a high percentage of mutations in chromatin regulating
genes that are epigenetic modifiers of gene activity (5,6).
Some of the genetic alterations uncovered by sequencing studies
corroborated the previous findings from molecular studies of ACC,
such as KIT overexpression (7–10).
Notably, several oncogenes and tumor suppressor genes that are
altered at high frequency in other types of solid tumors, such as
CDKN2A, TP53, EGFR, ERBB2 and PTEN (11), appear unaffected or rarely altered
in ACC (5,6,11).
The FGF-IGF-PI3K pathway is among these; the Stephen et al
found no genetic mutations in this pathway (6), while Ho et al (5) found recurrent mutations in the
FGF-IGF-PI3K pathway in only 30% of ACCs. Furthermore, similarly to
that found previously by next-generation sequencing in 24 ACCs
(6), Stephens et al
recently found similar, low frequency of genomic alterations in 28
cases of the relapsed and metastatic ACCs by the same sequencing
technique (12). Again, like in
the 24 primary ACCs (6), these
genetic alterations found in the relapsed and metastatic ACCs were
also low frequency events, compared with these same genetic
alterations seen in the more common solid tumors (12). This suggests that the low frequency
of genomic alterations may not account for the relapse and
metastasis of ACCs. Taken together, although some novel and known
genetic alterations have been identified in ACCs and these genomic
alterations may contribute to the molecular pathogenesis of ACC,
the relatively low frequency of any genetic mutation uncovered in
primary, relapsed, and metastatic ACCs suggests that epigenetic
alterations may also contribute in an important way to the
pathogenesis of ACC (11).
The molecular pathogenesis of ACC still remains
unclear. The most common molecular alterations found in ACC are the
t(6;9)(q22-23;p23-24) translocation resulting in the MYB-NFIB
fusion gene, which occurs in 29 to 86% of ACCs (3,5,6,13–16),
and overexpression of the MYB protein, observed in 89–97% of ACCs
(15,16). The role of these two molecular
alterations in ACC pathogenesis is not well understood. MYB
overexpression is often (15,17),
but not always (13–16,18),
associated with the MYB-NFIB fusion, multiple MYB-NFIB fusion
variants due to the differential breakpoints have also been
reported (13), and NFIB has been
found to fuse with non-MYB partners in ACC (19), so that the relationship between
these two molecular events is also unclear. Neither MYB-NFIB fusion
nor MYB overexpression has consistently been found to be associated
with prognostic features. Therefore, while improved understanding
of these alterations is imperative for elucidating the pathogenesis
of ACC, it is also necessary to explore additional aspects of the
unique pathology of ACC.
In the present study, we utilized a global
demethylating agent, 5-aza-2′-deoxycytidine (5-AZA), to unmask
silencing of putative TSGs in ACC xenograft models and a DNA
methylation array to identify oncogene and TSG candidates under the
control of promoter methylation in ACC. Our approach was to
circumvent the lack of viable ACC cell lines (20) by using primary xenograft tumor
models, in an attempt to identify relevant genes exhibiting
differential promoter methylation.
Materials and methods
Genomic DNA extraction from mouse
xenografts of ACC tumors
Freshly resected ACC tumors from three different
patients were transplanted in nude mice to establish ACC
xenografts. The establishment of mouse xenografts with ACC tumor
has been reported (21). When the
xenografts reached 125–250 mm3, mice were randomly
assigned into two groups, control and treatment. The control group
had 2 mice; the treatment group had 3 mice. The control group of 2
mice received no injection. In contrast, the mice in the treatment
group received a daily subcutaneous injection of
5-aza-2′-deoxycytidine (5-Aza) at a dose of 1 mg/kg of body weight
for 28 days by the South Texas Accelerated Research Therapeutics
Co. (San Antonio, TX, USA). 5-Aza is a nucleoside analog that has
been shown to cause hypomethylation of genes at a low dose
(22–24) by depleting DNA methyltransferase 1
(25,26) and this demethylation at CpG islands
in the gene promoter can often re-activate gene expression of the
once methylation-silenced gene (27).
After the termination of the treatment, these
xenografts of ACC tumors were harvested, snap-frozen in liquid
nitrogen, and stored at −80°C before genomic DNA extraction. Two
samples were harvested from each xenograft from each mouse.
The DNA was extracted from the same xenograft using
AllPrep DNA/RNA Mini kit (Qiagen, Valencia, CA, USA), according to
the manufacturer's instructions.
Methylation array and expression array of
mouse xenografts of ACC tumors
The extracted DNAs from mouse xenografts of ACC
tumors were submitted to the Johns Hopkins University core facility
for genome-wide DNA methylation screening using the Illumina
Infinium HumanMethylation27 BeadChip array (Illumina, Inc., San
Diego, CA, USA), according to the manufacturer's instructions. The
quality of all DNA samples was subjected to quality control by
Agilent's Bioanalyzer before methylation array.
Human tissue samples
Formalin-fixed, paraffin-embedded (FFPE) samples
were obtained from 32 patients treated surgically by the Department
of Otolaryngology-Head and Neck Surgery of the Johns Hopkins
Medical Institutions between the years 1999 and 2009. Tissue blocks
with high tumor yield were selected after all blocks had been
reviewed by an experienced head and neck pathologist (J.A.B.) to
confirm the diagnosis of ACC. Four sections of 20 μm thickness were
cut from each block and manually micro-dissected to yield ~80%
tumor purity. In addition, 20 FFPE parotid or submandibular glands
that were resected either for benign disease or as part of other
surgical procedures from 20 patients treated surgically by the
Department of Otolaryngology-Head and Neck Surgery of the Johns
Hopkins Medical Institutions between the years 1999 and 2004 were
included in the present study after histologic confirmation that
the tissues to be used were distant from any benign or inflammatory
lesion. All tissues were obtained with the approval of the Johns
Hopkins Institutional Review Board.
DNA extraction from frozen and FFPE
tissues
Genomic DNA was extracted from the frozen samples of
6 normal control salivary gland tissues and 6 ACC primary tumor
tissues and from microdissected FFPE tissue sections on glass
slides (normal and ACC tumor) using the SDS-PK method as described
previously (28,29). Briefly, a small piece of each
frozen tissue (normal and ACC tumor) was cut-off using a disposable
single-edge razor blade, and then collected in a 1.5-ml Eppendorf
tube.
Each FFPE tissue section on a glass slide was
scraped off by a disposable single-edge razor blade and then
collected in a 1.5-ml Eppendorf tube to undergo deparaffinization
in xylenes on a heat block in the fume hood. Xylenes were removed
when deparaffinization was complete. Residual amount of xylene in
each sample was washed 3 times by 100% ethanol.
Each frozen or FFPE sample was then digested in
0.02% (50 μg/ml) proteinase K (Roche, Indianapolis, IN, USA)
reconstituted in 1% sodium dodecyl sulfate (SDS) at 48°C for up to
72 h until no visible tissue was seen. DNA was then purified by
phenol-chloroform extraction and ethanol precipitation. DNA was
subsequently resuspended in LoTE buffer (10 mM Tris-HCl and 2.5 mM
EDTA), and the DNA concentration was quantified using the NanoDrop
ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA,
USA). DNA was stored at −20°C until further use.
Bisulfite treatment and bisulfite genomic
sequencing (BS-seq) of ACC and control genomic DNAs
The EpiTect Bisulfite kit (Qiagen) was used to
convert unmethylated cytosines in genomic DNA extracted from frozen
samples and FFPE samples (normal and ACC tumor) to uracils,
according to the manufacturer's instructions (29). Bisulfite converted xenograft
genomic DNA samples were stored at −80°C until use.
