Introduction
Colorectal cancer (CRC) is among the leading causes
of cancer-related mortality. In the United States, it was estimated
that 142,570 individuals were diagnosed with CRC in 2010, of whom
51,370 succumbed to the disease (1).
In Thailand, CRC is the third most common type of cancer in men and
the fifth in women. Furthermore, CRC statistics from 4 regional
registries in the country, which have been collecting data for
>20 years, clearly demonstrated that the incidence rates of CRC
are continuously increasing (2).
Between 2001 and 2003, the estimated incidence rates of CRC in
Thailand were 11.3/100,000 men and 7.9/100,000 women (2), while in 2008, the respective incidence
rates were 14.7/100,000 and 11.8/100,000 (3). In Southern Thailand, where this study
was conducted, the statistics between 2004 and 2007 reported CRC
incidences of 12.6/100,000 for men and 9.2/100,000 for women
(4).
Although surgery is the mainstay of treatment for
early-stage CRC, there is an increasing use of multidisciplinary
treatment. As adjuvant treatments are considered based on
individual patient risk, the identification of prognostic factors
is crucial for risk stratification. Well-known factors affecting
outcome in CRC patients are tumor invasion, nodal status,
metastatic status and carcinoembryonic antigen level (5,6).
Various histological parameters and biological
markers have been investigated for possible associations with CRC.
Biological markers, such as the DNA mismatch repair genes DCC,
NM23-H1 and K-Ras have been verified as being able to predict
disease relapse (7–10). The application of those molecular
markers at the clinical level remains limited, however, due to
limitations in their reproducibility and independency from other
major prognosticators. Over several years, our research team has
been focusing on the prognostic role of Wilms' tumor 1 gene
(WT1) expression in CRC and have found that high WT1
expression is correlated with poor survival, regardless of tumor
stage (11). These findings prompted
us to investigate the genetic epidemiology of WT1 in the
Southern Thailand population and its association with the
occurrence, severity and prognosis of CRC.
WT1 is among the biological markers that have
been proven to act as oncogenes in various human cancers (12). WT1, located on chromosome
11p13, encodes a zinc finger transcription factor that plays an
important role in cell growth and differentiation. In humans, WT1
is mainly expressed in the embryonic genitourinary system.
Pathologically high WT1 expression has been reported in acute
leukemic cells (13), lung cancer
(14), breast cancer (15) and CRC (11,16).
Immunotherapy against the WT1 antigen as an adjuvant biological
treatment for cancer is currently actively investigated globally
(12,17).
The aim of this study was to investigate the
single-nucleotide polymorphism (SNP) rs16754 of WT1 in CRC
patients. This variant was previously extensively investigated in
childhood hematological malignancies (18–20), but
has never been investigated in CRC. This study was undertaken to
determine whether rs16754 is associated with the occurrence of CRC,
as well as its correlation with pathological parameters and
clinical outcomes following surgery, particularly survival
probability.
Subjects and methods
Patients and controls
Patients aged 15–65 years with histologically proven
colorectal adenocarcinoma, who underwent definitive surgery at
Songklanagarind Hospital (Hat Yai, Thailand) between January, 2006
and June, 2013, were asked to participate in this study. A cut-off
of 65 years was selected to reduce interference caused by other
carcinogenic factors associated with ageing. CRC patients with
familial adenomatous polyposis coli syndrome were also excluded.
Blood samples were collected under informed consent. Subjects who
had no history of cancer were recruited from community-based
volunteers residing in the Songkhla province to be used as
controls. Age-matched controls were selected on an individual basis
without prior knowledge of the WT1 rs16754 genotypes, with
an age difference of ≤2 years between each case and the matched
controls. In order to increase the power of the study, the number
of controls was twice that of the CRC cases.
