1. Introduction
Brain tumors are the most common solid neoplasms and
the leading cause of death from cancer in children. Tumors of the
central nervous system (CNS) account for 20% of childhood cancers
and are second only to leukemia in frequency. Embryonal tumors with
multilayered rosettes (ETMR) are highly aggressive embryonic brain
tumors classified as World Health Organization (WHO) grade IV that
mainly occur in children under the age of four years (1). The five-year overall survival (OS)
rate of patients with ETMR remains <30% (2). Embryonic tumors (95%), ependymomas
(90%) and medullary epitheliomas (75%) are characterized by a
hallmark amplification of the Chromosome 19q13.42 miRNA cluster
(C19MC) (3). In the 2021
edition of the WHO classification of tumors of the CNS, ETMR
include the most common C19MC type, DICER1
alteration, not elsewhere classified or not otherwise specified
(4). ETMR exhibit unique
histopathological features which, alongside the detection of
C19MC or DICER alterations and enrichment of LIN28A,
are necessary for diagnosis. Currently, the diagnosis of ETMR
relies mainly on molecular biological detection. C19MC
amplification and TTYH1 fusion are signature gene changes in
ETMR, which are observed in ~90% of ETMR cases (5). Biallelic mutation in DICER1 is
the second most common genetic event and is present in ~5% of
patients with ETMR, occurring exclusively in tumors lacking
C19MC amplification (6).
Both C19MC amplification and DICER1 mutations may
have common downstream mechanisms, the LIN28A/let-7 pathway, and
miRNAs belonging to the let-7 miRNA family are considered
oncogenes (2). The current
treatment options for ETMR include maximum surgical excision and
adjuvant chemotherapy, high-dose chemotherapy with stem cell
salvage, and focal or whole-brain whole-spinal radiation therapy.
Most treatments for ETMR have failed under this high-intensity,
multi-modal treatment. For patients receiving chemotherapy, the
median OS time was 7.4 months, whereas that of those who did not
receive it was 1.2 months (7). For
patients with non-brainstem tumor location, the five-year
progression-free survival (PFS) and OS were 35 and 47%,
respectively, after treatment with intensified chemotherapy
(8). With further ETMR genome and
epigenetic studies, a new understanding of the ETMR molecular
biological mechanism was gained and potential therapeutic targets
were identified. However, current research on ETMR remains limited.
The treatment of patients with ETMR has only slightly improved, and
the five-year OS rate remains <30% (2). Therefore, there is an urgent need to
translate the molecular biology of ETMR into effective therapeutic
measures. The present review discusses the molecular biological
characteristics and treatment progress of ETMR to further elucidate
the pathogenesis of ETMR and lay a foundation for molecular therapy
of ETMR.
2. ETMR molecular mechanism
ETMR were distinguished from other pediatric
intracranial tumors by analyzing DNA methylation and transcriptome
sequencing data (9). ETMR are
molecularly similar with or without C19MC amplification
(6) and have unique
transcriptional and epigenetic backgrounds (10). Besides C19MC amplification,
few other gene changes or recurrent gene mutations occur (6,9,10),
suggesting that ETMR are mainly driven by epigenetic mechanisms
