Introduction
Attention deficit hyperactivity disorder (ADHD) is
the most commonly diagnosed neurobehavioral disorder in children
and adolescents, and is characterized by several symptoms,
particularly inattention, hyperactivity and impulsivity (1). It has been estimated that over 5% of
all school-aged children and adolescents meet criteria for ADHD.
ADHD has become a public health problem causing a significant
financial cost, stress to families, impact on academic and
vocational activities, as well as negative effects on self-esteem
(2).
Although the etiology of ADHD is unknown, family,
twin and adoption studies have demonstrated that ADHD is highly
heritable. It is widely accepted that several genes, which include
dopamine transporter gene (DAT1), dopamine D4 receptor gene (DRD4),
synaptosomal-associated protein of 25 kDa gene (SNAP-25), DRD5,
serotonin transporter gene (5-HTT) and serotonin receptor 1B
(5-HTR1B), each contributing a small fraction of the total genetic
variance, are implicated in ADHD (3). Animal, pharmacological and imaging
studies have indicated dopamine system dysfunction in the
pathogenesis of ADHD; therefore, dopaminergic system genes may be
candidate genes for ADHD (2).
Recently, polymorphisms at dopamine-related genes, such as
catechol-O-methyltransferase (COMT), DAT1 and DRD4, many of which
cause observed alterations in protein function or structure, have
been identified, prompting researchers to test their role in
increasing the risk of ADHD (4–5).
Studies of ADHD have focused less on GABAergic dysfunction than on
dopaminergic dysfunction. A study found that the γ-aminobutyric
acid (GABA) level in the blood increased and the ratio of
excitatory/inhibitory mediatory amino acids decreased significantly
in children with minimal cerebral dysfunction (6); moreover, the GABAergic agent
extended-release valproate (EVA) could decrease ADHD symptoms
(7–8). Whether GABAergic dysfunction has
direct or indirect importance in ADHD is unclear, and an
association study between the GABAergic genes and ADHD may be able
to clarify its role in the susceptibility to ADHD. To date, the
correlation of polymorphisms in GABAergic genes to ADHD has not
been reported.
Despite many efforts, the search for susceptibility
genes for ADHD has yielded largely inconsistent results. To
evaluate the involvement of a series of functional allelic variants
of dopaminergic/GABAergic genes in ADHD using polymerase chain
reaction-restriction fragment length polymorphism (PCR-RFLP), we
genotyped five functional single nucleotide polymorphisms (SNPs),
rs6275 polymorphism in the DRD2 gene, rs5320 polymorphism in the
dopamine beta-hydroxylase (DBH) gene, rs5326 polymorphism in the
DRD1 gene, rs4680 polymorphism in the COMT gene and rs1805057
polymorphism in the gamma-aminobutyric acid B receptor 1 (GABBR1)
gene, in 54 cases of ADHD and 67 healthy controls in Chinese Han
children.
Materials and methods
Sample
In total, 54 ADHD cases (40 males and 14 females)
were recruited from the outpatient clinic of Qilu Children’s
Hospital of Shandong University between January 2009 and June 2010.
To be included in the study, children had to meet the following
three criteria: i) have a diagnosis of ADHD classified according to
the DSM-IV (9); ii) be between the
ages of 7 and 16 years and have a full scale IQ above 70 according
to the Wechsler Intelligence scale for Chinese children
(standardized by Gong Yaoxian); iii) originate from the Han
population. Individuals were excluded for any evidence of major
neurological conditions or a primary diagnosis of schizophrenia,
affective disorder, pervasive developmental disorder or epilepsy.
In all patients, 85.7% had the combined subtype and 14.3% the
inattentive subtype of ADHD.