The current clinical practice guideline for CRC
patients, including preoperative investigation and postoperative
follow-up in our institution, was published in our previous study
(21). Briefly, adjuvant chemotherapy
was considered for stage III colon cancer patients, and adjuvant
chemoradiation was considered for stage II and III rectal cancer
cases. Patients who received neoadjuvant treatment were not
included in this study. All the patients were evaluated for ≥1 year
following surgery, or until death. Follow-up visits were scheduled
at 1-month intervals during the first year following surgery, every
3 months during the second year, and every 6 months thereafter.
Tumor staging was performed according to the sixth edition of the
TNM staging system of the American Joint Committee on Cancer,
version 5 (22). Our request for
access to pathological samples and clinical records was approved by
the Institutional Research Ethics Committee of Prince of Songkhla
University (Songkhla, Thailand).
Extraction of DNA and genotyping of
WT1 rs16754
Genomic DNA was isolated from peripheral blood
leukocytes using the QIAamp DNA Mini kit (Qiagen, Hilden, Germany),
following the manufacturer's protocol. Genotype determination of
the SNP rs16754, a synonymous SNP within exon 7, was performed
using the TaqMan SNP genotyping system (Applied Biosystems, Foster
City, CA, USA) (Table I). The details
of this method were previously reported (23). The assay mixes, including unlabeled
polymerase chain reaction (PCR) primers, carboxyfluorescein (FAM)
and VIC dye-labeled TaqMan minor groove binder probes of the
Assays-by-Design system, were designed by Applied Biosystems. The
reaction system contained 50 ng genomic DNA, 5 µl 2X TaqMan™
Genotyping Master Mix, 0.25 µl 40X Assay Mix, adjusted with Milli-Q
H2O to reach a total volume of 10 µl. The reaction
conditions included an initial step at 95°C for 10 min, followed by
40 cycles at 95°C for 15 sec, and at 60°C for 60 sec in a 96-well
plate including negative (no DNA) and positive controls to ensure
genotyping accuracy using Prism®7500 Fast Real-Time PCR and
GeneAmp® PCR system 7500 (both from Applied Biosystems). The
genotyping results were analyzed using the Applied Biosystems 7500
software, version 2.0.5, and randomly selected to be confirmed by
direct sequencing. Quality control was set at a call rate of
>95% and an accuracy rate of >99%.
 | Table I.Primers and TaqMan probe used for
Wilms' tumor 1 gene rs16754 genotyping. |
Table I.
Primers and TaqMan probe used for
Wilms' tumor 1 gene rs16754 genotyping.
| Primers | Sequence |
|---|
| Forward | GCC TCC CTT CCT CTT
ACT CTC T |
| Reverse | GATG CCG ACC GTA CAA
GAG T |
| Reporter-1 (T) | VIC-CAC ACG TCG CAC
ATC-NFQ |
| Reporter-1 (G) | FAM-CAC GCC GCA CAT
C-NFQ |
Study of WT1 expression at the mRNA
and protein level
Our study of WT1 mRNA expression among the
rs16754 genotypes used reverse transcription quantitative PCR
(RT-qPCR). RNA was extracted from 66 frozen tumor tissue samples
(31 GG, 20 GA and 15 AA) and 59 normal tissue samples (23 GG, 20 AG
and 6 AA) with known rs16754 genotypes. RT-qPCR was performed for
the WT1 gene with GAPDH as an internal control using
the TaqMan probe technique. The primers and probes used in our
study were previously described (24). A relative expression of the WT1
gene was calculated as log10 (copy number of
WT1/copy number of GAPDH).