The criteria used to select genes and a region in a
given gene for validation by BS-seq from the list of methylated
genes detected by methylation array (data not shown) were that: i)
the region selected for validation by BS-seq had to be in the
promoter region (5 kb) or 5′-UTR exons. We did not necessarily
validate the region corresponding to the probe set ID in
methylation array. ii) There was at least one CpG island in the
promoter region (5 kb) and the 5′-UTR exons were detected by
MethPrimer (30) in a given gene.
iii) Twenty genes selected for validation were among top 100
candidates in the gene list; the rest (12 genes) were not among top
candidates, but also included due to general interest.
Validation of the 32 selected candidate genes from
the methylation array was performed by BS-seq in a separate cohort
of 6 ACC primary tumors and 6 normal control parotid salivary gland
tissues. In brief, bisulfite-treated DNA was amplified with their
primers designed using MethPrimer (30) to span areas of CpG island(s) in
their promoters or with β-actin primers.
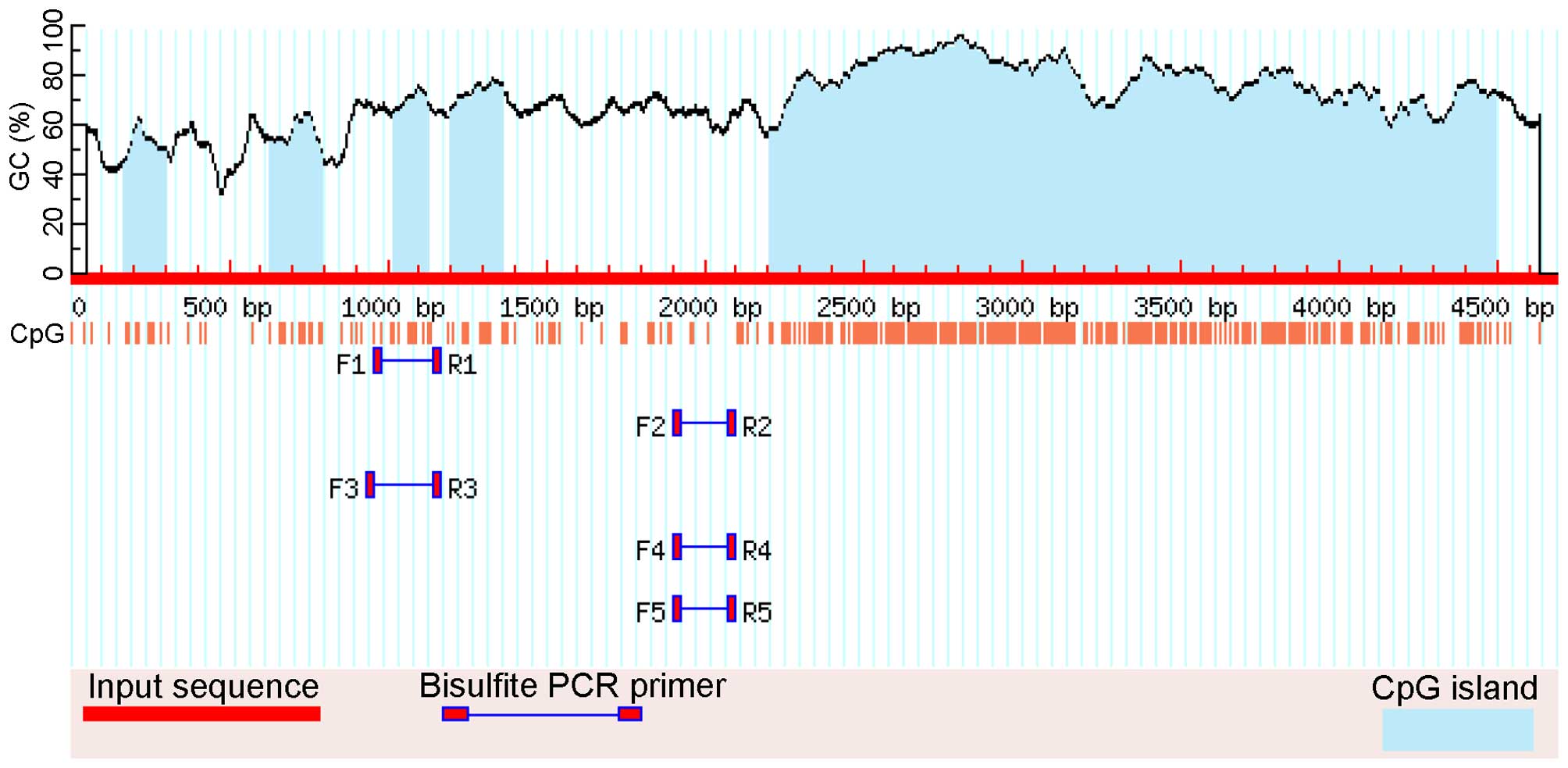
Specifically for HCN2, there are several CpG islands
in its proximal promoter and 5′-UTR exon (53 bp), but bisulfite
sequencing primer sets that span areas of the CpG island(s) were
not available due to the high density of CpGs. The amplified area
evaluated by BS-seq for the HCN2 promoter is illustrated in
Figs. 1 and 2. HCN2 and β-actin primer sequences were
specifically designed to contain no CG dinucleotides. The HCN2
primer sequences were: forward, 5′-GGA GGT ATT GGG GGT ATA GTT GTA
T-3′, which is located at -901 bases to -877 bases upstream of ATG
(the start codon) and reverse, 5′-CCA ACC AAA CAA AAA AAA CTA AAA
A-3′, which is located at -656 bases to -632 bases upstream of ATG.
β-actin primers were: forward, 5′-TGG TGA TGG AGG AGG TTT AGT AAG
T-3′ and reverse, 5′-AAC CAA TAA AAC CTA CTC CTC CCT TAA-3′.
Touch-down PCR was performed for all converted DNA samples. All PCR
products were purified using the QIAquick Gel Extract PCR
Purification kit (Qiagen), according to the manufacturer's
instructions (29). Purified PCR
products were then submitted to GeneWiz Inc. (Frederick, MD, USA)
for sequencing with one of the touch-down PCR primers. The criteria
to determine methylation in the electropherograms of the purified
PCR products were used as previously reported (31).
Quantitative methylation-specific PCR
(qMSP) by TaqMan assay
qMSP conditions and data interpretation were
previously described (28,32). Leukocyte DNA from a healthy
individual was first methylated in vitro with excess
SssI methyltransferase (New England Biolabs, Ipswich, MA,
USA) to generate universally methylated genomic DNA according to
the manufacturer's instruction, and then this universally
methylated genomic DNA was treated with the EpiTect Bisulfite kit
(Qiagen) to convert unmethylated cytosines in leukocyte DNA to
uracil, according to the manufacturer's instructions (29). These universally methylated,
bisulfite converted leukocyte DNA was made in a serial of 10-fold
dilutions (90–0.009 ng) to construct two calibration curves (HCN2
and β-actin) for each 384-well microtiter plate on each run. All
samples were within the range of sensitivity and reproducibility of
the TaqMan assay based on the amplification of the internal
reference standard, β-actin (threshold cycle value for β-actin of
≤40), as previously reported (31). The relative level of methylated DNA
in each sample was determined as a ratio of qMSP-amplified HCN2
gene to β-actin (reference gene) and then multiplied by 100 for
easier tabulation (average value of triplicates of the gene of
interest divided by the average value of triplicates of β-actin x
100). The HCN2 qMSP primer sequences were designed by
MethPrimer and their sequences were: forward, 5′-GTA TAG TTG TAT
TCG GAG TTC G-3′, which is located at -889 bases to -868 bases
upstream of ATG and reverse, 5′-AAC AAT ACC CTA AAA AAC CGT ACG-3′,
which is located at -742 bases to -719 bases upstream of ATG. The
HCN2 probe (sense) was manually designed and its sequence
was 5′-/56-FAM/TCG GGG AAA GGA GGT AAT TTC GGG/36-TAMSp/-3′, which
is located at -862 bases to -839 bases upstream of ATG (Figs. 1 and 2).
qMSP was carried out using the following conditions:
95°C for 5 min, followed by 45 cycles at 95°C for 15 sec and 60°C
for 1 min.
