Introduction
According to global cancer statistics 2018, gastric
cancer remains an important cancer type worldwide, as it is the
fifth most frequently diagnosed cancer and the third leading cause
of cancer-associated mortality in the world (1). Previously, the only treatment choice
for patients with advanced gastric cancer was chemotherapy
(2); however, the efficacy of
chemotherapy is limited, and increased evidence suggests that the
growth, invasion and metastasis of tumor rely on tumor
angiogenesis, which regulated by the vascular endothelial growth
factors (VEGFs) and corresponding receptors (3). Bevacizumab is an anti-VEGF monoclonal
recombinant humanized antibody, which is approved by the Food and
Drug Administration (FDA) to treat colorectal, breast, lung, renal,
ovarian cancers and glioblastoma (4). As a monoclonal antibody that
recognizes and binds to VEGF receptor 2 (VEGFR2), ramucirumab is
the only anti-angiogenic agent approved by the FDA for the
treatment of patients with advanced gastric cancer (5).
BC001 is a novel fully humanized monoclonal antibody
of VEGFR2. Our previous study showed that BC001 inhibited the
growth of gastric cancer in vitro and in vivo
(6); however, an AVAGAST trial
showed that bevacizumab combined with chemotherapy could not
significantly improve overall survival of patients with advanced
gastric cancer, indicating that it is important to identify an
effective BC001-combined therapeutic regimen for the treatment of
gastric cancer (7).
Numerousstudies have demonstrated that certain anti-angiogenic
drugs could induce autophagy (8-910).
Hydroxychloroquine (HCQ), an anti-malarial drug,
efficiently inhibits cellular lysosomal functions, and enhances the
anticancer effects of other therapeutic agents (11,12).
Evidence has suggested that therapies combined with HCQ are better
at producing positive anticancer effects than HCQ or therapy alone
(13). Although, whether HCQ
promotes the anticancer effect of BC001 in gastric cancer and
whether BC001 induces autophagy remain unclear.
In the present study, we reported that HCQ enhanced
the antiproliferative and proapoptotic properties of BC001 in
vitro, and promoted the antitumor effects of BC001 on a BGC823
cell-based xenograft tumor in vivo. Our data also revealed
that BC001 did not influence autophagy, whereas HCQ could inhibit
autophagy by impairing autophagosome fusion with lysosomes and
induced severe ultrastructural changes, which may contribute to the
impaired fusion. To increase our understanding of the mechanisms,
RNA-sequencing (RNA-Seq) was used to analyze alterations in gene
expression following combined treatments and single drug treatment.
The results showed that the expression of numerous
autophagy-associated genes were altered in HCQ-treated cells,
including tumor protein p53-inducible nuclear protein 1 (TP53INP1),
interleukin (IL)1B, tumor necrosis factor (TNF), Mediterranean
fever (MEFV), ubiquitin specific peptidase 36 (USP36), IL6, and
lysosome-associated genes also were changed, such as neuraminidase
(NEU)1, ATP-binding cassette subfamily A member 1 (ABCA1),
proprotein convertase subtilisin/kexin type 9 (PCSK9), myelin basic
protein (MBP) and NEU3. In addition, HCQ also affects multiple
pathways, including negative regulation of endothelial cell
proliferation, blood vessel remodeling, cell surface receptor
signaling pathways and notch receptor processing in signal
transduction, cancers and immune system. C-X-C motif chemokine
ligand 8 (CXCL8), TNF, IL6, ICAM1 and FOS may be 'hub' genes.
Therefore, our findings suggested that HCQ may enhance the
anticancer effects of BC001 in gastric cancer via complex
mechanisms.
Materials and methods
Cell culture
The human gastric cancer cell line BGC823 was
obtained from the American Type Culture and Collection, and was
cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo
Fisher Scientific, Inc.), supplemented with 10% fetal bovine serum
(FBS, Gibco; Thermo Fisher Scientific, Inc.). BGC823 cells were
cultured at 37°C in a humidified atmosphere containing 5%
CO2.
Reagents and antibodies
The antibodies for microtubule-associated light
chain 3 (LC3) (cat. no. 12741s) and cleaved-caspase-3 (cat. no.
9664s) were purchased from Cell Signaling Technology, Inc. p62
(cat. no. Ab109012), Ki67 (cat. no. Ab16667), and caspase-3 (cat.
no. Ab32351) antibodies were obtained from Abcam. Anti-CD31 (cat.
no. AF6191) was obtained from Affinity. Hydroxychloroquine (HCQ)
was purchased from Sigma-Aldrich (Merck KGaA); Annexin
V-fluorescein isothiocyanate (FITC) Assay kit was from BD
Biosciences, Cell Counting Kit-8 (CCK-8, cat. no. CK04) was
purchased from Dojindo Molecular Technologies, Inc., and
TRIzol® (cat. no. 9109) was purchased from Thermo Fisher
Scientific, Inc.
CCK-8 cell viability assay
A CCK-8 kit was used to detect the effects of HCQ on
the proliferation of BGC823 cells. Briefly, cells
(2.0x104 cells/ml) were seeded onto 96-well plate. After
incubation at 37°C for 24 h, cells were treated with
different concentrations of HCQ (0, 2, 4, 6, 8, 16, 32, 64, 128,
and 256 µg/ml) for 24 h. Then, 10 µl CCK-8 reagent
was added to each well and incubated at 37°C for another 1
h. The optical density (OD) was measured at 450 nm using a
microplate reader. Cell proliferation inhibition rate = (1-OD of
the treatment group/OD of the control group) x 100%.