The study of WT1 protein expression and localization
on tumor tissue used a WT1 immunohistochemical staining method, as
described in our previous publication (11). Briefly, formalin-fixed,
paraffin-embedded tissue samples from CRC patients were stained
overnight at 4°C with a mouse monoclonal anti-WT1 antibody (cat.
no. clone 6FH2; Dako, Carpinteria, CA, USA), which was used as the
primary antibody at a dilution of 1:100. The WT1 protein was
detected by the EnVision™+ system (Dako) according to the
manufacturer's instructions. Finally, the tissue sections were
incubated with 3,3′-diaminobenzidine (Sigma, St. Louis, MO, USA)
until a brown color developed, and then counterstained with Harris'
modified hematoxylin. In negative controls, the primary antibodies
were omitted. For evaluation of WT1 expression, slides of cancer
specimens were analyzed by a pathologist specialised in
gastroenterology (S.K.), who was blinded to the clinical
information and genotyping results. To grade the tissue expression
of WT1, we used the Allred scoring system, which is the standard
system used to evaluate staining intensity and staining pattern.
The numerical value for overall intensity (intensity score) is
based on a 4-point system: 0, 1, 2 and 3 (for none, light, medium,
or dark staining, respectively), while the numerical value for
percentage of stained cells (proportion score) is determined by a
geometric rather than linear division; no cells stained = 0; ≤1/100
cells stained = 1; ≤1/10 cells stained = 2; ≤1/3 cells stained = 3,
≤2/3 cells stained = 4; and all cells stained = 5. The sum of the
two values yields the total Allred's score, which ranges between 0
and 8.
Statistical analysis
Conformation to the Hardy-Weinberg equilibrium for
rs16754 was tested by its formula. A statistical analysis of the
association between the genotype frequency of each SNP and the
occurrence of disease was performed using the Chi-square test. Odds
ratios were calculated using univariate logistic regression
analysis. A survival analysis was performed using the log-rank
test. Evidence of disease progression, new metastases, second
primary disease and recurrence were defined as ‘progression’ in the
progression-free survival (PFS) analysis, while cancer-related
deaths were considered as failures in the overall survival (OS)
analysis. Unless otherwise stated, P<0.05 was considered to
indicate a statistically significant difference. All the
statistical analyses used the statistical package Stata Release 13
(StataCorp LP, College Station, TX, USA).
Results
Clinicopathological
characteristics
A total of 104 CRC cases, 41 women and 63 men, with
a mean age of 51 years (range, 16–63 years) were included in this
study. A total of 208 controls were used (67 men and 141 women),
with a mean age of 51 years (range, 17–63 years), all of whom
resided in the same geographic area as the patients (Table II).
 | Table II.Allele and genotype distribution of
Wilms' tumor 1 gene single-nucleotide polymorphism rs16754 in the
CRC and age-matched control groups. |
Table II.
Allele and genotype distribution of
Wilms' tumor 1 gene single-nucleotide polymorphism rs16754 in the
CRC and age-matched control groups.
|
| Subject group |
|
|---|
|
|
|
|
|---|
| Variables | CRC cases | Controls | P-value |
|---|
| Total no. | 104 | 208 |
|
| Mean age (range),
year | 51.0 (16–63) | 51.0 (17–63) | 0.92 |
| Allelotype (alleles,
%) |
|
| 0.03 |
| G | 126 (60.6) | 289 (69.5) |
|
| A | 82 (39.4) | 127 (30.5) |
|
| Genotype (cases,
%) |
|
| 0.03 |
| GG | 41 (39.4) | 98 (47.1) |
|
| GA | 44 (42.3) | 93 (44.7) |
|
| AA | 19 (18.3) | 17 (8.2) |
|
| Minor allele genotype
(cases, %)a |
|
| <0.01 |
|
GG/GA | 85 (81.7) | 191 (91.8) |
|
| AA | 19 (8.3) | 17 (8.2) |
|
Genotyping by the TaqMan-SNP genotyping assay
yielded a call rate of >99% and a reproducibility (accuracy)
rate of 100%. The genotype distribution conformed to the
Hardy-Weinberg equilibrium (P=0.80). The minor allele frequency
(MAF) of the rs16754 (allele A) was 0.33. The MAF among CRC cases
(0.39) was significantly higher compared with that in controls
(0.31) (P=0.03). The AA genotype was significantly associated with
CRC (odds ratio = 2.51, 95% confidence interval: 1.24–5.07,
P=0.01).