The healthy control group included 67 healthy
children (49 males and 18 females) who underwent a health
examination in Qilu Children’s Hospital of Shandong University. The
group was selected from the Han population and aged from 7 to 16
years. Subjects with relevant psychiatric/psychological or
developmental illnesses were excluded. The mean ages of the ADHD
and control groups were 9.14 (SD=1.556) and 9.06 (SD=1.732),
respectively. This study was approved by the ethics committee of
Qilu Children’s Hospital of Shandong University. All subjects were
unrelated individuals, and all subjects and their guardians
provided written informed consent.
Genotyping
Genomic DNA was extracted from the venous blood of
each participant using a TIANamp blood DNA kit (Tiangen Biotech Co.
Ltd., Beijing, China). PCR cycling was performed in a DNA thermal
cycler with a 2X Hotstart Taq PCR Mastermix kit (Tiangen Biotech
Co. Ltd.). The sequences of the primers and reaction conditions
used in this study are shown in Table
I.
 | Table IThe sequences of the primers and
reaction conditions for dopaminergic/GABAergic genes. |
Table I
The sequences of the primers and
reaction conditions for dopaminergic/GABAergic genes.
| SNPs | Sequences of
primers | Fragment size
(bp) | Annealing (°C) | Incision enzyme |
|---|
| rs4680 | F (5′-3′)
GGGGCCTACTGTGGCTACTC | 173 | 58 | NlaIII |
| R (5′-3′)
TTTTCCAGGTCTGACAACGG | | | |
| rs5326 | F (5′-3′)
AAGGGAGTCAGAAGACAGAT | 317 | 56 | BfaI |
| R (5′-3′)
CAGGCAGTGAGGATACGAAC | | | |
| rs6275 | F (5′-3′)
GGGAGCTGGAGATGGAGATGC | 358 | 59.5 | NcoI |
| R (5′-3′)
ATGGGACCTTTCACAGACCG | | | |
| rs1805057 | F (5′-3′)
AACAGTAACACAAACCATCC | 439 | 53 | EagI |
| R (5′-3′)
GCATGTTTGTAGAAGGTGCC | | | |
| rs5320 | F (5′-3′)
CACTGTCCACTTGGTCTACG | 218 | 56 | PmlI |
| R (5′-3′)
GCTCCTTAATGTAGCACCAG | | | |
In a 25 μl reaction, 200 ng of genomic DNA was used
with 1.0 μl of each primer (10 μmol/l) and 12.5 μl of 2X Hotstart
Taq PCR Mastermix. Samples were amplified using the following
cycles: initial denaturation of 3 min at 94°C, then 35 sec of
denaturation at 94°C, 35 sec at the annealing temperature, and 50
sec of extension at 72°C, repeated for 31 cycles and followed by
another 5 min period of extension at 72°C. PCR products were
further purified, and then digested by NlaIII, BfaI,
NcoI, EagI and PmlI, respectively. The
digested PCR products were confirmed by electrophoresis on 2.5%
agarose gels. Genotypes were determined by gel image processing
software.
Statistical analysis
The data were statistically processed with SPSS 11.0
software (SPSS, USA). Allele and genotype frequencies were compared
between two groups using the Chi-square test. Observed and expected
allele frequencies within populations were compared by means of a
Hardy-Weinberg test. To determine high- and low-risk genotype
combinations, a multifactor dimensionality reduction (MDR) method
was used. This procedure was specifically developed to detect
higher order interactions between polymorphisms to predict a
dichotomous trait variation, even when the marginal effects were
very small. The fitness of an MDR model was assessed by estimating
the testing accuracy; models that were true positive would have an
estimated testing accuracy of 0.5.