Real-time cell analyzer (RTCA)
assays
Briefly, 50 µl of cell culture medium
supplemented with FBS was added into each well of the E-plate 96
that was then connected to the system to obtain background
impedance readings. Cell suspensions (50 µl,
3×104 cells/ml) were seeded in E-plate 96 followed by
incubation at 37°C with 5% CO2. When the cells
reached the logarithmic growth phase, BC001 (20 µg/ml) or
HCQ (5 µg/ml) alone or in combination, were added to the
wells of E-plate 96. The control group (CK) received no treatment.
The plate was then incubated at room temperature for 30 min and
then placed on the RTCA SP Station for continuous impedance
recording every 2 min (14).
Flow cytometry
The cells were seeded in 6-well plates and were
treated with BC001 (20 µg/ml) or HCQ (5 µg/ml) alone
or in combination for 24 h. Untreated cells were used as a negative
control. After washing with PBS twice and subsequent by
trypsinization, the cells were resuspended in 500 µl of
binding buffer (BD Biosciences) supplemented with 5 µl of
Annexin V-FITC and 5 µl of propidium iodide (PI) according
to the manufacturer's recommendations. Finally, the fluorescence
intensity of the samples was determined by flow cytometry (EPICS
XL-MCL, Beckman Coulter, Inc.), and the number of apoptotic cells
in each sample was analyzed using FCS Express version 3.0 (De Novo
Software). The whole experiment was performed in triplicate.
Western blotting
Proteins of cells treated with BC001 (20
µg/ml) or HCQ (5 µg/ml) alone or in combination for
24 h, were extracted using radioimmunoprecipitation assay lysis
buffer (Roche Diagnostics), and protein concentrations were
measured using a BCA assay. The samples (20 µg/lane) were
subjected to 10% SDS-PAGE gels, and then transferred to a PVDF
membrane. After blocking with 5% non-fat milk ~1 hat room
temperature, the membrane was incubated with indicated antibodies
(LC3B, 1:2,000; p62, 1:1,000; GAPDH, 1:10,000) for 2 h at room
temperature. Then, the membrane was washed three times with TBST
buffer, and incubated with the horseradish peroxidase-conjugated
goat anti-rabbit secondary antibodies (cat. no. 7074, Cell
Signaling Technology, Inc., 1:10,000) at room temperature for 1 h.
The reaction was visualized using ECL (cat. no. 170-5060, Bio-Rad
Laboratories, Inc.) and detected by exposure to autoradiographic
film. Immunoreactive products were visualized using ECL and
quantified by densitometry using ImageJ software (version 1.50,
National Institutes of Health).
mCherry-enhanced green fluorescent
protein (EGFP)-LC3 immunofluorescence
To further analyze how HC001 or HCQ affected the
stepwise progression of autophagy, BGC823 cells were transfected
with 2.5 µg the plasmid expressing mCherry-EGFP-LC3 using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) in 6-well plate according to the manufacturer's
instructions. Then, BGC823 cells expressing mCherry-EGFP-LC3 were
added to plates. After 24 h, these cells were treated with BC001
(20 µg/ml) in the absence or presence of HCQ (5
µg/ml) at 37°Cfor 24 h. After treatment, cells on the
coverslips were fixed with 1% PFA in PBS for 15 min in the dark at
room temperature, and washed with PBS thoroughly. The autophagic
flux was measured using a laser scanning confocal microscope
(LSM880, Zeiss AG) (15,16).
Transmission electron microscopy
After treatment with 20 µg/ml BC001 and/or 5
µg/ml HCQ, BGC823 cells were washed with PBS twice and fixed
in glutaraldehyde (3.5% in 0.1 mol/l cacodylate buffer, pH 7.4) at
4°C for 24 h, respectively. Then, BGC823 cells were
post-fixed with 1% osmium tetroxide (OsO4) at 4°C
for 30 min and embedded in epoxy resins. Uranyl acetate (saturated
uranyl acetate in 50% alcohol) and lead citrate (1% lead citrate in
H2O) were applied for staining ultrathin sections, and
examined with transmission electron microscopy (H-7650, Hitachi
Ltd.) (17).
RNA-Seq and bioinformatics analysis
Total RNA (1 µg for each sample) was
extracted from BC001 or/and HCQ-treated BGC823 cel1ls using TRIzol
according to the manufacturer's instructions. cDNA libraries were
prepared, and the library products were then ready for sequencing
analysis via an BGISEQ-500 platform (Beijing Genomics Institute).
After raw reads were subjected to quality control testing using
Soapnuke software (version 1.4.0, clean reads were matched to the
reference genome. Following alignment, the RSEM tool (version
2.2.5, https://deweylab.github.io/RSEM/) was used for
transcript quantification, and the fragments per kilobase million
(FPKM) method was performed to calculate the expression level.