The WT1 genotype groups were not
significantly associated with any clinicopathological parameters in
the CRC cases (Table III). The mean
follow-up period was 34 months (range, 6–90 months). When the
genotypes were analyzed against survival, it was found that the AA
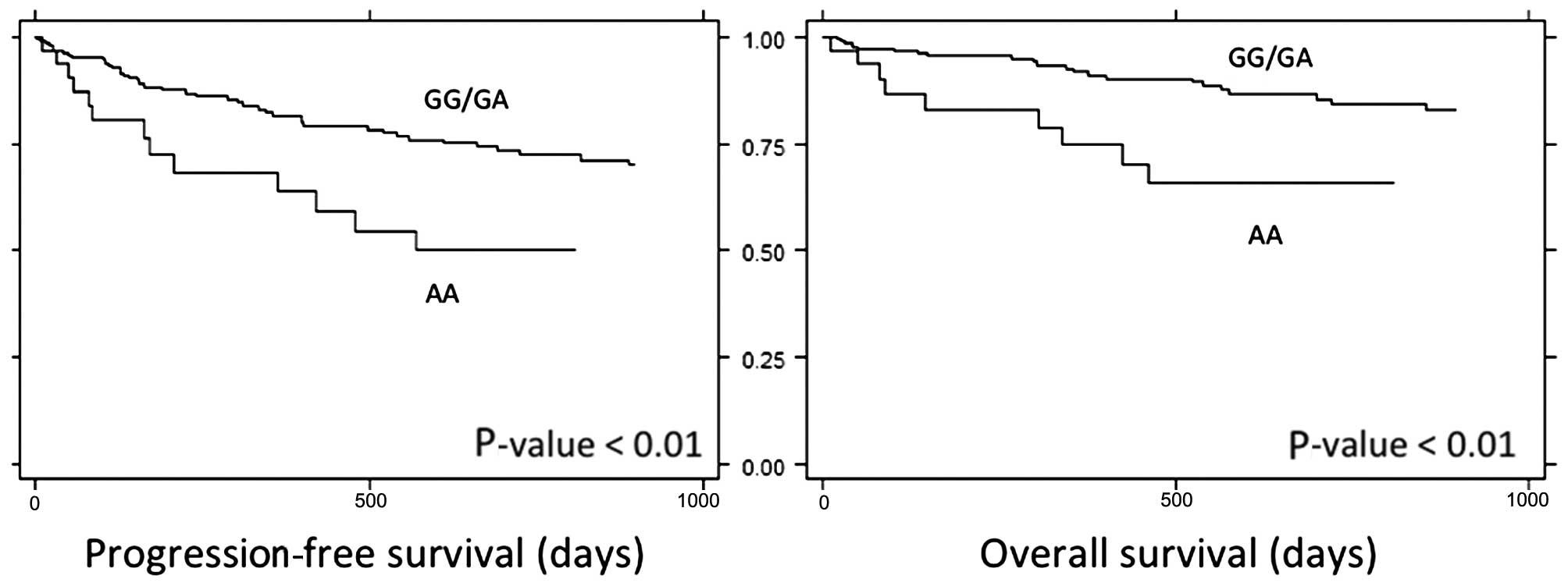
genotype was associated with poorer PFS and OS (Fig. 1). The 3-year PFS in the AA genotype
group was 43%, compared with 67% in other genotypes (P<0.01).
Furthermore, the 3-year OS in the AA genotype group was 60%,
compared to 81% in other genotypes (P<0.01).
 | Table III.Clinical and pathological parameters
of colorectal cancer patients and their correlation with the Wilms'
tumor 1 gene (WT1) rs16754 genotype. |
Table III.