(10).
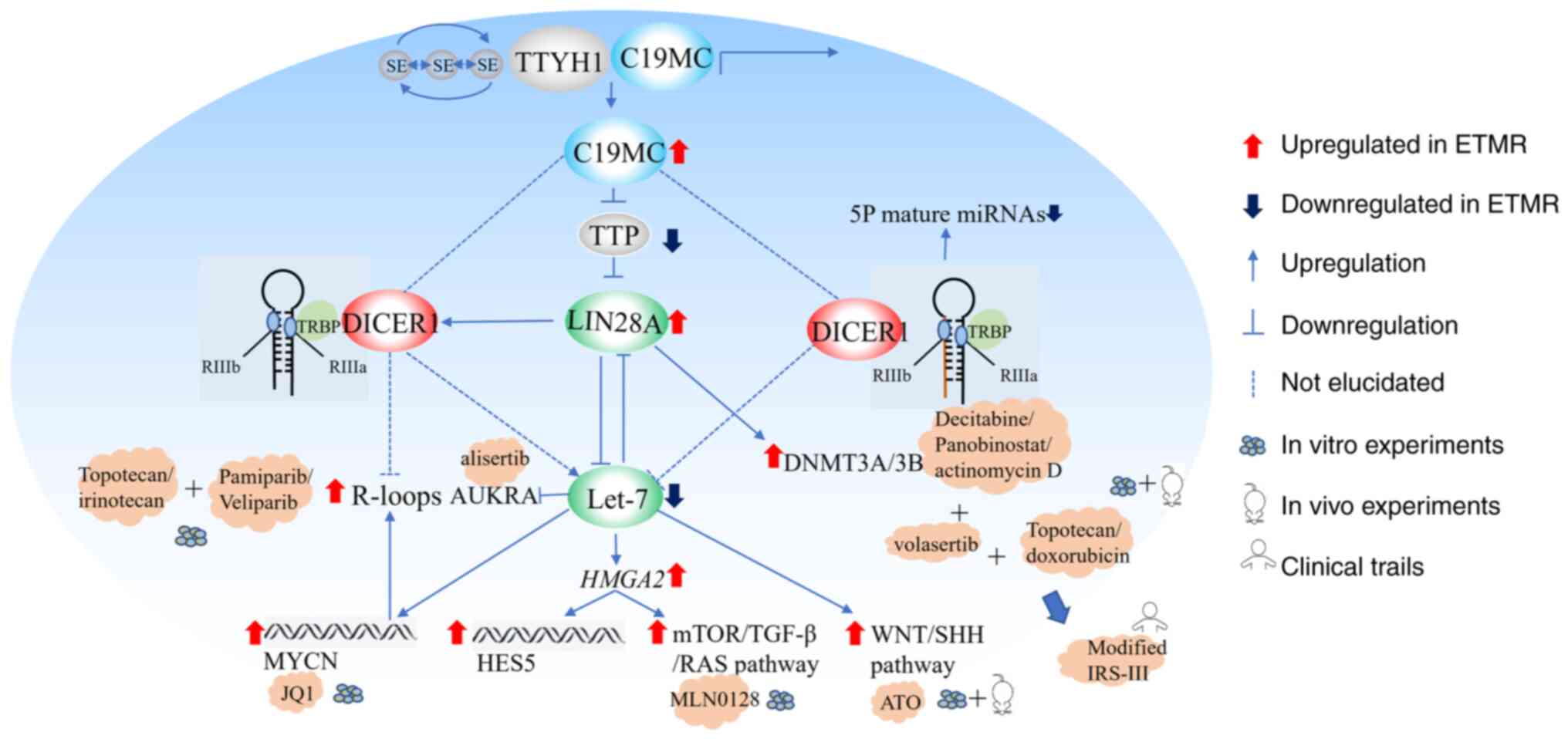
C19MC amplification-the signature gene
change in ETMR
C19MC, first reported in 2009(11), is the largest microRNA (miRNA or
miR) cluster unique to primates identified thus far, with a length
of >100 kb and encoding ~62 miRNAs (12,13).
The specific number of functional miRNAs remains unclear owing to
the conserved nature of the C19MC structure (12). C19MC is expressed only in
the placenta, testis and human embryonic stem cells, and its
expression decreases gradually with stem cell differentiation
(14). C19MC amplification
or fusion was found in 90% of patients with ETMR (5,12,15,16),
and is considered the only major recurrent genomic alteration
reported in ETMR to date (5,11,15-19).
C19MC is also expressed in ETMR without amplification,
albeit at an ~10-fold lower level, but not in the normal brain or
other brain tumors, which may be related to structural variations
(6). Regardless of C19MC
amplification, ETMR exhibit high molecular similarities (6). However, C19MC overexpression
is not unique to ETMR. C19MC amplification and tumor
suppressor p53 deletion are significant factors that drive
undifferentiated hepatic embryonic sarcoma development (20). In estrogen receptor-positive breast
cancer cells and hepatocellular carcinoma, C19MC
overexpression promotes cell cycle progression and induces
chemotherapy resistance, thereby increasing tumor cell viability.
Furthermore, C19MC overexpression is associated with poor
patient survival (21,22). Except for differences in tumor
distribution, C19MC amplification has no significant
relationship with the other clinical features of ETMR (6). Presently, it remains unclear whether
the absence or presence of C19MC amplification influences
the disease outcome (2).
ETMR are molecularly distinct entities based on the
analysis of miRNA data of ETMR and other pediatric intracranial
tumors (6,23). However, the differentially
expressed miRNAs in ETMR are similar. Transcriptional analyses
confirmed that mature miRNA expression, including C19MC
miRNAs and miR-17-92 miRNA clusters, increases whereas the
expression of let-7 miRNAs and miR-15 family miRNAs
decreases (23). However,
C19MC oncomiRs promote ETMR formation by synergistically
acting on the corresponding target genes. Enrichment analysis
revealed that the genes affecting neural stem cell maintenance,
epigenetic regulation and miRNA processes were upregulated, whereas
those affecting apoptosis, mRNA stability and neurogenesis were
downregulated. C19MC binds to the TTYH1 promoter to
facilitate tumor cell proliferation by targeting the cell
cycle-related tumor suppressors p21, P27 and RBL2. C19MC
stabilizes LIN28A and MYCN by targeting TTP, whereas LIN28A and
MYCN regulate tumor cells via DNMT3 and MAZ, respectively (23).
DICER1 as a potential ETMR driver
C19MC amplification, DICER1 mutation,
and MIR17HG amplification are involved in miRNA processes
(2,6). DICER1 is an RNase III kernel
enzyme involved in miRNA cytoplasmic processing. DICER1
deletion in mice inhibits cell proliferation and differentiation,
leading to early embryo death. DICER1 mutations in human
germlines, known as DICER1 syndrome, are associated with
tumor susceptibility and are characterized by early childhood tumor
development (24-28).