Results
Detection of genotypes of
dopaminergic/GABAergic genes
The A260/A280 ratios of
genomic DNA from blood specimens were 1.7–1.9; this meant that the
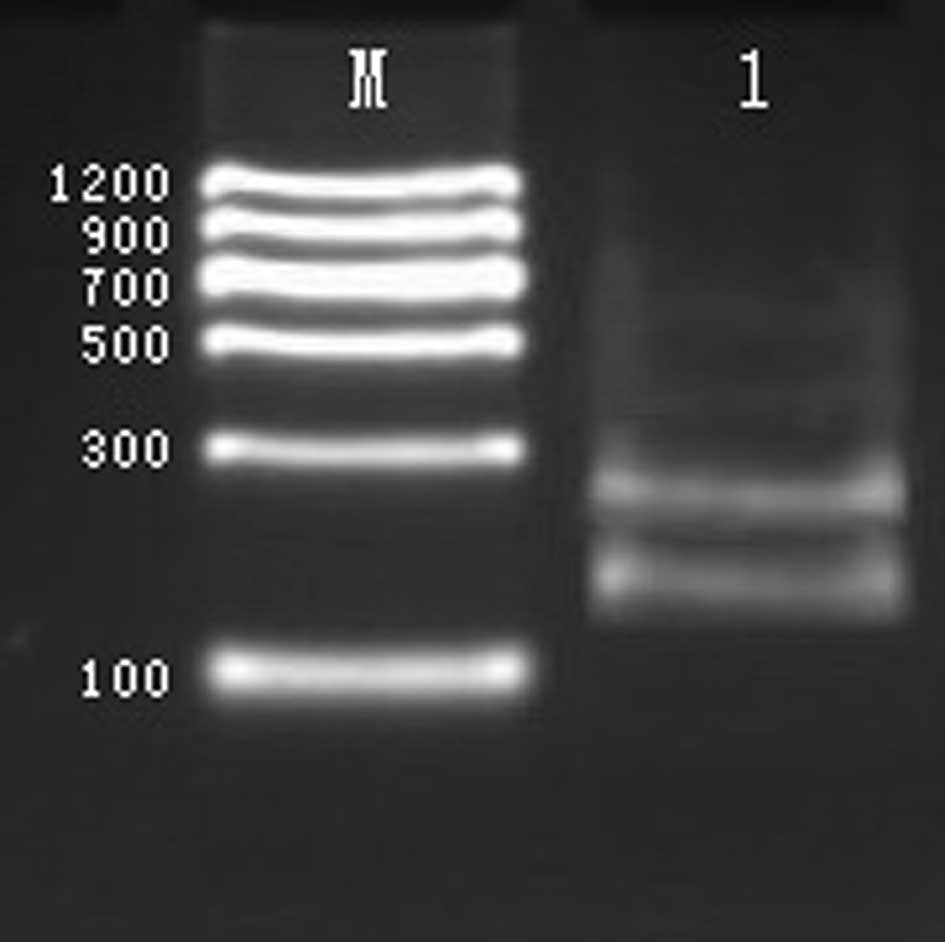
genomic DNA was eligible to be used as a template for PCR. By PCR,
a 173 bp fragment was amplified from the COMT gene, a 439 bp
fragment from the GABBR1 gene, a 218 bp fragment from the DBH gene,
a 317 bp fragment from the DRD1 gene, and a 358 bp fragment from
the DRD2 gene, as shown in Fig.
1.