ACC tissue microarray (TMA), detection of
MYB-NFIB fusion by fluorescence in situ hybridization (FISH) and
immunohistochemistry (IHC) staining
An ACC TMA was constructed in house. This TMA
comprises of the cores of 93 ACC tumors and 10 normal salivary
gland tissues. As previously reported (3), FISH was performed on formalin-fixed
paraffin-embedded sections in this ACC TMA to detect MYB-NFIB
fusion. For IHC, a mouse monoclonal HCN2 antibody (S71-37) was
obtained from Novus Biologicals LLC (Littleton, CO, USA). HCN2
antibody was validated in house using positive control tissue
sections (heart and brain) before it was used for IHC staining on
ACC TMA sections. IHC staining was evaluated by a pathologist with
expertise in the area of head and neck (J.A.B.) As previously
reported (33), each sample was
assigned an H-score, which is the product of percentage of cells in
each sample with positive staining (range, 0–100%) multiplied by
the intensity of staining (range, 0–3).
Statisticalanalysis, methods, and
bioinformatics
Retrospective medical record abstraction was carried
out to ascertain clinical and pathologic variables of interest.
Methylation level for HCN2 and IHC H-scores were
considered as continuous variables and reported as median and
interquartile range (IQR), or as binary variables categorized as
<median or ≥median. Median methylation level and H-score were
compared in ACC cases vs. controls, and then across
clinicopathological variables of interest among cases, using the
non-parametric Wilcoxon rank-sum test for binary variables and the
non-parametric Kruskall Wallis test for categorical variables.
Overall survival (OS) was calculated from the date of diagnosis to
the date of death from any cause. Disease-free survival (DFS) was
calculated from the date of diagnosis to the date of any (local,
regional or distant) recurrence. Patients with distant metastases
at the time of diagnosis, or who died of incident disease without
recurrence, were excluded from DFS analysis. OS and DFS by binary
methylation status of HCN2 gene (categorized as < median or ≥
median) were reported as median and standard error (SE) and
estimated using the Kaplan-Meier method and compared using the
log-rank test for equality of survival functions. Pairwise
correlation analysis was used to determine correlation between
methylation level and H-score.
Data analysis was performed using STATA 11.2
(StataCorp LP, College Station, TX, USA). P-values <0.05 were
considered statistically significant.
Results
Differentially methylated genes
identified by methylation array
The genome-wide methylation array identified 3,481
genes (data not shown) that were significantly differentially
methylated in ACC tumors that were treated with 5-Aza, compared
with control ACC tumors that received no treatment. Among these
genes, 32 candidate genes (Table
I) were selected for validation by BS-seq in a small set
composed of 6 ACC and 6 normal frozen tissues.
 | Table ITop 32 genes detected by methylation
array. |
Table I
Top 32 genes detected by methylation
array.
| Gene symbol | Rank in the gene
list | Probe Set ID | Normal | ACC |
|---|
| RGPD3 | 1 | cg06148997 | 6/6 | 6/6 |
| ABLIM2 | 9 | cg18665513 | 0/6 | 0/6 |
| ZNF653 | 11 | cg13798986 | 0/6 | 0/6 |
| ATF4 | 12 | cg13462160 | 0/6 | 0/6 |
| BCL2 | 16 | cg06881186 | 0/6 | 0/6 |
| PCMT1 | 20 | cg07671221 | 0/6 | 0/6 |
| TNFRSF11A | 23 | cg19524723 | 0/6 | 0/6 |
| ZNF527 | 28 | cg09011348 | 0/6 | 0/6 |
| SIM2 | 31 | cg21697851 | 0/6 | 0/6 |
| PINX1 | 39 | cg26027776 | 0/6 | 0/6 |
| ZNF703 | 45 | cg25487404 | 0/6 | 0/6 |
| HCN2 | 47 | cg25367758 | 6/6a | 0/6 |
| POU4F3 | 49 | cg04701505 | 0/6 | 0/6 |
| EXOSC2 | 52 | cg14638609 | 0/6 | 0/6 |
| ZNF749 | 53 | cg23077461 | 0/6 | 0/6 |
| DUSP4 | 55 | cg05972070 | 0/6 | 0/6 |
| LGI3 | 62 | cg18317494 | 0/6 | 0/6 |
| PEX5 | 68 | cg07748017 | 0/6 | 0/6 |
| FBXO41 | 87 | cg02063488 | 0/6 | 0/6 |
| GPR39 | 97 | cg19309079 | 0/6 | 0/6 |
| RUNX3 | 128 | cg27360282 | 0/6 | 0/6 |
| BHLHE41 | 130 | cg19243777 | 0/6 | 0/6 |
| GPR123 | 196 | cg20559403 | 0/6 | 0/6 |
| SOX21 | 287 | cg18368297 | 0/6 | 0/6 |
| TBX2 | 292 | cg27470066 | 0/6 | 0/6 |
| EHD1 | 652 | cg21739289 | 0/6 | 0/6 |
| TOLLIP | 2012 | cg12308164 | 0/6 | 0/6 |
| FRMD6 | 4423 | cg09410986 | 6/6 | 6/6 |
| BSG | 7360 | cg10362365 | 0/6 | N.D. |
| MALT1 | 9548 | cg21074092 | 0/6 | 0/6 |
| MORN1 | 10003 | cg22045975 | 0/6 | 0/6 |
| CT62 | 26514 | cg13125884 | 0/6 | 0/6 |
Among these 32 genes, FRMD6 and RGPD3, were
methylated in both normal and ACC tumor samples by BS-seq (Table I). Using BS-seq, 10 CpG
dinucleotides were reliably sequenced in HCN2 (NM_001194), as
illustrated in Figs. 1 and
2. The differential methylation
status was confirmed between normal and ACC (Table II). Among these 10 CpGs, 5 CpG
sites were methylated in all normal samples, whereas other 5 CpG
sites were methylated in most of the normal samples. No
methylation, except 1 CpG (CpG #4) in 2 samples, was found in all 6
ACC tumor tissues (Table II). The
differential methylation status detected by the methylation array
for the rest of the top 32 genes was found to be without
differential methylation between normal and ACC tumor samples on
BS-seq analysis (Table I). These
data indicate that these specific CpGs on the promoter region of
HCN2 could be demethylated during the carcinogenesis of ACC.
 | Table IIThe schematic illustration of 10 CpG
dinucleotides in the proximal region of HCN2 promoter and their
methylation status validated by bisulfite sequencing in a small
validation cohort of 6 normal and 6 ACC tumor samples. |
Table II
The schematic illustration of 10 CpG
dinucleotides in the proximal region of HCN2 promoter and their
methylation status validated by bisulfite sequencing in a small
validation cohort of 6 normal and 6 ACC tumor samples.