DICER1 biallelic mutations were found in 5% of ETMR cases,
which mainly occurred in cases without C19MC and
MIR17-92 miRNA amplification, including germline and somatic
mutations, which mainly occurred in the RNASEIII region and could
increase the proportion of 3p/5p miRNAs (2,6),
affecting miRNA processing. Currently, DICER1 is considered
the first ETMR susceptibility gene and a potential ETMR driver
(6). The MIR-17-92 miRNA
cluster is located on chromosome 13. The MIR17HG cluster is
a carcinogenic miRNA cluster associated with cancer proliferation
and increased invasiveness (29).
miR-17-92 miRNA cluster amplification was found in ~1% of
ETMR cases, with one case of C19MC amplification and two
cases of ETMR without C19MC amplification reported thus far.
During placental development, C19MC is co-expressed in
MIR17HG miRNA clusters (30), and several ‘seed’ sequences are
identical to the mature miRNA sequences encoded by C19MC,
suggesting that these two miRNA clusters may be co-regulated and
act on the same target genes (30,31).
In addition, CTNNB1 and TP53 mutations
were found in certain ETMR, and CTNNB1 mutations occurred in
10% ETMR (6,32,33),
which activated Wnt signaling by inhibiting β-catenin protein
degradation (34). TP53
mutations occur in 7% of ETMR (6).
Generally, there are relatively few frequent gene mutations
affecting the miRNA pathway, including the amplification of the
C19MC and miR-17-92 clusters and mutation of the
miRNA-processing gene, DICER1. Their distribution is
relatively exclusive (6).
Common downstream LIN28/let-7 pathway
of C19MC amplification or DICER1 mutation
LIN28A is an RNA-binding protein, which is involved
in regulating the development and self-renewal of embryonic stem
cells as a post-transcriptional regulator affecting the maturation
of miRNA (35-37).
Nearly all C19MC-amplified ETMR exhibit significant
enrichment of LIN28A (19,38), and LIN28A upregulation is
characteristic of aggressive malignant tumors (39). However, it is not unique to ETMR
and is also observed in 25% of atypical teratoid/rhabdoid tumor and
20% of high-grade gliomas, with LIN28A-positive tumor histology
tending to be ependymoblastoma, medulloepithelioma, or recurrent
cases, suggesting high sensitivity and low specificity of LIN28A
(19,40). Therefore, LIN28A positivity
supports the diagnosis of ETMR and is currently used as a marker
for ETMR.
LIN28A inhibits the maturation of let-7
miRNAs by binding to the terminal loop of the let-7
pre-miRNA and recruiting 3' terminal uridylyl transferase 4, which
causes reduced levels of mature let-7 miRNAs in the cell,
and further promotes the proliferation and metabolism of ETMR cells
by activating the mTOR signaling pathway (41-44).
The self-renewal capacity of cells is regulated by affecting
Wnt/Shh and NOTCH signaling (6,33,45).
Moreover, the proliferation and transcription processes of cells
are affected by promoting MYCN expression (46), whereas the DNA methylation state in
ETMR is affected by directly regulating DNMT3B and DNMT3A (23). Cumulative studies discovered
alternative targets for ETMR therapy (6,23,33,44,47-51).
DICER1 mutations may also affect LIN28A/let7
signaling. In ovarian Sertoli-Leydig cell tumors, Wilms tumors and
Uterine Corpus Endometrial Carcinoma with DICER1 RNase IIIb
mutations, let-7 targets are significantly upregulated
compared with other tumors that lack DICER1 RNase IIIb
mutations (52-54).
The phenotype resulting from DICER1 mutations can be
partially rescued by re-expressing let-7 miRNAs (53,54).
An increase in WNT and MYCN signaling was detected downstream of
the DICER1 mutations, which affected the RNase IIIb domain,
suggesting that DICER1 mutations have downstream effects
similar to those of LIN28A expression (52).
Role of ubiquitination in ETMR
Proteomics is rarely studied in ETMR, but holds out
the prospect to reflect functionally relevant tumor features more
closely. Ubiquitination is the second most common
post-translational modification of proteins following
phosphorylation (55).
Ubiquitination plays a crucial regulatory role in the modulation of
tumors, impacting cellular survival, proliferation and
differentiation. Integrated proteomics showed that histomorphology
stipulates the proteome signatures of ETMR; proteasome regulatory
proteins are highly abundant in ETMR, which indicates proteasome
inhibition as a promising therapeutic option in ETMR (47). Generally, C19MC
amplification is an indication of a gene change. DICER1
mutation and MIR17HG amplification mainly occur in
C19MC cases and influence the miRNA process as well as
C19MC. DICER1 is regarded as the first ETMR
susceptibility gene and a potential ETMR driver. High LIN28A
expression is a diagnostic marker of ETMR. LIN28A is involved in
the activation of multiple signaling pathways and presents a
potential therapeutic target. Proteasome regulatory particle
abundance is a distinctive, histology independent feature of ETMR;
proteasome inhibition represents a promising therapeutic
vulnerability in ETMR (47).