Genes with FPKM<10 were identified for differential expression
analysis. Based on the detection results, differentially expressed
genes (DEGs) were also analyzed by hierarchical clustering using
pheatmap function in R software(version 3.5.1, https://www.r-project.org/); the hypergeometric test
and false discovery rate (FDR) correction methods were also
employed. Finally, we used Gene Ontology (GO; http://geneontology.org/) and Kyoto Encyclopedia of
Genes and Genomes (KEGG; https://www.genome.jp/kegg/) pathway enrichment
analyses to study the mechanism underlying the HCQ-enhanced
anticancer effects of BC001 in gastric cancer (18,19).
In our analysis, we identified DEGs between samples using the
following criteria: FDR≤0.001 and a log2-fold change ratio ≥1.
Animal experiments
BGC823 cells (1.5×106 cells/ml) were
subcutaneously injected into the right flanks of the mice (nu/nu,
female, 6-8 weeks, 18-22 g, Vital River Laboratories Co., Ltd.).
Mice were housed in the laboratory under specific pathogen-free
conditions: Temperature, 22-25°C; humidity, 50-60%; 12-h
light/dark cycle. Mice had free access to water and Purina 5L79
rodent chow (Nestlé Purina PetCare Company). Once the tumors
reached a measurable size, these mice were randomly assigned to one
of the following four groups: i) Control, mice received an
intraperitoneal injection of PBS five times a week; ii) BC001, mice
received an intraperitoneal injection of BC001 5 mg/kg two times a
week; iii) HCQ, mice received an intraperitoneal injection of HCQ 5
mg/kg five times a week; and iv) combination, mice were treated
with intraperitoneal injection of BC001 (5 mg/kg two times a week)
and HCQ (HCQ 5 mg/kg five times a week). After 4 weeks of
treatment, tumor size was measured via two perpendicular dimensions
with calipers, then tumor volume was calculated using the formula:
(ab2)/2; 'a' represents the length and 'b' represents
the width of tumor, respectively. Finally, mice were euthanized
with an intraperitoneal injection of pentobarbital sodium (200
mg/kg) at the end of the experiment, then tumors were harvested and
weighed. The expression of Ki67, caspase-3, cleaved-caspase-3, and
CD31 in xenograft tumors was also detected by immunohistochemistry
(IHC) staining (6).
Statistical analysis
The results were presented as the mean ± standard
deviation of at least 3 independent experiments, and statistical
comparisons between groups were determined by one-way analysis of
variance or Student's t-test followed by a Tukey's post hoc test to
determine the significant differences of means in two or multiple
groups (n>2) comparisons. P<0.05 was considered to indicate a
statistically significant difference.
Results
HCQ promotes the antitumor effects of
BC001 in vitro
The effects of combined therapy of HCQ and BC001
against gastric cancer remains unknown. Here, we investigated the
effects of HCQ alone or together with BC001 on gastric cancer cell
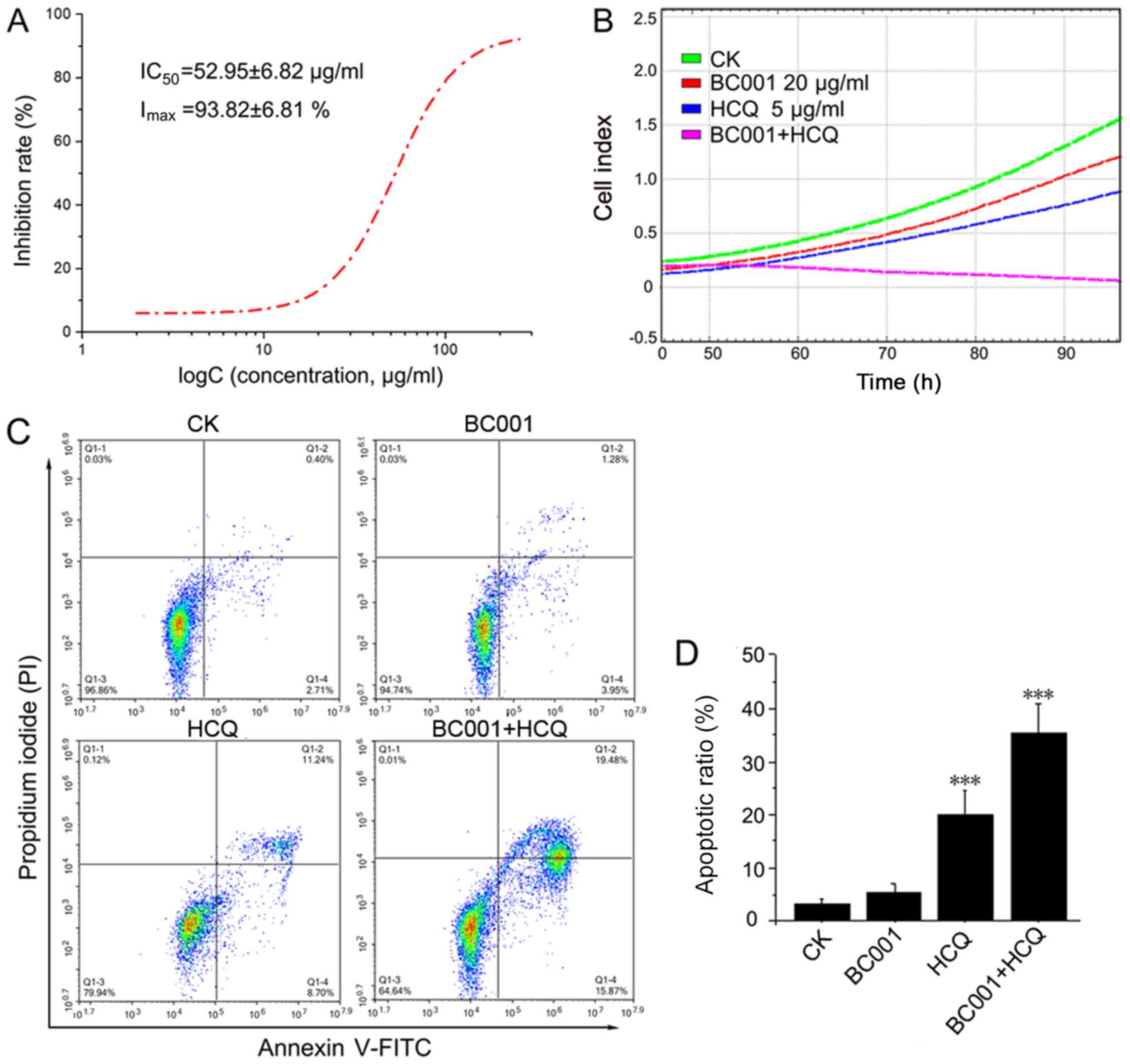
line, BGC823. As shown in Fig. 1A,
HCQ alone could inhibit BGC823 cells in a dose-dependent manner,
with a half-inhibitory concentration of 52.95±6.82 µg/ml. In
addition, the results from RTCA data showed that BC001 (20
µg/ml) or HCQ (5 µg/ml) alone could decrease the cell
index; the cell index of cells treated by HCQ and BC001 combined
was the lowest, indicating that BC001 and HCQ could inhibit BCG823
proliferation. Of note, HCQ significantly enhanced the
anti-proliferative effect of BC001 (Fig. 1B). In addition, we investigated
whether HCQ could promote BGC823 apoptosis together with BC001.