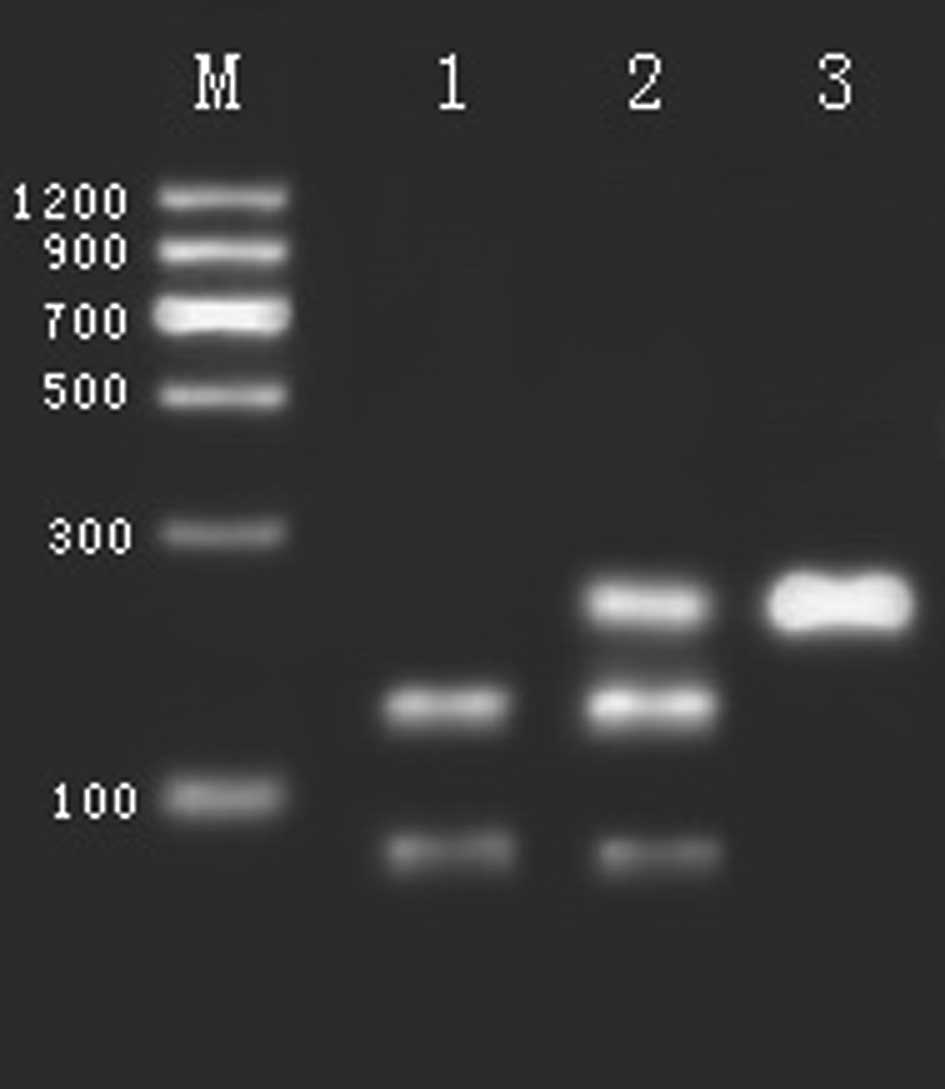
At the rs4680 polymorphism in the COMT gene digested
by NlaIII, there were three genotypes: a) the predominant
homozygote, GG, indicated by three fragments (26, 33 and 114 bp);
b) the heterozygote, GA, indicated by five fragments (18, 26, 33,
96 and 114 bp); and c) the rare homozygote, AA, as shown in
Fig. 2 (18, 26 and 33 bp cannot be
seen). At the rs5326 polymorphism in the DRD1 gene digested by
BfaI, there were three genotypes: a) the predominant
homozygote, GG, indicated by one fragment (317 bp); b) the
heterozygote, GA, indicated by three fragments (139, 178 and 317
bp; and c) the rare homozygote, AA, as shown in Fig. 3. At the rs6275 polymorphism in the
DRD2 gene digested by NcoI, there were three genotypes: a)
the CC genotype, indicated by one fragment (358 bp); (b) the
heterozygote, CT, indicated by three fragments (106, 252 and 358
bp); and c) the TT genotype, as shown in Fig. 4. At the rs1805057 polymorphism in
the GABBR1 gene digested by EagI, only the GG genotype was
observed in all patients and controls, as shown in Fig. 5. At the rs5320 polymorphism in the
DBH gene digested by PmlI, there were three genotypes: a)
the predominant homozygote, GG, indicated by one fragment (218 bp);
b) the heterozygote, GA, indicated by three fragments (77, 141 and
218 bp; and c) the rare homozygote, AA, as shown in Fig. 6.