3. ETMR treatment progress
Traditional treatment regimens have provided only
limited improvement in patients with ETMR, and new treatment option
development and design of new targeted therapies remain
challenging. The fusion of TTYH1 with C19MC forms a
super enhancer-dependent original transcription and epigenetic
state (23). High C19MC,
MYCN and LIN28A expression was further promoted under the action of
the super enhancer. Several drugs have been identified as treatment
options in response to these molecular changes (Fig. 1).
Gualano et al (48) reported an ETMR patient with
long-term survival (>5 years), whose post-treatment
histopathology revealed maturation of undifferentiated embryonal
cells into mature neuronal and ganglionic phenotypes. This finding
suggests the notion of differentiation as a promising therapeutic
approach toward novel drug development for the treatment of deadly
pediatric brain tumors (48).
Targeting the ETMR metabolic pathway shows promise
for in vitro assays. Small-scale drug screens of 73
small-molecule inhibitors showed that ETMR cells were particularly
sensitive to mTOR, IGF1R, PI3K and topoisomerase inhibition, while
showing little to no sensitivity to other receptor tyrosine kinase
inhibitors (44). The BT183 ETMR
cell line was screened using 35 different compounds in another
study and was sensitive to a variety of drugs, including
topoisomerase inhibitors such as topotecan and daunorubicin,
epigenetic regulatory agents such as decitabine and Panobinostat,
actinomycin D, and some targeted drugs, including the PLK1
inhibitor volasertib, auroral kinase inhibitor alisertib, and mTOR
inhibitor MLN0128. In vivo verification indicated that
topotecan, volasertib and actinomycin D alone prolonged the
survival of mice and significantly inhibited tumor growth at
treatment initiation; however, complete remission was not achieved.
Further study of the potential therapeutic effects of topotecan and
daunorubicin in a multi-drug setting, when combined with
vincristine and methotrexate, respectively, demonstrated that
topotecan combined with chemotherapy resulted in improved survival
than daunorubicin; however, the study lacked long-term treatment or
disease control (49). Cocito
et al (50) generated two
novel patient-derived ETMR cell lines from resected patient derived
tumor samples and created three patient-derived xenograft models.
They further conducted high-throughput drug screening utilizing
2,480 approved and investigational drugs, against the
patient-derived ETMR cell lines. A total of 1,953 combinations were
selected; however, the subsequent data are currently being
validated in vivo (50).
Integrated proteomics showed that ETMR and BT183 cells harbor
proteasome regulatory proteins in abundance. Further, in
vitro assays using BT183 highlighted that ETMR tumor cells are
highly vulnerable toward treatment with the CNS penetrant
proteasome inhibitor Marizomib. However, the study is limited by
rather small case numbers. Consequently, not all molecular subtypes
may be sufficiently represented (47).
Based on the aforementioned in vitro
experimental results, the Dana-Farber Cancer Institute's modified
IRS-III protocol incorporates preclinical active agents, such as
doxorubicin and actinomycin D, into the treatment regimen for ETMR.
Hanson et al (51) included
five patients with ETMR, all of whom underwent complete tumor
resection and were treated with IRS-III for 12-51 weeks. A total of
four patients received local radiation therapy and the fifth
received high-dose chemotherapy with an autologous stem cell rescue
cycle. The results showed that the PFS rate of all four patients
was >18 months. A total of five patients with mild sinusoidal
obstructive syndrome and one patient with grade 3 peripheral
neuropathy tolerated chemotherapy (51). However, the small number of cases
included in the study and inconsistencies in the various additional
treatments also influenced the findings.
Genetic instability has also been exploited as a
potential therapeutic strategy. ETMR are highly sensitive to
topoisomerase inhibitors that dissolve the R-loop (56,57).
Topotecan and irinotecan act as TOP1 inhibitors via the covalent
binding of TOP1 to DNA (58), and
PARP1 can release TOP1 through adenosine diphosphate ribose. The
combination of TOP1 and Irinotecan has been shown to increase
R-loop formation and causes DNA damage. ETMR cell viability was
inhibited and this synergistic effect was dependent on sufficient
topoisomerase inhibition. However, this study requires in
vivo investigation (6).
LIN28A overexpression promotes Shh and Wnt signal
activation by down-regulating let7-miRNA in ETMR (6,33).
The SHH inhibitor arsenic trioxide (ATO) inhibits ETMR cell
proliferation and GLI expression in ETMR-xenografted mice, and
prolongs mice survival (33). ATO
also inhibits cell differentiation by acting on corresponding
targets (33). Thus, ETMR growth
decrease may be achieved by inducing cell differentiation, rather
than by specifically inhibiting SHH.