Cell apoptosis was analyzed by fluorescence-activated cell sorting
using Annexin V-FITC/PI staining after treatment with HCQ, BC001
alone or in combination. The results revealed that BC001 did not
appear to significantly promote apoptosis compared with the
control, while HCQ could enhance apoptosis induced by BC001
(Fig. 1C and D).
HCQ enhances the anticancer activity of
BC001 in vivo
To investigate the in vivo efficacy of the
combined treatment of HCQ and BC001 in gastric cancer, a BGC823
xenograft tumor model was established in nude mice. As presented in
Fig. 2, significant tumor growth
suppression was observed in the HCQ and BC001 treatment groups
compared with the control. In addition, the tumor volume and size
of the combination group were significantly reduced, compared with
the control (Fig. 2A and B). We
also analyzed the expression of Ki67, caspase-3, cleaved-caspase-3
and CD31 in tumor tissues using IHC. Compared with the untreated
and single drug-treated groups, cleaved-caspase-3 expression was
increased, while Ki67 and CD31 expression was reduced in the
combination group (Fig. 2C). This
indicated that HCQ also increased the anticancer effects of BC001
in vivo by inhibiting cell growth and promoting
apoptosis.
Autophagy is not influenced by BC001, but
is affected by HCQ, which leads to ultrastructural changes of
BGC823 cells
Next, we examined the role of BC001 on autophagy in
BGC823 cells. Firstly, we evaluated the expression of Beclin1 and
LC3II, which are indicators of autophagosome formation (12). As shown in Fig. 3A, no notable changes were reported
in the expression of Beclin1 and LC3II in BC001 (0.1, 1.0, 10, 100
µg/ml)-treated BGC823 cells. In addition, the expression of
autophagy-related protein P62 (a hallmark protein of autophagy) was
also similar to that of the control group (Fig. 3B). These data indicated that BC001
has no effect on the autophagy in BGC823 cells. In contrast, in
BGC823 cells treated with HCQ at 5 µg/ml, the conversion of
LC3-I to LC3-II was promoted. In addition, the combination
treatment of BC001 and HCQ had similar effects as HCQ treatment
alone (Fig. 3C-E). To further
analyze how HCQ or BC001 affected the stepwise progression of
autophagy, we constructed an mCherry-EGFP-LC3 reporter to observe
the progression of autophagy flux. As shown in Fig. 4A, few yellow regions were observed
in the untreated BGC823 cells. However, after 12 h of HCQ
treatment, red and yellow regions were observed in the cells as
compared with the control. Collectively, these results demonstrated
that HCQ inhibited autophagy in BGC823 cells. Additionally,
ultrastructural changes of BGC823 cells treated with HCQ and/or
BC001 were investigated to identify morphological alterations of
cell organelles and compartments. The results revealed swelling of
the mitochondrial outer chambers in BGC823 cells treated with 5
µg/ml HCQ after 24 h. We also observed large fields of
vacuoles and the dilatation of rough endoplasmic reticulum (rER)
with formation of reticular rER clusters in cells. Furthermore,
membrane-bound vesicles containing cytosolic materials or
organelles were observed; degradative autophagic vacuoles were more
abundant after HCQ treatment. However, BC001 had no notable effects
on ultrastructural changes in BGC823 cells, while compared with HCQ
group, combined treatment revealed no marked alterations (Fig. 4B). Collectively, BC001 (20
µg/ml) neither induced nor inhibited the autophagy in BGC823
cells, yet HCQ could notably induce ultrastructural changes, which
may contribute to the impairment of cellular lysosomal
functions.