| CpG 1 | CpG 2 | CpG 3 | CpG 4 | CpG 5 | CpG 6 | CpG 7 | CpG 8 | CpG 9 | CpG 10 |
|---|
| N1 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● |
| N2 | ● | ● | ● | ● | ● | ○ | ● | ● | ○ | ○ |
| N3 | ○ | ● | ● | ● | ● | ○ | ● | ○ | ○ | ● |
| N4 | ● | ● | ● | ● | ● | ○ | ● | ● | ● | ● |
| N5 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● |
| N6 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● |
| T1 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ |
| T2 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ |
| T4 | ○ | ○ | ○ | ● | ○ | ○ | ○ | ○ | ○ | ○ |
| T4 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ |
| T5 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ |
| T6 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ |
Validation of the differential
methylation of HCN2 by quantitative methylation-specific PCR
(qMSP)
A separate cohort of normal (n=20) and ACC tumor
(n=32) FFPE samples were used to further validate by qMSP the
differential methylation status of HCN2 in the same proximal
promoter region detected by BS-seq. Further qMSP validation of HCN2
promoter methylation levels confirmed the hypomethylation of the
same region in this cohort of ACC tumor FFPE samples (Table III and Fig. 3; P=0.04). These data indicate that
promoter demethylation of HCN2 could be a frequent event in the
carcinogenesis of ACC.
 | Table IIIThe correlation of the clinical
variables and patient outcomes with the median HCN2 promoter
methylation status in a proximal region detected by the methylation
array. |
Table III
The correlation of the clinical
variables and patient outcomes with the median HCN2 promoter
methylation status in a proximal region detected by the methylation
array.
| Variables | n | Median methylation
level of HCN2 (IQR) | P-value |
|---|
| Tumor vs. control
(n=52) | | | 0.04 |
| Control | 20 | 14.1 (0–28.3) | |
| Case | 32 | 0.4 (0–9.5) | |
| Tumor only
(n=31) | | | |
| Gender | | | 0.25 |
| Male | 11 | 0.7 (0–25.9) | |
| Female | 20 | 0.09 (0–8.1) | |
| Age (years) | | | 0.68 |
| <50 | 17 | 0.7 (0–9.5) | |
| 50+ | 14 | 0.3 (0–7.1) | |
| Smoking
history | | | 0.89 |
| No | 16 | 1.0 (0–9.5) | |
| Yes | 13 | 0.2 (0–10.5) | |
| Site | | | 0.48 |
| Major | 14 | 0.6 (0–9.4) | |
| Minor | 17 | 0.6 (0–20.0) | |
| T-stage | | | 0.76 |
| I–II | 11 | 4.8 (0–9.5) | |
| III–IV | 16 | 1.0 (0–15.3) | |
| N-stage | | | 0.44 |
| 0 | 21 | 4.8 (0–16) | |
| 1+ | 6 | 0.4 (0–7.1) | |
| M-stage | | | 0.53 |
| 0 | 24 | 1.0 (0–10.0) | |
| 1 | 3 | 7.1 (0–96.4) | |
| Overall stage | | | 0.79 |
| I–II | 9 | 5.4 (0–9.5) | |
| III–IV | 18 | 0.6 (0–10.5) | |
| Margin | | | 0.34 |
|
Negative/close | 6 | 0.10 (0–9.5) | |
| Positive | 21 | 4.8 (0–10.5) | |
| Pattern | | | 0.10 |
| Cribriform or