 | Figure 2Restriction endonuclease analysis of
polymorphism rs4680 in the COMT gene. M, DNA marker; Lane 1, GA 18,
26, 33, 96 and 114 bp. Lane 2, GG genotype 26, 33 and 114 bp. Lane
3, AA 18, 26, 33 and 96 bp (18, 26 and 33 bp cannot be seen). |
Hardy-Weinberg equilibrium
No significant differences were found between the
ADHD and control groups in the genotypic frequencies of the rs6275
polymorphism (χ2=0.601, 0.071, P>0.05), rs5320
polymorphism (χ2=0.888, 0.023, P>0.05), rs5326
polymorphism (χ2=0.184, 0.207, P>0.05) and rs4680
polymorphism (χ2=0.264, 0.071, P>0.05), and the
results showed goodness of fit for Hardy-Weinberg equilibrium,
indicating that population stratification was not a factor in the
results.
Genotypes and allele frequencies of four
polymorphisms in dopaminergic/GABAergic genes in two groups
The distribution of the AA genotype and A allele
frequencies of the rs5320 polymorphism in the DBH gene in ADHD
children differed significantly from that in healthy controls
(P=0.016, 0.014; OR=9.484, 2.222, respectively). The genotype
distribution and allele frequencies of the rs6275 polymorphism in
the DRD2 gene, the rs5326 polymorphism in the DRD1 gene and the
rs4680 polymorphism in the COMT gene showed no significant
difference between ADHD children and healthy controls
(χ2=2.716, 2.003; χ2=2.045, 1.554;
χ2=0.503, 0.258, respectively, P>0.05), as shown in
Table II.
 | Table IIComparisons of genotypes and allele
frequencies of four polymorphisms in dopaminergic/GABAergic genes
in two groups. |
Table II
Comparisons of genotypes and allele
frequencies of four polymorphisms in dopaminergic/GABAergic genes
in two groups.
| Groups | n | | Genotypes [n
(%)] | Allele frequencies
(%) |
|---|
| rs5320 | | AA | AG | GG | A | G |
| ADHD group | 54 | 6 (11.1)a | 17 (31.5) | 31 (57.4) | 26.9b | 73.1 |
| Control group | 67 | 1 (1.5) | 17 (25.4) | 49 (73.1) | 14.2 | 85.8 |
| rs6275 | | TT | TC | CC | T | C |
| ADHD group | 54 | 10 (18.5)c | 31 (57.4) | 13 (24.1) | 47.2c | 52.8 |
| Control group | 67 | 21 (31.3) | 34 (50.8) | 12 (17.9) | 56.7 | 43.3 |
| rs5326 | | GG | GA | AA | G | A |
| ADHD group | 54 | 25 (46.3)d | 22 (40.7) | 7 (13.0) | 66.7d | 33.3 |
| Control group | 67 | 39 (58.2) | 23 (34.3) | 5 (7.5) | 75.4 | 24.6 |
| rs4860 | | AA | AG | GG | A | G |
| ADHD group | 54 | 4 (7.4)e | 18 (33.3) | 32 (59.3) | 24.1e | 75.9 |
| Control group | 67 | 3 (4.5) | 22 (32.8) | 42 (62.7) | 20.9 | 79.1 |
MDR analysis
Multilocus analysis was performed using the MDR
method. Of all the models analyzed, one four-locus model had a
maximum cross-validation consistency of ten out of ten, a maximum
prediction accuracy of 61.98%, and a maximum sensitivity of 64.61%.