HCQ actives multiple pathways to promote
the anticancer effects of BC001 in BGC823 cells
To understand how HCQ or BC001 inhibits
proliferation and promotes cell death in BGC823 cells, we treated
BGC823 cells with 5 µg/ml HCQ or 20 µg/ml BC001 for
48 h and performed global profiling of the transcriptome of HCQ or
BC001-treated cells via RNA-Seq to identify the candidate genes.
Following RNA-Seq analysis, a heatmap was generated to display the
hierarchical clustering of genes in HCQ- and/or BC001-treated cells
(Fig. 5A). Our results revealed
679 significantly upregulated and 601 downregulated DEGs in
HCQ-treated cells compared with control cells. In addition, 319
upregulated and 479 downregulated DEGs were reported for HCQ +
BC001-treated cells compared with BC001-treated cells (Fig. 5B). Notably, a previous study have
indicated an important role for lysosomes or the lysosome-dependent
pathway after HCQ treatment (8).
In the present study, we found that HCQ influenced certain
autophagy and lysosomal genes, such as TP53INP1, IL1B, TNF, MEFV,
USP36, IL6, NEU1, ABCA1, PCSK9, MBP, NEU3 (Fig. 5C and D). To further investigate the
DEG-related pathways to reveal the potential mechanisms of HCQ, we
performed enrichment analyses to identify possible associated
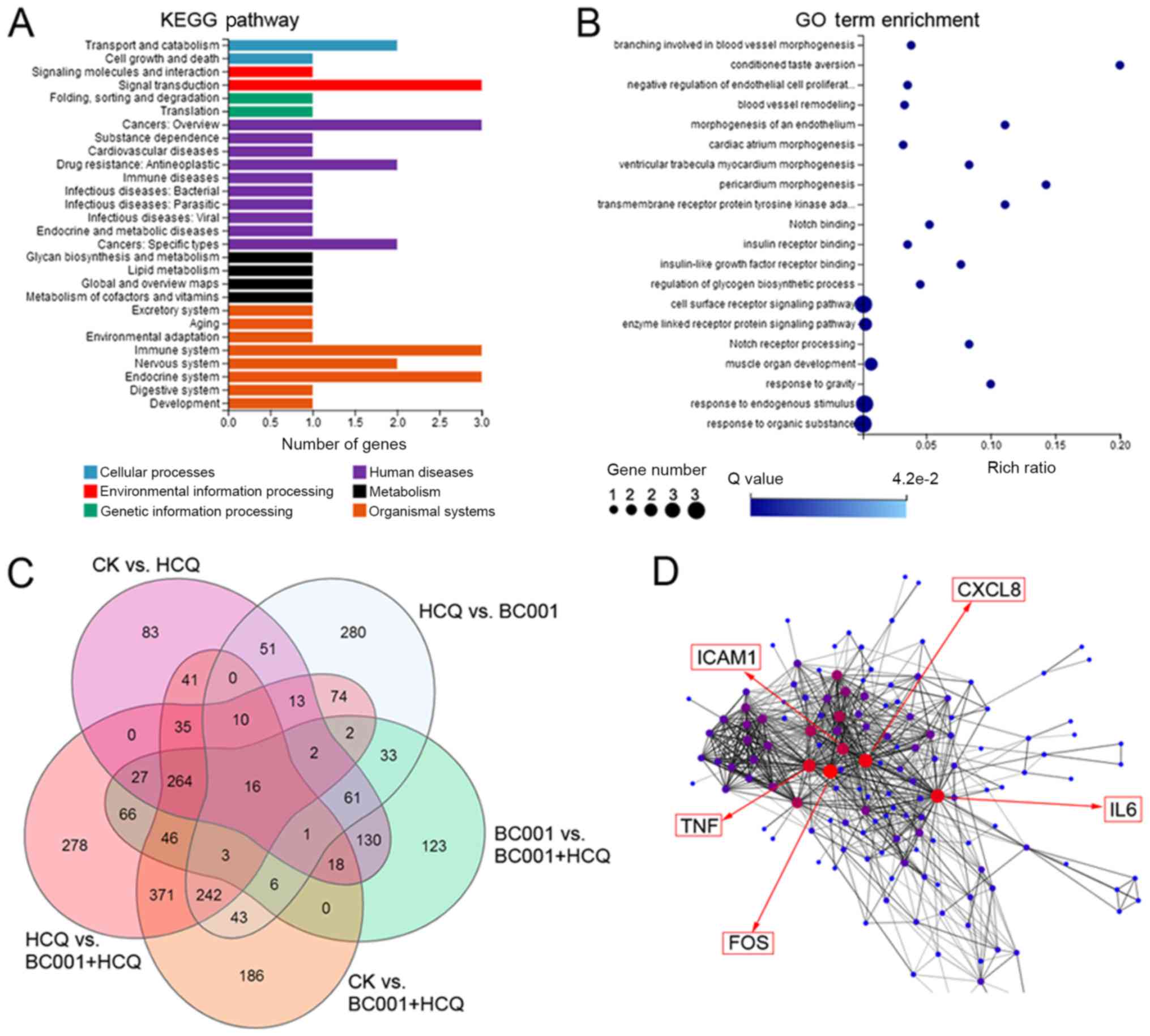
pathways. The results showed that HCQ affected BGC823 cells via
multiple pathways, including 'signal transduction', 'cancers' and
'immune system' (Fig. 6A). In
addition, the combination of HCQ with BC001 was determined to
mediate DEGs associated with 'negative regulation of endothelial
cell proliferation', 'blood vessel remodeling', 'cell surface
receptor signaling pathway', and 'notch receptor processing'
associated with 'signal transduction', 'cancers' and 'immune
system' (Fig. 6B). When analyzing
the DEGs in different groups via a Venn diagram, 16 genes were
identified (Table I and Fig. 6C). Compared with the BC001 group,
we found 470 upregulated DEGs and 367 downregulated DEGs in the
combined group, and highly interconnected 'hub' genes, including
CXCL8, TNF, IL6, ICAM1 and FOS, were revealed by gene co-expression
network analysis (Fig. 6D). These
finding indicates HCQ promotes the anticancer effect of BC001 in
BGC823 cells partially via regulating immune responses.