tubular | 26 | 3.0 (0–10.5) | |
| Solid | 2 | 0 (0–0) | |
| Nodal metastases on
neck dissection | | | 0.63 |
| No | 9 | 4.8 (0–9.4) | |
| Yes | 6 | 0.4 (0–7.1) | |
| Perineural
invasion | | | 0.09 |
| No | 1 | 39.7 (n/a) | |
| Yes | 18 | 0.6 (0–5.4) | |
| MYB-NFIB
translocation | | | 0.22 |
| Negative | 11 | 6.7 (0–16.0) | |
| Positive | 14 | 0.4 (0–7.1) | |
| Any
recurrencea | | | 0.02 |
| No | 14 | 5.4 (0.2–16.0) | |
| Yes | 14 | 0 (0–4.8) | |
| Local
recurrence | | | 0.03 |
| No | 22 | 3.0 (0–10.5) | |
| Yes | 9 | 0 (0–0) | |
| Regional
recurrence | | | 0.30 |
| No | 30 | 0.6 (0–9.5) | |
| Yes | 1 | 0 (n/a) | |
| New distant
metastases | | | 0.05 |
| No | 20 | 4.0 (0–13.3) | |
| Yes | 11 | 0 (0–4.8) | |
| Vital status at
last follow-up | | | 0.10 |
| Alive | 14 | 7.1 (0–16.0) | |
| Expired | 17 | 0 (0–5.4) | |
Comparison of HCN2 promoter methylation
with clinico-pathological parameters
After confirming that the promoter region of HCN2 is
significantly hypomethylated in ACC compared to normal tissue, we
investigated whether the quantitative methylation status of the
HCN2 promoter in ACC specimens was associated with
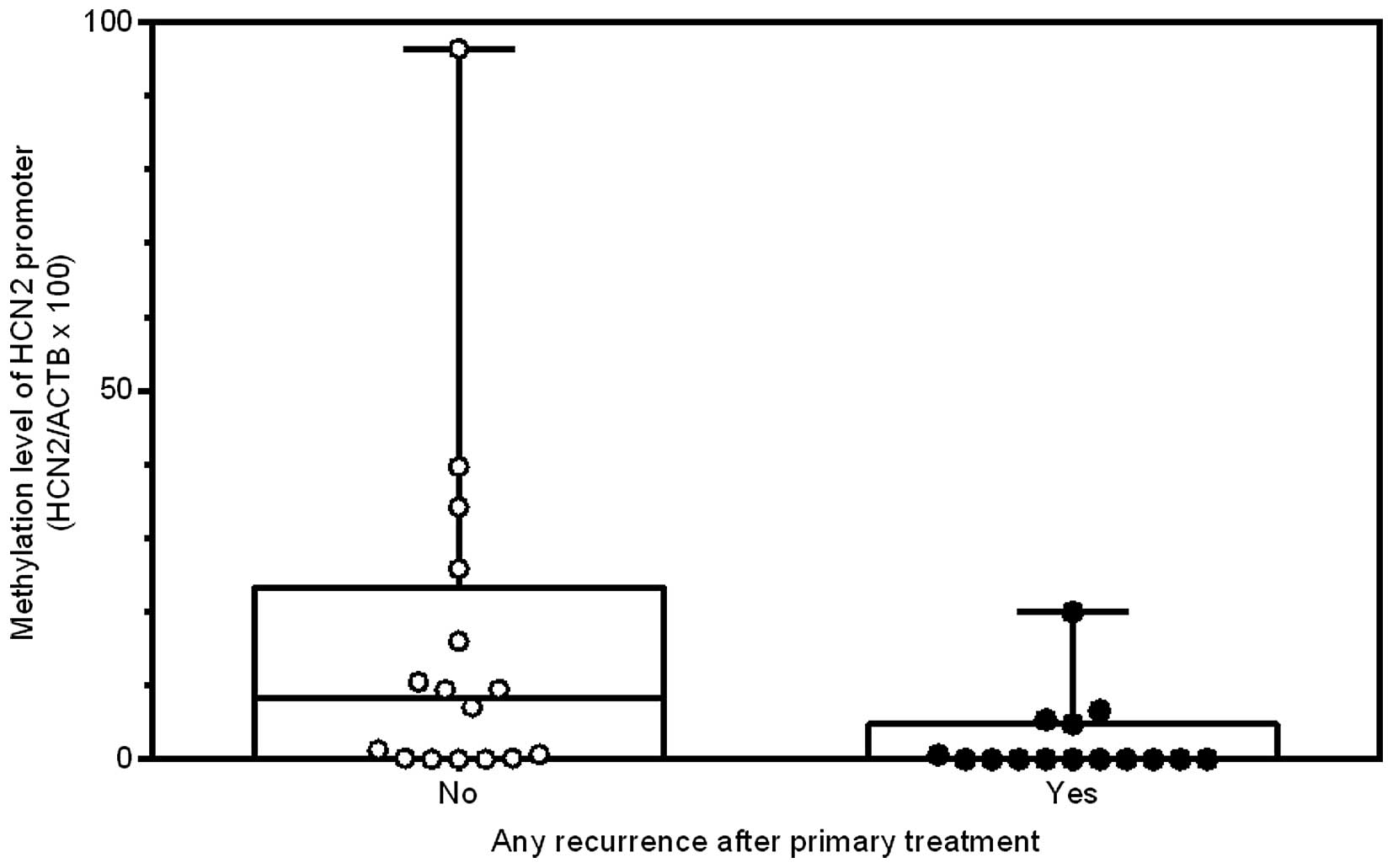
clinicopathological characteristics (Table III). Patients who developed
disease recurrence had significantly lower HCN2 promoter
methylation levels than those who did not (0 vs. 5.4, P=0.02;
Table III and Fig. 4). The same was true when
considering only local recurrence (0 vs. 3.0, P=0.03) and only new
distant metastases (0 vs. 4.0, P=0.05). Thus, hypomethylation of
the HCN2 promoter region may increase the risk of local and distant
tumor recurrence. In addition, as previously reported (3), the local (9 out of 31 cases) and
distant (11 out of 31 cases) recurrences were the predominant
recurrences and the regional (1 out of 31 cases) recurrence was
rare in this cohort of ACCs.
Since the MYB-NFIB fusion gene and MYB
over-expression are found in most ACC patients (13,19,34),
the presence of the MYB-NFIB fusion gene was examined. There were
11 MYB-NFIB translocation-negative patients and 14 MYB-NFIB
translocation-positive patients (Table III). No correlation was found
between MYB overexpression and HCN2 promoter hypomethylation
(Table III, P=0.22), indicating
that HCN2 promoter demethylation occurred independently of MYB-NFIB
translocation in this cohort.
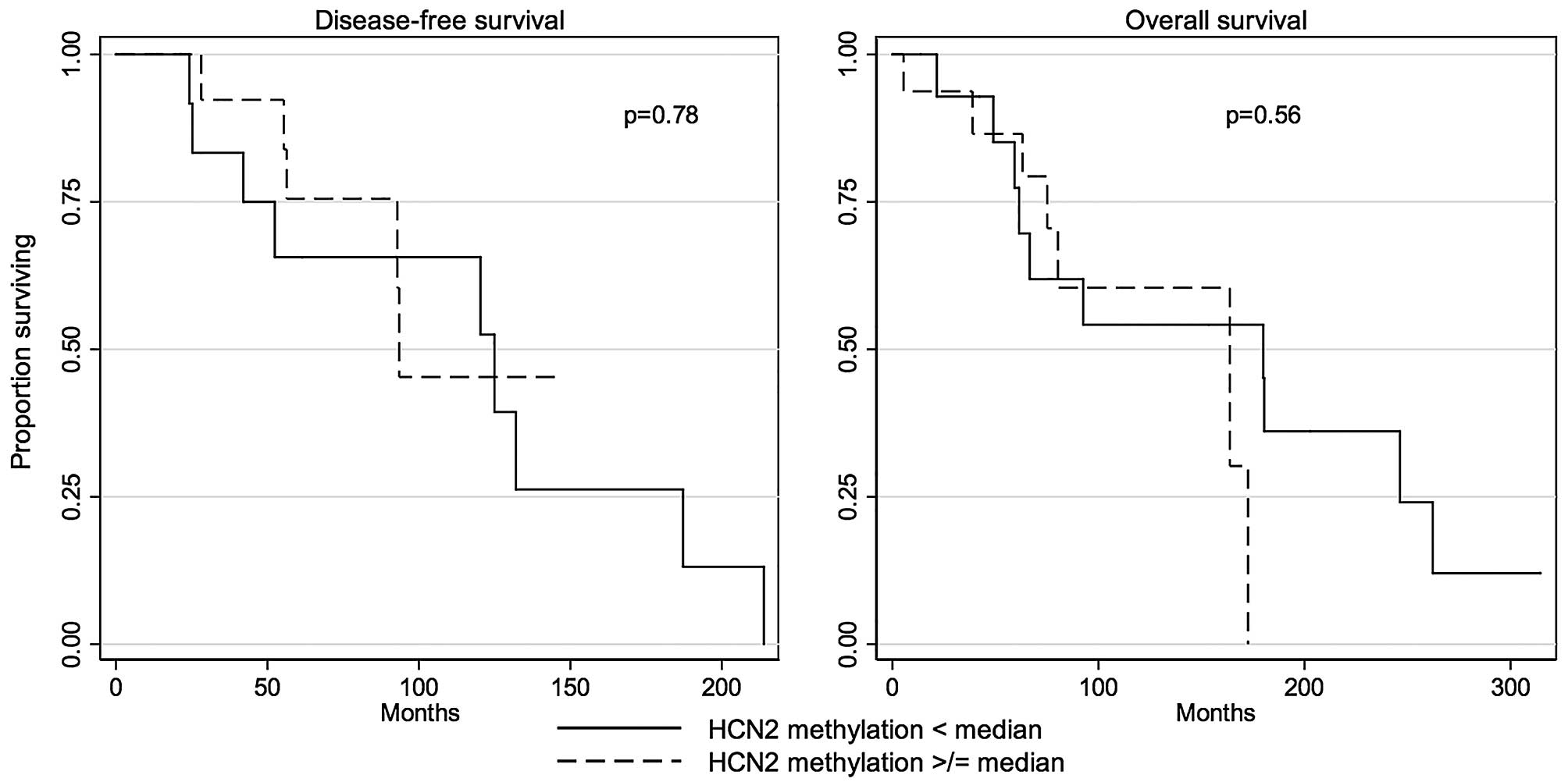
The differential methylation status in the proximal
region of HCN2 promoter was not correlated with DFS (P=0.78,
log-rank test; Table IV and
Fig. 5) or OS (P=0.56, log-rank
test; Table IV and Fig. 5).
 | Table IVSurvival by promoter methylation
level of HCN2 gene among ACC cases. |
Table IV
Survival by promoter methylation
level of HCN2 gene among ACC cases.