It also had the smallest P-value in the sign test for
cross-validation (P<0.0001). The best model contains four-locus
SNPs consisting of rs5320, rs6275, rs5326 and rs4680, as shown in
Table III.
 | Table IIIGene-gene interaction model and
characteristics. |
Table III
Gene-gene interaction model and
characteristics.
| Model |
rs5320/rs6275/rs5326 |
rs5320/rs6275/rs5326/rs4680 |
|---|
| Training
accuracy | 0.6731 | 0.741 |
| Test accuracy | 0.4959 | 0.6198 |
| χ2 | 12.2003 | 24.3338 |
| Significance test
(P) | 0.0005 | <0.0001 |
| CV consistency | 9/10 | 10/10 |
| Sensitivity | 0.5988 | 0.6461 |
| Specificity | 0.733 | 0.8176 |
| Odds ratio | 4.0969 | 8.1819 |
| 95% CI | 1.8243–9.2006 | 3.3967–19.7084 |
Discussion
ADHD is associated with impaired school performance,
peer relationships, parent-child relationships, behavior and
emotions (10–11). The incidence has been increasing
year by year. Although the etiology of ADHD is not fully
understood, convincing data support the hypothesis that genetic
factors play a fundamental role in the etiology of ADHD. The
estimated heritability of ADHD is 70–90% (12).
Several lines of evidence indicate dopamine system
dysfunction in the pathogenesis of ADHD (13). It is widely accepted that
dopaminergic genes, encoding for enzymes, receptors and
transporters, each contributing a small fraction of the total
genetic variance, are implicated in ADHD (5,14).
DBH is responsible for conversion of dopamine to
norepinephrine. It is released along with catecholamines from the
adrenal medulla and from sympathetic nerve endings, and its coding
gene has been mapped to chromosome 9q34 (15). Numerous polymorphisms in the DBH
gene have been found to independently exert their direct effect on
the enzyme activity. Kopecková et al found that the risk of
ADHD was significantly increased in the presence of the A allele of
polymorphism G444A as well as in the presence of the T allele of
polymorphism C1603T in the DBH gene (16). Another study found that the -1021
C/T polymorphism, accounting for up to 50% of the enzymatic
activity, was associated with ADHD (17). Further study is required to
determine whether other polymorphisms are involved in the
development of ADHD. In this study, a functional polymorphism,
rs5320 in the DBH gene, was observed with PCR-RFLP analysis in 54
cases of ADHD and 67 healthy controls, and the distribution of the
AA genotype and A allele frequencies of this polymorphism in ADHD
children differed significantly from that in healthy controls. Our
finding introduces a novel exonic SNP in the DBH gene showing a
putative association with ADHD, which adds further support for the
involvement of the DBH gene in ADHD. However, the sample size in
this study is relatively small, which decreased statistical power,
so the results may be due to chance although nominally significant.
Moreover, this finding requires replication in independent
populations of various ethnic origins.
Evidence from human and animal studies suggested
that the DRD1 gene was a good candidate for involvement in ADHD
(18). Bobb et al found
that ADHD probands were more likely than controls to have the C
allele of rs4532 and the A allele of rs265981 in the DRD1 gene
(19). Another polymorphism -94
G/A (rs5326) in the DRD1 gene was evaluated in this study, and the
genotype distribution and allele frequencies of this polymorphism
showed no significant difference between children with ADHD and
healthy controls. There are several possible explanations for this
negative result. First, this selected SNP has a very small main
effect or no direct effect on ADHD, or our sample lacked the power
to detect it. Second, it is feasible that environmental risk
factors may interact with genetic risk factors, reducing or
abolishing its main effect from genotype alone; in this case we
would have little chance of detecting such associations unless we
had also measured environmental risk factors.
DRD2-deficient mice displayed reduced locomotor
activity, as well as reduced spontaneous movements (20). The human DRD2 gene is located on
chromosome 11q22–23, and consists of 8 exons separated by 7 introns
(21). Several polymorphisms were
identified shortly after the gene was cloned; these SNPs include
the TaqI restriction site (TaqI sites ‘A’, ‘B’ and
‘D’) and one short tandem repeat polymorphism. Some research found
a highly positive correlation between ADHD and TaqI A
polymorphism (rs1800497) in the DRD2 gene (22–23).