 | Table ISixteen differentially expressed
genes in HCQ and/or BC001-treated BGC823 cells. |
Table I
Sixteen differentially expressed
genes in HCQ and/or BC001-treated BGC823 cells.
| Gene | Log2a | Log2b | Log2c | Log2d | Log2e |
|---|
| RBM14-RBM4 | −1.626 | −2.868 | −1.134 | 1.242 | −1.734 |
| CITED2 | 1.683 | −1.050 | 1.597 | 2.733 | −2.647 |
| NEU3 | −2.047 | −3.233 | −2.138 | 1.185 | −1.095 |
| SPINK5 | −1.144 | 1.720 | −1.328 | −2.864 | 3.048 |
| SPRED2 | 1.401 | −1.111 | 1.370 | 2.513 | −2.482 |
| FOS | 1.921 | −1.428 | 2.193 | 3.349 | −3.621 |
| ADGRD1 | 1.814 | −1.122 | 1.543 | 2.936 | −2.665 |
| SCHIP1 | 2.065 | 4.232 | 1.699 | −2.167 | 2.533 |
| IRS1 | 1.218 | −1.362 | 1.441 | 2.580 | −2.802 |
| DLL4 | 3.233 | −1.080 | 3.140 | 4.312 | −4.219 |
| CYP26B1 | 1.737 | −1.158 | 1.379 | 2.895 | −2.536 |
| SHROOM3 | 1.296 | −1.236 | 1.235 | 2.533 | −2.471 |
| SPRY4 | 3.244 | −1.039 | 3.244 | 4.284 | −4.284 |
| TMEM164 | −1.093 | −2.196 | −1.127 | 1.103 | −1.069 |
| ZBED3 | −1.788 | −3.010 | −1.804 | 1.221 | −1.206 |
| EIF4EBP3 | 1.567 | 5.033 | 1.178 | −3.466 | 3.855 |
Discussion
VEGFs and its receptors are important regulators of
tumor angiogenesis (20),
VEGFR2-targeting monoclonal antibody BC001 significantly inhibits
angiogenesis and tumor growth (6);
however, increasing evidence suggests that anti-angiogenic drugs
could induce autophagy within tumors to mediate resistance
(21), and HCQ can enhance the
potential of some anticancer therapies (22).
In this study, our results suggested that HCQ
inhibited the growth of BGC823 cells, and promoted the
antiproliferative properties of BC001. In addition, HCQ promoted
the apoptosis of BGC823 cells; the apoptotic rate was significantly
increased in cells treated with both BC001 and HCQ. Additionally,
the combination treatment exhibited higher inhibition potential
than BC001 or HCQ against the growth of gastric cancer in
vivo. Therefore, HCQ may enhance the anticancer activity of
BC001 in gastric cancer. In addition, we found that BC001 neither
induced nor inhibited the autophagy of BGC823 cells, whereas HCQ
promoted the conversion of LC3-I to LC3-II, induced swelling of
mitochondrial outer chambers, and promoted the formation of
reticular rER clusters and degradative autophagic vacuoles. Our
findings suggest that HCQ impaired the basal autophagic flux and
inhibited autophagy, but BC001 did not affect HCQ-inhibited
autophagy.