Clinical and pathological parameters
of colorectal cancer patients and their correlation with the Wilms'
tumor 1 gene (WT1) rs16754 genotype.
|
| WT1 rs16754
genotype |
|
|---|
|
|
|
|
|---|
| Parameters | GG/GA (%) | AA (%) | P-value |
|---|
| All (n=104) | 85 (81.7) | 19 (18.3) |
|
| Gender |
|
|
0.20 |
|
Female | 36 (87.8) | 5
(12.2) |
|
|
Male | 49 (77.9) | 14 (22.2) |
|
| Tumor location |
|
|
0.07 |
| Left
colon/rectum | 76 (84.4) | 14 (15.6) |
|
| Right
colon | 9
(64.3) | 5
(35.7) |
|
| Tumor size, cm
(range) |
5.6
(2.5–16.5) |
5.5 (2.5–12) |
0.82 |
| Age, years |
|
|
0.52 |
|
<40 | 14 (87.5) | 2
(12.5) |
|
|
>40 | 71 (80.7) | 17 (19.3) |
|
| T-stage |
|
|
0.90 |
|
1–2 | 8
(80.0) | 2
(20.0) |
|
|
3–4 | 76 (81.7) | 17 (18.3) |
|
| N stage |
|
|
0.51 |
| N0 | 30 (79.0) | 8
(21.0) |
|
|
N1-2 | 53 (84.1) | 10 (15.9) |
|
| M stage |
|
|
0.97 |
| M0 | 66 (81.5) | 15 (18.5) |
|
| M1 | 18 (81.8) | 4
(18.2) |
|
| AJCC stage |
|
|
0.60 |
|
0-2 | 30 (79.0) | 8
(21.0) |
|
|
3–4 | 54 (83.1) | 11 (16.9) |
|
| CEA, ng/dl |
|
|
0.31 |
|
<5 | 49 (86.0) | 8
(14.0) |
|
|
>5 | 36 (78.3) | 10 (21.7) |
|
|
Differentiation |
|
|
0.96 |
|
High | 45 (81.8) | 10 (18.2) |
|
|
Moderate/poor | 35 (81.4) | 8
(18.6) |
|
|
Lymphovascular/perineural invasion |
|
|
0.18 |
| No | 58 (85.3) | 10 (14.4) |
|
|
Yes | 23 (74.2) | 8
(25.8) |
|
| Lymph node
ratio |
|
|
0.07 |
|
<0.35 | 62 (86.1) | 10 (13.9) |
|
|
>0.35 | 19 (70.4) | 8
(29.6) |
|
| Survival
analysisa,% |
|
| <0.01 |
| 3-year
PFS | 67.00 | 42.90 |
|
| 3-year
OS | 80.60 | 59.20 |
|
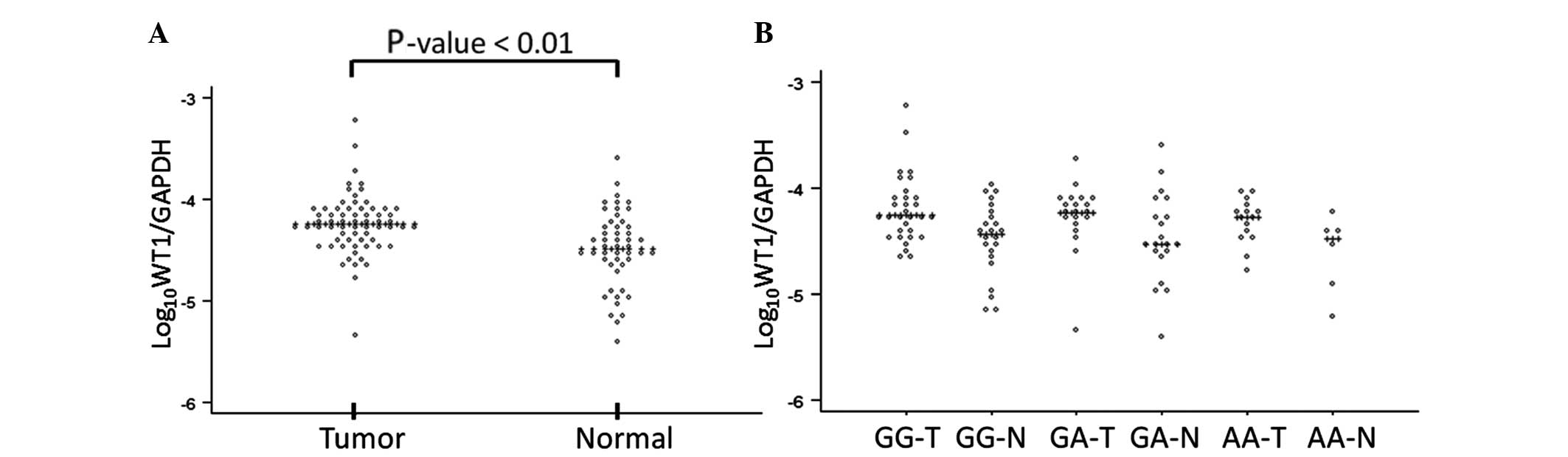
On RT-qPCR, the relative expression of WT1 in
the tumor tissue was significantly higher compared with that in the
adjacent normal colonic mucosa (P<0.01). However, there were no
significant differences in the relative expressions among the three
genotype groups (Fig. 2). On
immunohistochemical staining in the 101 cases for which
histological slides were obtained, the majority of CRC tumors (94
cases, 93%) exhibited positive staining results. Apart from one
case in the AA group, which displayed nuclear staining, all
immunoreactivity was localized in the cytoplasm. The average Allred
scores in the AA (6.4) and GA (6.7) genotype groups were higher
compared with those in the GG group (5.9); however, the differences
did not reach statistically significant levels (P=0.09).
Discussion
The oncologic role of WT1 has been validated
in various types of cancer, including CRC (13–16). Our
previous study demonstrated a correlation between high expression