However, results of a study by Todd et al did not support
the DRD2 gene TaqI A contributing to susceptibility for ADHD
(24). This study explored a
functional polymorphism, rs6275 in the DRD2 gene, to expound the
involvement of the gene in ADHD, but we failed to establish whether
the genotype distribution and allele frequencies of this
polymorphism were significantly different between children with
ADHD and healthy controls. The inconsistency of the previous
association studies between the DRD2 gene and ADHD may be due to
genetic heterogeneity, which is a significant challenge in the
genetic study of ADHD. It could also be argued that ethnic
differences, sample demographics, such as gender stratification due
to the predominance of male subjects, and study populations with
comorbidity in these studies accounted for the contradictory
results. Further studies including larger sample sizes are
certainly required to confirm whether the rs6275 polymorphism in
the DRD2 gene is associated with ADHD. Moreover, the association
between other DRD2 gene polymorphisms and ADHD requires further
investigations to determine the role of the DRD2 gene in ADHD.
COMT plays a crucial role in the metabolism of
catecholamines in the frontal cortex, and its encoding gene is on
chromosome 22q11.2. Interest in COMT comes from its involvement in
dopaminergic pathways. The most studied polymorphism in COMT is an
SNP, Val158Met (rs4680), which leads to either methionine (Met) or
valine (Val) at codon 158, resulting in a three- to four-fold
reduction in COMT activity. This polymorphism is functional with
the val/val genotype increasing enzyme activity. Pálmason et
al found that the Met allele of Val158Met was associated with
ADHD and increased ADHD symptom severity (25). Contrary to the previously observed
association, we failed to gain a positive result for rs4680 in
COMT. The possible explanations for this include the heterogeneity
between our sample and others, and the possibility that these
results are false-negative findings.
GABA is the principal inhibitory neurotransmitter in
the human brain. Since ADHD is a disorder of disinhibition, it is
reasonable to suggest that genetic defects in GABA could be
involved (26). GABA exerts its
effects mainly through two receptor subtypes termed A and B. GABA B
receptors, including subunit 1 and 2, are highly expressed in the
limbic system. In 1998, the gene encoding GABA B receptors subunit
1 (GABBR1) was cloned, and several polymorphisms of the human
GABBR1 gene were identified, but only two were missense mutations.
The missense mutation G1465A (rs1805057) led to an amino acid
substitution, Gly489Ser, within the N-terminal extracellular domain
of GABBR1, which was necessary for heteromeric assembly of such a
receptor and, therefore, may affect its ligand binding properties.
To explore the possible role of the rs1805057 polymorphism in the
etiology of ADHD, we examined the distribution of genotypes and
allele frequencies of this SNP; however, only genotype GG was
observed in all patients and healthy controls. This study failed to
document any association between the rs1805057 polymorphism and
ADHD, and this polymorphism may be ethno-geographically
localized.
In general, the dopamine receptors are found on
inhibitory GABA neurons (27).
Certain studies showed that dopaminergic receptors, such as the D2
receptor were involved in regulating GABA synthesis in the
striatonigral system. On the other hand, the GABA B subtype of
receptor, not the GABA A subtype, is possibly involved in reducing
the release of dopamine and controlling the activity of
nigrostriatal dopaminergic neurons by regulating the
cAMP-generating system (28–29).
Interactions of the dopaminergic and GABAergic system may play a
role in the pathogenesis of ADHD. In addition, ADHD is a multigene
disease, and the SNP association study in one gene was insufficient
to explain the complex genetic basis of ADHD susceptibility.
Therefore, in this study, we evaluated the multilocus genetic
interactions involved in the development of ADHD using MDR
analysis. Of the models detected using MDR, we chose the four-locus
model as the best one for determining ADHD susceptibility based on
its balanced accuracy and cross-validation consistency. This model
included four genotypes: rs5320 polymorphism, rs6275 polymorphism,
rs5326 polymorphism and rs4680 polymorphism in dopaminergic genes,
which may contribute more markedly to susceptibility to ADHD than
any single genetic polymorphism.