To understand how HCQ or BC001 inhibits the growth
of gastric cancer, we used RNA-Seq to identify DEGs in BGC823 cells
treated with HCQ and/or BC001. HCQ was determined to have
influenced the expression of certain autophagy genes (TP53INP1,
IL1B, TNF, MEFV, USP36, IL6) and lysosomal genes (NEU1, ABCA1,
PCSK9, MBP, NEU3). Additionally, the results of enrichment analyses
indicated that HCQ not only inhibited gastric cancer via regulating
autophagy and lysosomal genes, but also via multiple pathways,
including 'negative regulation of endothelial cell proliferation',
'blood vessel remodeling', 'cell surface receptor signaling
pathway' and 'notch receptor processing' associated with 'signal
transduction', 'cancers' and 'immune system'. Furthermore, 16 genes
(RBM14-RBM4, Cbp/p300-interacting transactivator 2, NEU3, serine
peptidase inhibitor, Kazal type 5, sprout related EVH1 domain
containing 2, FOS, adhesion G protein-coupled receptor D1,
Schwannomin-interacting protein 1, insulin receptor substrate 1,
δ-like protein 4 precursor, cytochrome P450 family 26 subfamily B
member 1, shroom family member 3, sprouty RTK signaling antagonist,
transmembrane protein 164, zinc finger BED domain-containing
protein 3, eukaryotic translation initiation factor 4E binding
protein 3) were significant DEGs between groups treated by HCQ or
HCQ + BC001. Consistent with recent reports, chloroquine modulates
the antitumor immune response (23,24);
in our study, co-expression networks showed that CXCL8, TNF, IL6,
ICAM1 and FOS may be highly interconnected 'hub' genes. IL-6 and
TNFα are pro-inflammatory cytokines. Emerging evidence has revealed
that the expression of IL-6 and TNFα was significantly increased in
gastric cancer, which promoted gastric cancer cell migration and
invasion (25). In addition, HCQ
disrupted the CXCR/CXCL axis which could induce invasion and
metastasis of malignant melanoma in an autophagy-independent manner
(26). ICAM1, which has been
related to the aggressive nature of gastric cancer, can be induced
by proinflammatory cytokines (27). FOS, a proto-oncogene, has been
implicated as a regulator of cell proliferation, cell death,
differentiation and transformation (28).
In conclusion, our results suggested that HCQ
increased anticancer activity of BC001 in gastric cancer,
suggesting that the combined treatment of HCQ and BC001 may be
considered as a promising approach for the treatment of gastric
cancer. However, further investigation is necessary to validate the
combined mechanisms.
Acknowledgments
We thank generous support from Leixiang Yang (Key
Laboratory of Tumor Molecular Diagnosis and Individualized Medicine
of Zhejiang Province & Clinical Research Institute, Zhejiang
Provincial People's Hospital, People's Hospital of Hangzhou Medical
College).
Funding
This work was supported by Natural Science
Foundation of Zhejiang Province (grant no. LY16H310009), Zhejiang
Province Public Welfare Technology Application Research Project
(grant no. LGF18H160022) and Zhejiang Provincial Project for
Medical and Health Science and Technology (grant no.
2016KYA028).
Availability of data and materials
All data are included in this published article.
Authors' contributions
WW and LL performed the experiments and wrote the
paper, YZ, XT, QY, FG, and JJ conceived and designed the
experiments and edited the manuscript, ZX, XT, ZY and XY analyzed
the data, GZ and QF revised the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
All animal experiments were performed following the
approval of the Ethical Committee of Zhejiang Provincial People's
Hospital (approval no. KY2015156), and according to the Guideline
for the Care and Use of Laboratory Animals (29).
Patient consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sharma MR, Joshi SS, Karrison TG, Allen K,
Suh G, Marsh R, Kozloff MF, Polite BN, Catenacci DVT and Kindler
HLA: A UGT1A1 genotype-guided dosing study of modified FOLFIRINOX
in previously untreated patients with advanced gastrointestinal
malignancies. Cancer. 125:1629–1636. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Viallard C and Larrivée B: Tumor
angiogenesis and vascular normalization: Alternative therapeutic
targets. Angiogenesis. 20:409–426. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gilbert MR, Dignam JJ, Armstrong TS, Wefel
JS, Blumenthal DT, Vogelbaum MA, Colman H, Chakravarti A, Pugh S,
Won M, et al: A randomized trial of bevacizumab for newly diagnosed
glioblastoma. N Engl J Med. 370:699–708. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Javle M, Smyth EC and Chau I: Ramucirumab:
Successfully targeting angiogenesis in gastric cancer. Clin Cancer
Res. 20:5875–5881. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xuan ZX, Li LN, Zhang Q, Xu CW, Yang DX,
Yuan Y, An YH, Wang SS, Li XW and Yuan SJ: Fully human VEGFR2
monoclonal antibody BC001 attenuates tumor angiogenesis and
inhibits tumor growth. Int J Oncol. 45:2411–2420. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Han K, Jin J, Maia M, Lowe J, Sersch MA
and Allison DE: Lower exposure and faster clearance of bevacizumab
in gastric cancer and the impact of patient variables: Analysis of
individual data from AVAGAST phase III trial. AAPS J.
16:105610632014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hu Y-L, Jahangiri A, De Lay M and Aghi MK:
Hypoxia-induced tumor cell autophagy mediates resistance to
anti-angiogenic therapy. Autophagy. 8:979–981. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Levy JMM, Towers CG and Thorburn A:
Targeting autophagy in cancer. Nat Rev Cancer. 17:528–542. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mokarram P, Albokashy M, Zarghooni M,
Moosavi MA, Sepehri Z, Chen QM, Hudecki A, Sargazi A, Alizadeh J,
Moghadam AR, et al: New frontiers in the treatment of colorectal
cancer: Autophagy and the unfolded protein response as promising
targets. Autophagy. 13:781–819. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cook KL, Warri A, Soto- Pantoja DR, Clarke
PA, Cruz MI, Zwart A and Clarke R: Hydroxychloroquine inhibits
autophagy to potentiate antiestrogen responsiveness in
ER+ breast cancer. Clin Cancer Res. 20:3222–3232. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rosenfeld MR, Ye X, Supko JG, Desideri S,
Grossman SA, Brem S, Mikkelson T, Wang D, Chang YC, Hu J, et al: A
phase I/II trial of hydroxychloroquine in conjunction with
radiation therapy and concurrent and adjuvant temozolomide in
patients with newly diagnosed glioblastoma multiforme. Autophagy.