ETMR progression and pluripotency maintenance in
tumor cells depend on the high level of transcriptional activity
catalyzed by MYCN. JQ1, a BET inhibitor, binds competitively to the
BRD4 fusion oncoprotein (BRD4-NUT), thereby separating BRD4-NUT
from chromatin and leading to cancer cell differentiation and
apoptosis (59,60). JQ1 can significantly reduce the
viability of ETMR cells by competitively binding to the bromine
domain and inhibiting transcriptional activity while downregulating
the levels of C19MC miRNA, LIN28A and DNTMT3B6(23). However, further in vivo
studies are required to confirm this hypothesis.
Despite the availability of various therapeutic
strategies, ETMR remains a highly lethal disease. With the
development of molecular biology, ETMR diagnosis is no longer
ambiguous and further validation of these drugs through in
vitro experiments and in vivo models is necessary.
However, some difficulties remain. First, there is a limited number
of ETMR cases and few long-term survivors. Second, large-scale drug
screening is hampered by a lack of suitable in vitro and
murine models. There are currently two cell lines containing the
amplification of C19MC (BT183 and NCH3602); however, the
establishment of suitable xenotransplantation models is
complicated, possibly because of the unique microenvironment in
which ETMR originate, making them different when replicated in
murine xenotransplantation models. In addition, C19MC is
unique to primates, and the resulting murine
knockout/overexpression model may fail to replicate the unique
epigenomic complexity of ETMR.
4. Conclusions
ETMR is a highly aggressive intracranial tumor in
children. With recent studies on C19MC amplification,
genomic and epigenetic studies of ETMR in children have improved
our understanding of its biological nature. ETMR molecular
diagnosis, genetic alteration identification, and possible
therapeutic target exploration are also progressing. Despite
preclinical and clinical studies, the survival rates of patients
with ETMR have only slightly improved. These results highlight the
limitations of these studies and the lack of prospective drug
combination experiments. Second, because of the relatively rare
disease itself, few long-term survivors, and unclear prognostic
factors, the two ETMR cell lines found so far contain C19MC
amplification but lack other ETMR molecular characteristics and
drug targets, which complicates the exploration of new therapeutic
strategies as preclinical models. Therefore, it is necessary to
improve understanding of the molecular biological changes in ETMR;
further explore the relationship between C19MC
amplification, DICER1 mutations, and the LIN28/let-7
pathway; determine the role of ubiquitination in ETMR as well as
establish reasonable animal models; and provide possibilities for
further treatment through reasonably designed targeted therapy and
comprehensive preclinical trials.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Science and
Technology Planning Project of Sichuan (Project: Construction of
accurate diagnosis and hierarchical treatment system for pediatric
hematological tumor diseases-grant no. 2021YFS0027).
Availability of data and materials
The data generated in the present study are included
in the figures and/or tables of this article.
Authors' contributions
WL conceived the study and conducted majority of the
literature search and drafting the initial text. JG and XG
contributed critical revision during the development of the
manuscript. All authors read and approved the final version of the
manuscript. Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Li BK, Al-Karmi S, Huang A and Bouffet E:
Pediatric embryonal brain tumors in the molecular era. Expert Rev
Mol Diagn. 20:293–303. 2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lambo S, Von Hoff K, Korshunov A, Pfister
SM and Kool M: ETMR: A tumor entity in its infancy. Acta
Neuropathol (Berl). 140:249–266. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Su Y and Ma XL: The characteristics and
treatment of rare embryonal tumors of central nervous system in
children. Chin J Appl Clin Pediatr. 36:168–171. 2021.
|
|
4
|
Louis DN, Perry A, Wesseling P, Brat DJ,
Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM,
Reifenberger G, et al: The 2021 WHO classification of tumors of the
central nervous system: A summary. Neuro Oncol. 23:1231–1251.
2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kleinman CL, Gerges N, Papillon-Cavanagh
S, Sin-Chan P, Pramatarova A, Quang DA, Adoue V, Busche S, Caron M,
Djambazian H, et al: Fusion of TTYH1 with the C19MC microRNA
cluster drives expression of a brain-specific DNMT3B isoform in the
embryonal brain tumor ETMR. Nat Genet. 46:39–44. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Lambo S, Gröbner SN, Rausch T, Waszak SM,
Schmidt C, Gorthi A, Romero JC, Mauermann M, Brabetz S, Krausert S,
et al: The molecular landscape of ETMR at diagnosis and relapse.