| | Disease-free
survival | Overall
survival |
|---|
| |
|
|
|---|
| Promoter
methylation level | n | Median months
(SE) | Log-rank
P-value | Median months
(SE) | Log-rank
P-value |
|---|
| HCN2, also see
Fig. 5 | | | 0.78 | | 0.56 |
| < median | 16 | 124.9 (20.5) | | 180.0 (62.5) | |
| ≥ median | 16 | 93.5 (33.6) | | 163.7 (13.5) | |
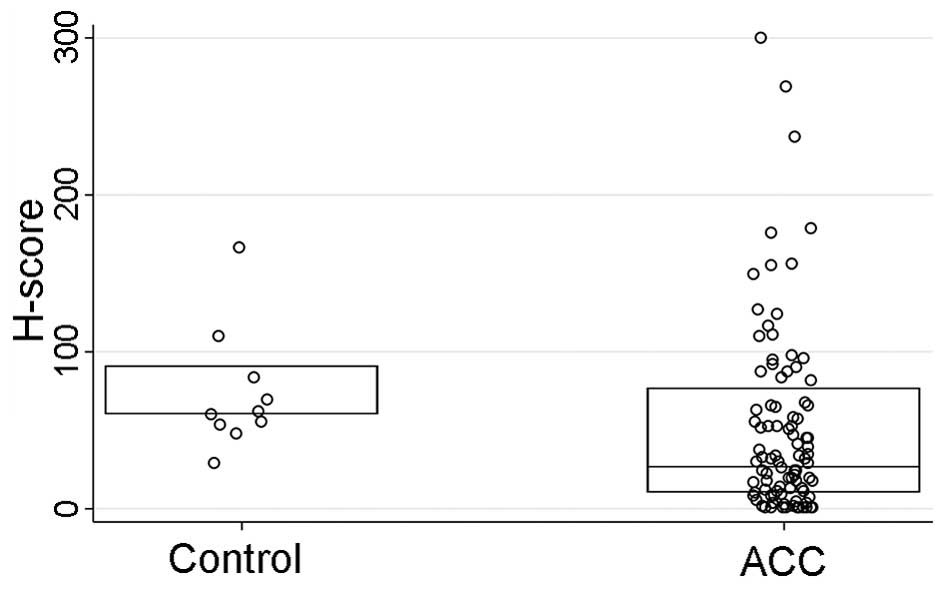
HCN2 IHC staining was performed on a tissue
microarray (TMA) sections comprised of ACC tumors (n=93) and normal
salivary gland tissue cores as controls (n=10). The HCN2 staining
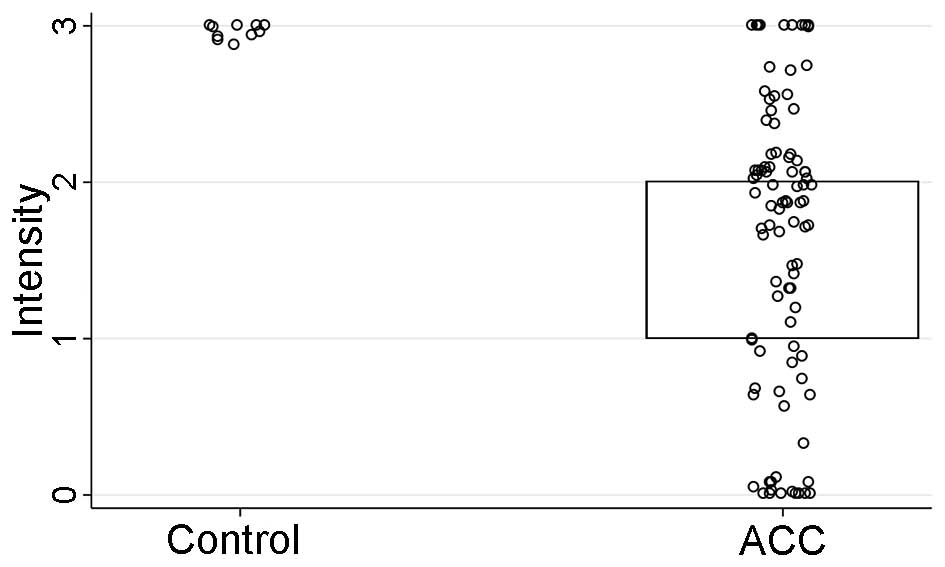
intensity (P<0.001) and H-score (P=0.01), but not the percentage
of the positively stained cells (P=0.10), were significantly
stronger in normal control tissues than those of ACC tissues
(Table V and Figs. 6Figure 7–8). However, IHC parameters were not
significantly correlated with HCN2 promoter methylation level
(P=0.67), clinicopathological characteristics (P>0.09 for all),
or survival (DFS P=0.74, OS P=0.53; log-rank test).
 | Table VHCN2 IHC analysis between normal
control and ACC cases. |
Table V
HCN2 IHC analysis between normal
control and ACC cases.
| Score | ACC cases median
(IQR) (n=93) | Controls median
(IQR) (n=10) | P-value |
|---|
| HCN2 H-score also
see Fig. 6 | 27 (10–77) | 60 (60–90) | 0.01 |
| HCN2 intensity also
see Fig. 7 | 2 (1–2) | 3 (3–3) | <0.001 |
| HCN2 percent
staining also see Fig. 8 | 13 (5–33) | 20 (20–30) | 0.10 |
Discussion
While there is increasing insight being gained into
the genetic alterations in ACC, the epigenetic landscape remains
somewhat unknown. Our objective was to use a novel method to
identify differentially methylated genes in ACC in an attempt to
broaden our understanding of its pathogenesis. Currently, there are
no FDA-approved chemotherapeutic drugs available for the treatment
of ACC, and there is a need to find targetable molecular
alterations to develop effective adjuvant treatment options.
The regulation of gene expression of oncogenes and
tumor suppressor genes (TSGs) through DNA promoter methylation
plays an important role in the carcinogenesis of many types of
human cancers (27) and may also
play a role in the carcinogenesis of ACC. In fact, the methylome of
ACC has been profiled (35) with
four genes validated (35).
We used a xenograft-based pharmacological
demethylation and genome-wide methylation array approach, as there
is a lack of viable cell lines (20) in which to perform these studies.
Through this non-biased screening for genes whose promoters were
under the control of methylation in ACC patients, we have
identified HCN2 as a putative oncogene whose promoter was
hypomethylated in ACC patients. The hypomethylation of HCN2
promoter suggests that HCN2 may act as an oncogene in the
pathogenesis of ACC. Furthermore, the hypomethylation in the HCN2
promoter is correlated with the any recurrence, local recurrence,
and distant metastasis of ACC primary tumors.
Hyperpolarization-activated cyclic nucleotide-gated
(HCN) 2 (HCN2) belongs to a non-selective cation channel family
that has 4 members, HCN1-4. The molecular identity of HCN channels
remained unknown until the first member of HCN was cloned from
mouse brain (36,37) and the other members were later
cloned (38–43). HCN isoforms were not originally
named as HCNs, but the HCN nomenclature was proposed and adopted
later (44,45). HCN channels were originally named
If (f for ‘funny’), Ih (h for
hyperpolarization activated), or Iq (q for queer in
neurons), based on the unique electrophysiological properties of
the current that HCN channels carry (44,46).
Furthermore, HCN1-4 share ~60% sequence identity of each other.
All four HCN channels share one unusual
property-activation by a hyperpolarized cell membrane potential.
HCN channels open upon hyperpolarization of the cell membrane
potential and close at a positive cell membrane potential. They all
also share another property, a cyclic nucleotide binding domain
(CNBD) for cAMP and cGMP in their C-termini. Because they contain a
CNBD, their activity can be furthermore modulated by hormones and
neurotransmitters that regulate the productions of cAMP and cGMP to
control heart rate and rhythm as well as neuronal pacemaking. The
binding of cAMP and cGMP make HCNs active even at positive cell
membrane potential (44,46). HCNs are slow activating, inward
non-selective cation channels. The current of HCNs is carried by
K+ and Na+ (ratio 4:1) (44,46).