10:1359–1368. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Selvakumaran M, Amaravadi RK, Vasilevskaya
IA and O'Dwyer PJ: Autophagy inhibition sensitizes colon cancer
cells to antiangiogenic and cytotoxic therapy. Clin Cancer Res.
19:2995–3007. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chiu CH, Lei KF, Yeh WL, Chen P, Chan YS,
Hsu KY and Chen AC: Comparison between xCELLigence biosensor
technology and conventional cell culture system for real-time
monitoring human tenocytes proliferation and drugs cytotoxicity
screening. J Orthop Surg Res. 12:1492017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Y, Zhang J, Huang Z-H, Huang X-H,
Zheng W-B, Yin X-F, Li Y-L, Li B and He Q-Y: Isodeoxyelephantopin
induces protective autophagy in lung cancer cells via Nrf2- p62-
keap1 feedback loop. Cell Death Dis. 8:e28762017. View Article : Google Scholar
|
|
16
|
Gump JM and Thorburn A: Sorting cells for
basal and induced autophagic flux by quantitative ratiometric flow
cytometry. Autophagy. 10:1327–1334. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Alirezaei M, Flynn CT, Wood MR, Harkins S
and Whitton JL: Coxsackievirus can exploit LC3 in both autophagy-
dependent and - independent manners in vivo. Autophagy.
11:1389–1407. 2015. View Article : Google Scholar :
|
|
18
|
Dong J-K, Lei H-M, Liang Q, Tang Y-B, Zhou
Y, Wang Y, Zhang S, Li W-B, Tong Y, Zhuang G, et al: Overcoming
erlotinib resistance in EGFR mutation-positive lung adenocarcinomas
through repression of phosphoglycerate dehydrogenase. Theranostics.
8:1808–1823. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Han J, Chen M, Wang Y, Gong B, Zhuang T,
Liang L and Qiao H: Identification of biomarkers based on
differentially expressed genes in papillary thyroid carcinoma. Sci
Rep. 8:99122018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pirola L, Ciesielski O and Balcerczyk A:
The methylation status of the epigenome: Its emerging role in the
regulation of tumor angiogenesis and tumor growth, and potential
for drug targeting. Cancers (Basel). 10:2682018. View Article : Google Scholar
|
|
21
|
Mulcahy Levy JM, Zahedi S, Griesinger AM,
Morin A, Davies KD, Aisner DL, Kleinschmidt-DeMasters BK,
Fitzwalter BE, Goodall ML, Thorburn J, et al: Autophagy inhibition
overcomes multiple mechanisms of resistance to BRAF inhibition in
brain tumors. eLife. 6:e196712017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao Z, Xia G, Li N, Su R, Chen X and
Zhong L: Autophagy inhibition promotes bevacizumab- induced
apoptosis and proliferation inhibition in colorectal cancer cells.
J Cancer. 9:3407–3416. 2018. View Article : Google Scholar :
|
|
23
|
Maes H, Kuchnio A, Carmeliet P and
Agostinis P: Chloroquine anticancer activity is mediated by
autophagy-independent effects on the tumor vasculature. Mol Cell
Oncol. 3:e9700972015. View Article : Google Scholar
|
|
24
|
Chen D, Xie J, Fiskesund R, Dong W, Liang
X, Lv J, Jin X, Liu J, Mo S, Zhang T, et al: Chloroquine modulates
antitumor immune response by resetting tumor-associated macrophages
toward M1 phenotype. Nat Commun. 9:8732018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang H, Wang Y, Cao Y, Wu B, Li Y, Fan L,
Tan Z, Jiang Y, Tang J, Hu J, et al: Interleukin- 6, tumor necrosis
factor- alpha and receptor activator of nuclear factor kappa ligand
are elevated in hypertrophic gastric mucosa of
pachydermoperiostosis. Sci Rep. 7:96862017. View Article : Google Scholar
|
|
26
|
Yin S, Xia C, Wang Y, Wan D, Rao J, Tang
X, Wei J, Wang X, Li M, Zhang Z, et al: Dual receptor recognizing
liposomes containing paclitaxel and hydroxychloroquine for primary
and metastatic melanoma treatment via autophagy-dependent and
independent pathways. J Control Release. 288:148–160. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Figenschau SL, Knutsen E, Urbarova I,
Fenton C, Elston B, Perander M, Mortensen ES and Fenton KA: ICAM1
expression is induced by proinflammatory cytokines and associated
with TLS formation in aggressive breast cancer subtypes. Sci Rep.
8:117202018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
van IJzendoorn DGP, Forghany Z, Liebelt F,
Vertegaal AC, Jochemsen AG, Bovée JVMG, Szuhai K and Baker DA:
Functional analyses of a human vascular tumor FOS variant identify
a novel degradation mechanism and a link to tumorigenesis. J Biol
Chem. 292:21282–21290. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
National Research Council: Guide for the
Care and Use of Laboratory Animals. 8th edition. The National
Academies Press; Washington, DC: 2011
|