Nature. 576:274–280. 2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Xu K, Sun Z, Wang L and Guan W: Embryonal
tumors with multilayered rosettes, C19MC-altered or not elsewhere
classified: Clinicopathological characteristics, prognostic
factors, and outcomes of 17 children from 2018 to 2022. Front
Oncol. 12(1001959)2022.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Juhnke BO, Gessi M, Gerber NU, Friedrich
C, Mynarek M, von Bueren AO, Haberler C, Schüller U, Kortmann RD,
Timmermann B, et al: Treatment of embryonal tumors with
multilayered rosettes with carboplatin/etoposide induction and
high-dose chemotherapy within the prospective P-HIT trial. Neuro
Oncol. 24:127–137. 2022.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Capper D, Jones DTW, Sill M, Hovestadt V,
Schrimpf D, Sturm D, Koelsche C, Sahm F, Chavez L, Reuss DE, et al:
DNA methylation-based classification of central nervous system
tumors. Nature. 555:469–474. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Raghuram N, Khan S, Mumal I, Bouffet E and
Huang A: Embryonal tumors with multi-layered rosettes: A disease of
dysregulated miRNAs. J Neurooncol. 150:63–73. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Li M, Lee KF, Lu Y, Clarke I, Shih D,
Eberhart C, Collins VP, Van Meter T, Picard D, Zhou L, et al:
Frequent amplification of a chr19q13.41 MicroRNA polycistron in
aggressive primitive neuroectodermal brain tumors. Cancer Cell.
16:533–546. 2009.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Bentwich I, Avniel A, Karov Y, Aharonov R,
Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al:
Identification of hundreds of conserved and nonconserved human
microRNAs. Nat Genet. 37:766–770. 2005.PubMed/NCBI View
Article : Google Scholar
|
|
13
|
Bortolin-Cavaille ML, Dance M, Weber M and
Cavaille J: C19MC microRNAs are processed from introns of large
Pol-II, non-protein-coding transcripts. Nucleic Acids Res.
37:3464–3473. 2009.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Bar M, Wyman SK, Fritz BR, Qi J, Garg KS,
Parkin RK, Kroh EM, Bendoraite A, Mitchell PS, Nelson AM, et al:
MicroRNA discovery and profiling in human embryonic stem cells by
deep sequencing of small RNA libraries. Stem Cells. 26:2496–2505.
2008.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Korshunov A, Remke M, Gessi M, Ryzhova M,
Hielscher T, Witt H, Tobias V, Buccoliero AM, Sardi I, Gardiman MP,
et al: Focal genomic amplification at 19q13.42 comprises a powerful
diagnostic marker for embryonal tumors with ependymoblastic
rosettes. Acta Neuropathol. 120:253–260. 2010.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Pfister S, Remke M, Castoldi M, Bai AHC,
Muckenthaler MU, Kulozik A, von Deimling A, Pscherer A, Lichter P
and Korshunov A: Novel genomic amplification targeting the microRNA
cluster at 19q13.42 in a pediatric embryonal tumor with abundant
neuropil and true rosettes. Acta Neuropathol. 117:457–464.
2009.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Korshunov A, Sturm D, Ryzhova M, Hovestadt
V, Gessi M, Jones DT, Remke M, Northcott P, Perry A, Picard D, et
al: Embryonal tumor with abundant neuropil and true rosettes
(ETANTR), ependymoblastoma, and medulloepithelioma share molecular
similarity and comprise a single clinicopathological entity. Acta
Neuropathol. 128:279–289. 2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Picard D, Miller S, Hawkins CE, Bouffet E,
Rogers HA, Chan TS, Kim SK, Ra YS, Fangusaro J, Korshunov A, et al:
Markers of survival and metastatic potential in childhood CNS
primitive neuro-ectodermal brain tumours: An integrative genomic
analysis. Lancet Oncol. 13:838–848. 2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Spence T, Sin-Chan P, Picard D, Barszczyk
M, Hoss K, Lu M, Kim SK, Ra YS, Nakamura H, Fangusaro J, et al:
CNS-PNETs with C19MC amplification and/or LIN28 expression comprise
a distinct histogenetic diagnostic and therapeutic entity. Acta
Neuropathol. 128:291–303. 2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Setty BA, Jinesh GG, Arnold M, Pettersson
F, Cheng CH, Cen L, Yoder SJ, Teer JK, Flores ER, Reed DR and Brohl
AS: The genomic landscape of undifferentiated embryonal sarcoma of
the liver is typified by C19MC structural rearrangement and
overexpression combined with TP53 mutation or loss. PLoS Genet.
16(e1008642)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Ward A, Shukla K, Balwierz A, Soons Z,
König R, Sahin Ö and Wiemann S: MicroRNA-519a is a novel oncomir
conferring tamoxifen resistance by targeting a network of
tumour-suppressor genes in ER + breast cancer. J Pathol.
233:368–379. 2014.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Fornari F, Milazzo M, Chieco P, Negrini M,
Marasco E, Capranico G, Capranico G, Mantovani V, Marinello J,
Sabbioni S, et al: In hepatocellular carcinoma miR-519d is
up-regulated by p53 and DNA hypomethylation and targets CDKN1A/p21,
PTEN, AKT3 and TIMP2. J Pathol. 227:275–285. 2012.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Sin-Chan P, Mumal I, Suwal T, Ho B, Fan X,
Singh I, Du Y, Lu M, Patel N, Torchia J, et al: A C19MC-LIN28A-MYCN
oncogenic circuit driven by hijacked Super-enhancers is a distinct
therapeutic vulnerability in ETMRs: A lethal brain tumor. Cancer
Cell. 36:51–67.e7. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Bernstein E, Kim SY, Carmell MA, Murchison
EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV and Hannon
GJ: Dicer is essential for mouse development. Nat Genet.