In addition, HCN channels exist as tetramers on cell membrane
(44,46).
HCN4 is predominantly expressed in the sinoatrial
(SA) node of heart, while HCN2 is predominantly expressed in
ventricular cardiomyocytes and also in the sinoatrial node. Overall
HCN channels are predominantly expressed in human heart, central
and peripheral neurons, and photoreceptors in retina (40). HCN channels account for the
rhythmic activity of cells in the sinoatrial node of heart and
neurons due to their spontaneous, repetitive depolarization
(47,48).
A variety of ion channels have been implicated in
malignancies and can function to regulate tumor cell survival,
proliferation, growth and metastasis (49), therefore, ion channels may be
employed as drug targets. However, it remains challenging to define
the role of an individual ion channel in malignancy. There is no
inherited HCN2 mutation reported in human diseases, although
acquired dysfunction of HCN2 has been reported in epilepsy and
neuropathic pain (50).
HCNs including HCN2 have not been studied in
salivary gland tumors and any other type of tumor, except that HCN2
was reported to be overexpressed in lung carcinoma cells in which
HCN2 acted as an upstream regulator of cell apoptosis induced by
Ca2+ overload by protein kinase C inhibitors (51). Although HCN2 normally conducts
K+ and Na+ (44,46),
it has been reported that HCN2 was permeable to Ca2+,
and it was suggested that they may participate in pathological
Ca2+ signaling when HCN2 is overexpressed (52).
Since there were no reliable HCN2 antibodies
available for immunohistochemical staining until now, it remained
unknown whether HCN2 is overexpressed in human ACC tissues and
whether these channels exist on the cell membrane. IHC staining was
done on our ACC TMA sections. We found that the hypomethylation of
the HCN2 promoter did not increase the number of cells with HCN2
expression in ACC tumor samples, though there did appear to be an
increase in staining intensity compared with the normal control
samples.
In conclusion, through our screening and sequencing,
we have identified hypomethylation of HCN2 as a potential biomarker
for ACC that may be associated with more aggressive disease. The
functional role of HCN2 remains to be seen, and as viable cell
lines of ACC are more available, this can be further explored.
Acknowledgements
Dr Patrick K. Ha is supported by the National
Institutes of Health (NIH)/National Institute of Dental and
Craniofacial Research grant (5R03DE022591 and 5R01DE023227).
References
|
1
|
Renehan A, Gleave EN, Hancock BD, Smith P
and McGurk M: Long-term follow-up of over 1000 patients with
salivary gland tumours treated in a single centre. Br J Surg.
83:1750–1754. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu J, Shao C, Tan ML, Mu D, Ferris RL and
Ha PK: Molecular biology of adenoid cystic carcinoma. Head Neck.
34:1665–1677. 2012. View Article : Google Scholar
|
|
3
|
Rettig EM, Tan M, Ling S, Yonescu R,
Bishop JA, Fakhry C and Ha PK: MYB rearrangement and
clinicopathologic characteristics in head and neck adenoid cystic
carcinoma. Laryngoscope. 125:E292–E299. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Moskaluk CA: Adenoid cystic carcinoma:
Clinical and molecular features. Head Neck Pathol. 7:17–22. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ho AS, Kannan K, Roy DM, Morris LG, Ganly
I, Katabi N, Ramaswami D, Walsh LA, Eng S, Huse JT, et al: The
mutational landscape of adenoid cystic carcinoma. Nat Genet.
45:791–798. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Stephens PJ, Davies HR, Mitani Y, Van Loo
P, Shlien A, Tarpey PS, Papaemmanuil E, Cheverton A, Bignell GR,
Butler AP, et al: Whole exome sequencing of adenoid cystic
carcinoma. J Clin Invest. 123:2965–2968. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Holst VA, Marshall CE, Moskaluk CA and
Frierson HF Jr: KIT protein expression and analysis of c-kit gene
mutation in adenoid cystic carcinoma. Mod Pathol. 12:956–960.
1999.PubMed/NCBI
|
|
8
|
Vila L, Liu H, Al-Quran SZ, Coco DP, Dong
HJ and Liu C: Identification of c-kit gene mutations in primary
adenoid cystic carcinoma of the salivary gland. Mod Pathol.
22:1296–1302. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tetsu O, Phuchareon J, Chou A, Cox DP,
Eisele DW and Jordan RCK: Mutations in the c-Kit gene disrupt
mitogen-activated protein kinase signaling during tumor development
in adenoid cystic carcinoma of the salivary glands. Neoplasia.
12:708–717. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sung JY, Ahn HK, Kwon JE, Jeong H, Baek
CH, Son YI, Ahn YC, Park K, Ahn MJ and Ko YH: Reappraisal of KIT
mutation in adenoid cystic carcinomas of the salivary gland. J Oral
Pathol Med. 41:415–423. 2012. View Article : Google Scholar
|
|
11
|
Frierson HF Jr and Moskaluk CA: Mutation
signature of adenoid cystic carcinoma: Evidence for transcriptional
and epigenetic reprogramming. J Clin Invest. 123:2783–2785. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ross JS, Wang K, Rand JV, Sheehan CE,
Jennings TA, Al-Rohil RN, Otto GA, Curran JC, Palmer G, Downing SR,
et al: Comprehensive genomic profiling of relapsed and metastatic
adenoid cystic carcinomas by next-generation sequencing reveals
potential new routes to targeted therapies. Am J Surg Pathol.
38:235–238. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mitani Y, Li J, Rao PH, Zhao YJ, Bell D,
Lippman SM, Weber RS, Caulin C and El-Naggar AK: Comprehensive
analysis of the MYB-NFIB gene fusion in salivary adenoid cystic
carcinoma: Incidence, variability, and clinicopathologic
significance. Clin Cancer Res. 16:4722–4731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
West RB, Kong C, Clarke N, Gilks T,
Lipsick JS, Cao H, Kwok S, Montgomery KD, Varma S and Le QT: MYB
expression and translocation in adenoid cystic carcinomas and other
salivary gland tumors with clinicopathologic correlation. Am J Surg
Pathol. 35:92–99. 2011. View Article : Google Scholar :
|
|
15
|
Brill LB II, Kanner WA, Fehr A, Andrén Y,
Moskaluk CA, Löning T, Stenman G and Frierson HF Jr: Analysis of
MYB expression and MYB-NFIB gene fusions in adenoid cystic
carcinoma and other salivary neoplasms. Mod Pathol. 24:1169–1176.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Persson M, Andrén Y, Moskaluk CA, Frierson
HF Jr, Cooke SL, Futreal PA, Kling T, Nelander S, Nordkvist A,
Persson F, et al: Clinically significant copy number alterations
and complex rearrangements of MYB and NFIB in head and neck adenoid
cystic carcinoma. Genes Chromosomes Cancer. 51:805–817. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Persson M, Andrén Y, Mark J, Horlings HM,
Persson F and Stenman G: Recurrent fusion of MYB and NFIB
transcription factor genes in carcinomas of the breast and head and
neck. Proc Natl Acad Sci USA. 106:18740–18744. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Stenman G: Fusion oncogenes in salivary
gland tumors: Molecular and clinical consequences. Head Neck
Pathol. 7(Suppl 1): S12–S19. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mitani Y, Rao PH, Futreal PA, Roberts DB,
Stephens PJ, Zhao YJ, Zhang L, Mitani M, Weber RS, Lippman SM, et
al: Novel chromosomal rearrangements and break points at the t(6;9)
in salivary adenoid cystic carcinoma: Association with MYB-NFIB
chimeric fusion, MYB expression, and clinical outcome. Clin Cancer
Res. 17:7003–7014. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Phuchareon J, Ohta Y, Woo JM, Eisele DW
and Tetsu O: Genetic profiling reveals cross-contamination and
misidentification of 6 adenoid cystic carcinoma cell lines: ACC2,
ACC3, ACCM, ACCNS, ACCS and CAC2. PLoS One. 4:e60402009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Moskaluk CA, Baras AS, Mancuso SA, Fan H,
Davidson RJ, Dirks DC, Golden WL and Frierson HF Jr: Development
and characterization of xenograft model systems for adenoid cystic
carcinoma. Lab Invest. 91:1480–1490. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Taylor SM and Jones PA: Multiple new
phenotypes induced in 10T1/2 and 3T3 cells treated with
5-azacytidine. Cell. 17:771–779. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jones PA and Taylor SM: Cellular
differentiation, cytidine analogs and DNA methylation. Cell.