of the WT1 protein and poor outcome in CRC (11). In the present study, we focused on a
genetic variant of WT1 and its association with this
disease. The rs16754 SNP, located in exon 7 of WT1, has been
found to exhibit a significant correlation between its major allele
and relatively favorable clinical outcomes in hematological
malignancies (18–20). According to the 1000 Genomes Browsers
(browser.1000genomes.org) (25), A is the major allele of rs16754 in
Western populations, while G is the major allele in East Asian
populations (26,27). The MAF of 0.31 in our study was
comparable with previous reports from other Asian populations in
the 1000 Genomes database and in reverse correlation to the MAFs
reported in European populations (18).
The CRC patients included were limited to those aged
<65 years, in order to exclude potential factors associated with
ageing that may affect disease development. In addition, there was
a certain difficulty in recruiting older age-matched controls in
our community. Our analysis demonstrated that the MAF of the
rs16754 in CRC cases significantly deviated from that in the
age-matched controls. The subsequent genotype analysis demonstrated
a significant association between the AA genotype and CRC
prognosis. As the WT1 protein expression has been identified as an
unfavorable prognostic marker in CRC (11), it is possible that this variant
exhibits a correlation with expression levels. Our finding of WT1
overexpression in CRC tissue was consistent with previous reports
(11,16). Although differences in WT1
expression in blood leukocytes among rs16754 genotypes have been
reported in a previous study (18), a
recent study on pediatric leukemia failed to detect any significant
association between expression and genotype (28). Consistent with the latter study, our
findings suggest that the expression level of WT1 in colonic tissue
is independent of the rs16754 genotype. As the rs16754 is a
synonymous SNP, the association may not indicate a direct causal
association between the gene and disease development. However, the
SNP may be in a linkage relationship with other disease-causal
variants of WT1.
In conclusion, our study identified a genetic
association of the rs16754 of WT1 in a group of Thai CRC
patients aged <65 years. The study also demonstrated a
significant correlation between risk genotype (AA) and poorer
outcome following multimodal treatment. The data suggest that this
variant may be a candidate disease marker in CRC.