35:215–217. 2003.PubMed/NCBI View
Article : Google Scholar
|
|
25
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Bio. 15:509–524. 2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
De Kock L, Priest JR, Foulkes WD and
Alexandrescu S: An update on the central nervous system
manifestations of DICER1 syndrome. Acta Neuropathol. 139:689–701.
2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Foulkes WD, Priest JR and Duchaine TF:
DICER1: Mutations, microRNAs and mechanisms. Nat Rev Cancer.
14:662–672. 2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Uro-Coste E, Masliah-Planchon J, Siegfried
A, Blanluet M, Lambo S, Kool M, Roujeau T, Boetto S, Palenzuela G,
Bertozzi AI, et al: ETMR-like infantile cerebellar embryonal tumors
in the extended morphologic spectrum of DICER1-related tumors. Acta
Neuropathol. 137:175–177. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Mogilyansky E and Rigoutsos I: The
miR-17/92 cluster: A comprehensive update on its genomics,
genetics, functions and increasingly important and numerous roles
in health and disease. Cell Death Differ. 20:1603–1614.
2013.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Gu Y, Sun J, Groome LJ and Wang Y:
Differential miRNA expression profiles between the first and third
trimester human placentas. Am J Physiol Endocrinol Metab.
304:E836–E843. 2013.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Malnou EC, Umlauf D, Mouysset M and
Cavaillé J: Imprinted MicroRNA gene clusters in the evolution,
development, and functions of mammalian placenta. Front Genet.
9(706)2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Gessi M, Zur Muehlen A, Lauriola L,
Gardiman MP, Giangaspero F and Pietsch T: TP53, β-Catenin and
c-myc/N-myc status in embryonal tumours with ependymoblastic
rosettes: TP53, β-Catenin, c-myc/N-myc in embryonal tumors with
ependymoblastic rosettes. Neuropathol Appl Neurobiol. 37:406–413.
2011.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Neumann JE, Wefers AK, Lambo S, Bianchi E,
Bockstaller M, Dorostkar MM, Meister V, Schindler P, Korshunov A,
von Hoff K, et al: A mouse model for embryonal tumors with
multilayered rosettes uncovers the therapeutic potential of
Sonic-hedgehog inhibitors. Nat Med. 23:1191–1202. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wu G, Xu G, Schulman BA, Jeffrey PD,
Harper JW and Pavletich NP: Structure of a β-TrCP1-Skp1-β-catenin
complex. Mol Cell. 11:1445–1456. 2003.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Viswanathan SR, Daley GQ and Gregory RI:
Selective blockade of MicroRNA processing by Lin28. Science.
320:97–100. 2008.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Viswanathan SR and Daley GQ: Lin28: A
MicroRNA regulator with a macro role. Cell. 140:445–449.
2010.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Yu J, Vodyanik MA, Smuga-Otto K,
Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA,
Ruotti V, Stewart R, et al: Induced pluripotent stem cell lines
derived from human somatic cells. Science. 318:1917–1920.
2007.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Korshunov A, Ryzhova M, Jones DTW,
Northcott PA, Van Sluis P, Volckmann R, Koster J, Versteeg R,
Cowdrey C, Perry A, et al: LIN28A immunoreactivity is a potent
diagnostic marker of embryonal tumor with multilayered rosettes
(ETMR). Acta Neuropathol. 124:875–881. 2012.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Viswanathan SR, Powers JT, Einhorn W,
Hoshida Y, Ng TL, Toffanin S, O'Sullivan M, Lu J, Phillips LA,
Lockhart VL, et al: Lin28 promotes transformation and is associated
with advanced human malignancies. Nat Genet. 41:843–848.
2009.PubMed/NCBI View
Article : Google Scholar
|
|
40
|
Rao S, Rajeswarie RT, Chickabasaviah Yasha
T, Nandeesh BN, Arivazhagan A and Santosh V: LIN28A, a sensitive
immunohistochemical marker for embryonal tumor with multilayered
Rosettes (ETMR), is also positive in a subset of atypical
teratoid/rhabdoid tumor (AT/RT). Childs Nerv Syst. 33:1953–1959.
2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Hagan JP, Piskounova E and Gregory RI:
Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in
mouse embryonic stem cells. Nat Struct Mol Biol. 16:1021–1025.
2009.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho
J, Yeom KH, Han J and Kim VN: TUT4 in Concert with Lin28 suppresses
MicroRNA biogenesis through Pre-MicroRNA uridylation. Cell.
138:696–708. 2009.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhu H, Shyh-Chang N, Segrè AV, Shinoda G,
Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG,
et al: The Lin28/let-7 Axis regulates glucose metabolism. Cell.