20:85–93. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cameron EE, Bachman KE, Myöhänen S, Herman
JG and Baylin SB: Synergy of demethylation and histone deacetylase
inhibition in the re-expression of genes silenced in cancer. Nat
Genet. 21:103–107. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jones PA and Taylor SM: Hemimethylated
duplex DNAs prepared from 5-azacytidine-treated cells. Nucleic
Acids Res. 9:2933–2947. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Taylor SM and Jones PA: Mechanism of
action of eukaryotic DNA methyltransferase. Use of
5-azacytosine-containing DNA. J Mol Biol. 162:679–692. 1982.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Herman JG and Baylin SB: Gene silencing in
cancer in association with promoter hypermethylation. N Engl J Med.
349:2042–2054. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shao C, Sun W, Tan M, Glazer CA, Bhan S,
Zhong X, Fakhry C, Sharma R, Westra WH, Hoque MO, et al:
Integrated, genome-wide screening for hypomethylated oncogenes in
salivary gland adenoid cystic carcinoma. Clin Cancer Res.
17:4320–4330. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shao C, Bai W, Junn JC, Uemura M,
Hennessey PT, Zaboli D, Sidransky D, Califano JA and Ha PK:
Evaluation of MYB promoter methylation in salivary adenoid cystic
carcinoma. Oral Oncol. 47:251–255. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li LC and Dahiya R: MethPrimer: Designing
primers for methylation PCRs. Bioinformatics. 18:1427–1431. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim MS, Louwagie J, Carvalho B, Terhaar
Sive Droste JS, Park HL, Chae YK, Yamashita K, Liu J, Ostrow KL,
Ling S, et al: Promoter DNA methylation of oncostatin m receptor-β
as a novel diagnostic and therapeutic marker in colon cancer. PLoS
One. 4:e65552009. View Article : Google Scholar
|
|
32
|
Durr ML, Mydlarz WK, Shao C, Zahurak ML,
Chuang AY, Hoque MO, Westra WH, Liegeois NJ, Califano JA, Sidransky
D, et al: Quantitative methylation profiles for multiple tumor
suppressor gene promoters in salivary gland tumors. PLoS One.
5:e108282010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ling S, Chang X, Schultz L, Lee TK, Chaux
A, Marchionni L, Netto GJ, Sidransky D and Berman DM: An
EGFR-ERK-SOX9 signaling cascade links urothelial development and
regeneration to cancer. Cancer Res. 71:3812–3821. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bell D, Roberts D, Karpowicz M, Hanna EY,
Weber RS and El-Naggar AK: Clinical significance of Myb protein and
downstream target genes in salivary adenoid cystic carcinoma.
Cancer Biol Ther. 12:569–573. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bell A, Bell D, Weber RS and El-Naggar AK:
CpG island methylation profiling in human salivary gland adenoid
cystic carcinoma. Cancer. 117:2898–2909. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Santoro B, Grant SGN, Bartsch D and Kandel
ER: Interactive cloning with the SH3 domain of N-src identifies a
new brain specific ion channel protein, with homology to eag and
cyclic nucleotide-gated channels. Proc Natl Acad Sci USA.
94:14815–14820. 1997. View Article : Google Scholar
|
|
37
|
Santoro B, Liu DT, Yao H, Bartsch D,
Kandel ER, Siegelbaum SA and Tibbs GR: Identification of a gene
encoding a hyperpolarization-activated pacemaker channel of brain.
Cell. 93:717–729. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ludwig A, Zong X, Jeglitsch M, Hofmann F
and Biel M: A family of hyperpolarization-activated mammalian
cation channels. Nature. 393:587–591. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gauss R, Seifert R and Kaupp UB: Molecular
identification of a hyperpolarization-activated channel in sea
urchin sperm. Nature. 393:583–587. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ludwig A, Zong X, Stieber J, Hullin R,
Hofmann F and Biel M: Two pacemaker channels from human heart with
profoundly different activation kinetics. EMBO J. 18:2323–2329.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Vaccari T, Moroni A, Rocchi M, Gorza L,
Bianchi ME, Beltrame M and DiFrancesco D: The human gene coding for
HCN2, a pacemaker channel of the heart. Biochim Biophys Acta.
1446:419–425. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Seifert R, Scholten A, Gauss R, Mincheva
A, Lichter P and Kaupp UB: Molecular characterization of a slowly
gating human hyperpolarization-activated channel predominantly
expressed in thalamus, heart, and testis. Proc Natl Acad Sci USA.
96:9391–9396. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ishii TM, Takano M, Xie LH, Noma A and
Ohmori H: Molecular characterization of the
hyperpolarization-activated cation channel in rabbit heart
sinoatrial node. J Biol Chem. 274:12835–12839. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Clapham DE: Not so funny anymore: Pacing
channels are cloned. Neuron. 21:5–7. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hofmann F, Biel M and Kaupp UB:
International Union of Pharmacology. LI. Nomenclature and
structure-function relationships of cyclic nucleotide-regulated
channels. Pharmacol Rev. 57:455–462. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pape HC: Queer current and pacemaker: The
hyperpolarization-activated cation current in neurons. Annu Rev
Physiol. 58:299–327. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Stieber J, Hofmann F and Ludwig A:
Pacemaker channels and sinus node arrhythmia. Trends Cardiovasc
Med. 14:23–28. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
DiFrancesco D: The role of the funny
current in pacemaker activity. Circ Res. 106:434–446. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lang F and Stournaras C: Ion channels in
cancer: Future perspectives and clinical potential. Philos Trans R
Soc Lond B Biol Sci. 369:201301082014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Benarroch EE: HCN channels: Function and
clinical implications. Neurology. 80:304–310. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Norberg E, Karlsson M, Korenovska O,
Szydlowski S, Silberberg G, Uhlén P, Orrenius S and Zhivotovsky B:
Critical role for hyperpolarization-activated cyclic
nucleotide-gated channel 2 in the AIF-mediated apoptosis. EMBO J.
29:3869–3878. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Michels G, Brandt MC, Zagidullin N, Khan
IF, Larbig R, van Aaken S, Wippermann J and Hoppe UC: Direct
evidence for calcium conductance of hyperpolarization-activated
cyclic nucleotide-gated channels and human native If at
physiological calcium concentrations. Cardiovasc Res. 78:466–475.
2008. View Article : Google Scholar : PubMed/NCBI
|