Acknowledgements
This study was supported by the Thailand Research
Fund (grant no. MRG5208118). Surasak Sangkhathat is supported by
the Anandamahidol Foundation. Dave Patterson reviewed the English
in the manuscript.
References
|
1
|
Jemal A, Center MM, DeSantis C and Ward
EM: Global patterns of cancer incidence and mortality rates and
trends. Cancer Epidemiol Biomarkers Prev. 19:1893–1907. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Khuhaprema T and Srivatanakul P: Colon and
rectum cancer in Thailand: An overview. Jpn J Clin Oncol.
38:237–243. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Khuhaprema T, Attasara P, Sriplung H,
Wiangnon S and Sangrajrang S: Cancer in Thailand. 7:Bangkok,
Thailand: National Cancer Institute. 2007–2009. 2013.
|
|
4
|
Sriplung H, Bilheem S, Kuntipundee T and
Geater SL: Differences in cancer incidence among predominantly
Muslim and Buddhist subpopulations in Songkhla. Asian Pac J Cancer
Prev. 15:9979–9983. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kritsanasakul A, Boonpipattanapong T,
Wanitsuwan W, Phukaoloun M, Prechawittayakul P and Sangkhathat S:
Impact of lymph node retrieval on surgical outcomes in colorectal
cancers. J Surg Oncol. 106:238–242. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Boonpipattanapong T and Chewatanakornkul
S: Preoperative carcinoembryonic antigen and albumin in predicting
survival in patients with colon and rectal carcinomas. J Clin
Gastroenterol. 40:592–595. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu Z, Zhang Y, Niu Y, Li K, Liu X, Chen H
and Gao C: A systematic review and meta-analysis of diagnostic and
prognostic serum biomarkers of colorectal cancer. PLoS One.
9:e1039102014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Coppedè F, Lopomo A, Spisni R and Migliore
L: Genetic and epigenetic biomarkers for diagnosis, prognosis and
treatment of colorectal cancer. World J Gastroenterol. 20:943–956.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Belov L, Zhou J and Christopherson RI:
Cell surface markers in colorectal cancer prognosis. Int J Mol Sci.
12:78–113. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chaiyapan W, Duangpakdee P,
Boonpipattanapong T, Kanngern S and Sangkhathat S: Somatic
mutations of K-ras and BRAF in Thai colorectal cancer and their
prognostic value. Asian Pac J Cancer Prev. 14:329–332. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bejrananda T, Phukaoloun M,
Boonpipattanapong T, Wanitsuwan W, Kanngern S, Sangthong R and
Sangkhathat S: WT1 expression as an independent marker of poor
prognosis in colorectal cancers. Cancer Biomark. 8:35–42.
2010.2011-2011. PubMed/NCBI
|
|
12
|
Cheever MA, Allison JP, Ferris AS, Finn
OJ, Hastings BM, Hecht TT, Mellman I, Prindiville SA, Viner JL,
Weiner LM, et al: The prioritization of cancer antigens: A National
Cancer Institute pilot project for the acceleration of
translational research. Clin Cancer Res. 15:5323–5337. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu SY, Gu WY, Chen ZX, Wang XL, Cen JN, He
HL, Chai YH and Chen CS: The significance of detecting WT1
expression in childhood acute leukemias. Pediatr Hematol Oncol.