147:81–94. 2011.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Spence T, Perotti C, Sin-Chan P, Picard D,
Wu W, Singh A, Anderson C, Blough MD, Cairncross JG, Lafay-Cousin
L, et al: A novel C19MC amplified cell line links Lin28/let-7 to
mTOR signaling in embryonal tumor with multilayered rosettes. Dev
Oncol. 16:62–71. 2014.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Patterson M, Gaeta X, Loo K, Edwards M,
Smale S, Cinkornpumin J, Xie Y, Listgarten J, Azghadi S, Douglass
SM, et al: let-7 miRNAs can act through notch to regulate human
gliogenesis. Stem Cell Rep. 3:758–773. 2014.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Molenaar JJ, Domingo-Fernández R, Ebus ME,
Lindner S, Koster J, Drabek K, Mestdagh P, van Sluis P, Valentijn
LJ, van Nes J, et al: LIN28B induces neuroblastoma and enhances
MYCN levels via let-7 suppression. Nat Genet. 44:1199–1206.
2012.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Dottermusch M, Biabani A, Lempertz T,
Schumann Y, Navolic J, Godbole S, Obrecht D, Frank S, Dorostkar MM,
Voß H, et al: Integrated proteomics spotlight the proteasome as a
therapeutic vulnerability in embryonal tumors with multilayered
rosettes. Neuro Oncol. 26:935–949. 2024.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Gualano FM, Hassoun P, Carter CL and
Hanson D: Embryonal tumor with multilayered rosettes:
Post-treatment maturation and implications for future therapy.
Cancer Reports. 6(e1812)2023.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Schmidt C, Schubert NA, Brabetz S, Mack N,
Schwalm B, Chan JA, Selt F, Herold-Mende C, Witt O, Milde T, et al:
Preclinical drug screen reveals topotecan, actinomycin D, and
volasertib as potential new therapeutic candidates for ETMR brain
tumor patients. Dev Oncol. 19:1607–1617. 2017.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Cocito C, Arias-Stella EU, Zhang X,
McKnight C, Itkin Z, Klumpp-Thomas C, Cruzeiro GA, Chi SN, Pisapia
DJ, Filbin MG and Dahmane N: ATRT-11. development of novel
preclinical models and therapeutic strategies for etmr. Neuro
Oncol. 25 (Suppl 1)(i3)2023.
|
|
51
|
Hanson D, Hoffman LM, Nagabushan S,
Goumnerova LC, Rathmann A, Vogel T, Ziegler DS and Chi S: A
modified IRS-III chemotherapy regimen leads to prolonged survival
in children with embryonal tumor with multilayer rosettes.
Neurooncol Adv. 2(vdaa120)2020.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Rakheja D, Chen KS, Liu Y, Shukla AA,
Schmid V, Chang TC, Khokhar S, Wickiser JE, Karandikar NJ, Malter
JS, et al: Somatic mutations in DROSHA and DICER1 impair microRNA
biogenesis through distinct mechanisms in Wilms tumors. Nat Commun.
5(4802)2014.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Vedanayagam J, Chatila WK, Aksoy BA,
Majumdar S, Skanderup AJ, Demir E, Schultz N, Sander C and Lai EC:
Cancer-associated mutations in DICER1 RNase IIIa and IIIb domains
exert similar effects on miRNA biogenesis. Nat Commun.
10(3682)2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Wang Y, Chen J, Yang W, Mo F, Senz J, Yap
D, Anglesio MS, Gilks B, Morin GB and Huntsman DG: The oncogenic
roles of DICER1 RNase IIIb domain mutations in ovarian
sertoli-leydig cell tumors. Neoplasia. 17:650–660. 2015.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Antao AM, Tyagi A, Kim KS and Ramakrishna
S: Advances in deubiquitinating enzyme inhibition and applications
in cancer therapeutics. Cancers (Basel). 12(1579)2020.PubMed/NCBI View Article : Google Scholar
|
|
56
|
El Hage A, French SL, Beyer AL and
Tollervey D: Loss of topoisomerase I leads to R-loop-mediated
transcriptional blocks during ribosomal RNA synthesis. Gene Dev.
24:1546–1558. 2010.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Staker BL, Hjerrild K, Feese MD, Behnke
CA, Burgin AB and Stewart L: The mechanism of topoisomerase I
poisoning by a camptothecin analog. Proc Natl Acad Sci.
99:15387–15392. 2002.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Das SK, Rehman I, Ghosh A, Sengupta S,
Majumdar P, Jana B and Das BB: Poly(ADP-ribose) polymers regulate
DNA topoisomerase I (Top1) nuclear dynamics and camptothecin
sensitivity in living cells. Nucleic Acids Res. 44:8363–8375.
2016.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Smith SG and Zhou MM: The bromodomain: A
new target in emerging epigenetic medicine. ACS Chem Biol.
11:598–608. 2016.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Filippakopoulos P, Qi J, Picaud S, Shen Y,
Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et
al: Selective inhibition of BET bromodomains. Nature.
468:1067–1073. 2010.PubMed/NCBI View Article : Google Scholar
|