27:581–591. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Oji Y, Miyoshi S, Maeda H, Hayashi S,
Tamaki H, Nakatsuka S, Yao M, Takahashi E, Nakano Y, Hirabayashi H,
et al: Overexpression of the Wilms' tumor gene WT1 in de novo lung
cancers. Int J Cancer. 100:297–303. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Domfeh AB, Carley AL, Striebel JM,
Karabakhtsian RG, Florea AV, McManus K, Beriwal S and Bhargava R:
WT1 immunoreactivity in breast carcinoma: Selective expression in
pure and mixed mucinous subtypes. Mod Pathol. 21:1217–1223. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Oji Y, Yamamoto H, Nomura M, Nakano Y,
Ikeba A, Nakatsuka S, Abeno S, Kiyotoh E, Jomgeow T, Sekimoto M, et
al: Overexpression of the Wilms' tumor gene WT1 in colorectal
adenocarcinoma. Cancer Sci. 94:712–717. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Oka Y and Sugiyama H: WT1 peptide vaccine,
one of the most promising cancer vaccines: Its present status and
the future prospects. Immunotherapy. 2:591–594. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Damm F, Heuser M, Morgan M, Yun H,
Grosshennig A, Göhring G, Schlegelberger B, Döhner K, Ottmann O,
Lübbert M, et al: Single nucleotide polymorphism in the mutational
hotspot of WT1 predicts a favorable outcome in patients with
cytogenetically normal acute myeloid leukemia. J Clin Oncol.
28:578–585. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Becker H, Maharry K, Radmacher MD, Mrózek
K, Metzeler KH, Whitman SP, Schwind S, Kohlschmidt J, Wu YZ, Powell
BL, et al: Clinical outcome and gene- and microRNA-expression
profiling according to the Wilms tumor 1 (WT1) single nucleotide
polymorphism rs16754 in adult de novo cytogenetically normal acute
myeloid leukemia: A Cancer and Leukemia Group B study.
Haematologica. 96:1488–1495. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen X, Yang Y, Huang Y, Tan J, Chen Y,
Yang J, Dou H, Zou L, Yu J and Bao L: WT1 mutations and single
nucleotide polymorphism rs16754 analysis of patients with pediatric
acute myeloid leukemia in a Chinese population. Leuk Lymphoma.
53:2195–2204. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chalieopanyarwong V, Boonpipattanapong T,
Prechawittayakul P and Sangkhathat S: Endoscopic obstruction is
associated with higher risk of acute events requiring emergency
operation in colorectal cancer patients. World J Emerg Surg.
8:342013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Compton CC and Greene FL: The staging of
colorectal cancer: 2004 and beyond. CA Cancer J Clin. 54:295–308.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Phusantisampan T, Sangkhathat S, Phongdara
A, Chiengkriwate P, Patrapinyokul S and Mahasirimongkol S:
Association of genetic polymorphisms in the RET-protooncogene and
NRG1 with Hirschsprung disease in Thai patients. J Hum Genet.
57:286–293. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sangkhathat S, Kanngurn S, Chaiyapan W,
Gridist P and Maneechay W: Wilms' tumor 1 gene (WT1) is
overexpressed and provides an oncogenic function in pediatric
nephroblastomas harboring the wild-type WT1. Oncol Lett. 1:615–619.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Abecasis GR, Altshuler D, Auton A, Brooks
LD, Durbin RM, Gibbs RA, Hurles ME and McVean GA: 1000 Genomes
Project Consortium: A map of human genome variation from
population-scale sequencing. Nature. 467:1061–1073. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Altshuler DM, Gibbs RA, Peltonen L, et al:
International HapMap 3 Consortium: Integrating common and rare
genetic variation in diverse human populations. Nature. 467:52–58.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lauhakirti D, Sritana N, Boonthimat C,
Promsuwicha O and Auewarakul CU: WT1 mutations and polymorphisms in
Southeast Asian acute myeloid leukemia. Exp Mol Pathol. 91:682–686.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ho PA, Alonzo TA, Gerbing RB, Kuhn J,
Pollard JA, Hirsch B, Raimondi SC, Gamis AS and Meshinchi S: The
prognostic effect of high diagnostic WT1 gene expression in
pediatric AML depends on WT1 SNP rs16754 status: Report from the
Children's Oncology Group. Pediatr Blood Cancer. 61:81–88. 2014.
View Article : Google Scholar : PubMed/NCBI
|