In conclusion, our findings suggested that rs5320
polymorphism in the DBH gene was likely to be associated with the
susceptibility to ADHD in Chinese Han children, but rs6275
polymorphism in the DRD2 gene, rs5326 polymorphism in the DRD1
gene, rs4680 polymorphism in the COMT gene, and rs1805057
polymorphism in the GABBR1 gene, were not confirmed to be directly
related to ADHD. A multilocus SNP, including rs5320, rs6275, rs5326
and rs4680 polymorphisms in dopaminergic/GABAergic genes, acted in
combination to affect the susceptibility to ADHD, and the potential
gene-gene interaction existed among dopaminergic/GABAergic genes in
the genetic pathology of ADHD.
Acknowledgements
The authors gratefully extend their gratitude to all
subjects for their participation and cooperation in this study, as
well as to Da-wei Wang for rewriting the manuscript.
References
|
1
|
Hong Q, Wang YP, Zhang M, et al: Homer
expression in the hippocampus of an animal model of
attention-deficit/hyperactivity disorder. Mol Med Report.
4:705–712. 2011.PubMed/NCBI
|
|
2
|
Genro JP, Kieling C, Rohde LA and Hutz MH:
Attention-deficit/hyperactivity disorder and the dopaminergic
hypotheses. Expert Rev Neurother. 10:587–601. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Elia J and Devoto M: ADHD genetics: 2007
update. Curr Psychiatry Rep. 9:434–439. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sharp SI, Mcquillin A and Gurling HM:
Genetics of attention-deficit hyperactive disorder (ADHD).
Neuropharmacology. 57:590–600. 2009. View Article : Google Scholar
|
|
5
|
Gizer IR, Ficks C and Waldman ID:
Candidate gene studies of ADHD: a meta-analytic review. Hum Genet.
126:51–90. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kolesnichenko LS, Kulinskiĭ VI and Gorina
AS: Amino acids and their metabolites in blood and urine of
children with minimal cerebral dysfunction. Vopr Med Khim.
45:58–64. 1999.(In Russian).
|
|
7
|
Miyazaki M, Ito H, Saijo T, et al:
Favorable response of ADHD with giant SEP to extended-release
valproate. Brain Dev. 28:470–472. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Powell SG, Thomsen PH, Frydenberg M and
Rasmussen H: Long-term treatment of ADHD with stimulants: a large
observational study of real-life patients. J Atten Disord.
15:439–451. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
American Psychiatric Association (APA).
Diagnostic and statistical manual of mental disorders. 4th ed.
American Psychiatric Press; Washington DC: 1994
|
|
10
|
Li J, Kang C, Zhang H, et al: Monoamine
oxidase A gene polymorphism predicts adolescent outcome of
attention-deficit/hyperactivity disorder. Am J Med Genet B.
144B:430–433. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Barnard-Brak L, Sulak TN and Fearon DD:
Coexisting disorders and academic achievement among children with
ADHD. J Atten Disord. 15:506–515. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Domschke K, Sheehan K, Lowe N, et al:
Association analysis of the monoamine oxidase A and B genes with
attention deficit hyperactivity disorder (ADHD) in an Irish sample:
preferential transmission of the MAO-A 941G allele to affected
children. Am J Med Genet B Neuropsychiatr Genet. 134B:110–114.
2005. View Article : Google Scholar
|
|
13
|
DiMaio S, Grizenko N and Joober R:
Dopamine genes and attention-deficit hyperactivity disorder: a
review. J Psychiatry Neurosci. 28:27–38. 2003.PubMed/NCBI
|
|
14
|
Fisher SE, Francks C, McCracken JT, et al:
A genomewide scan for loci involved in
attention-deficit/hyperactivity disorder. Am J Hum Genet.
70:1183–1196. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Craig SP, Buckle VJ, Lamouroux A, Mallet J
and Craig IW: Localization of the human dopamine beta hydroxylase
(DBH) gene to chromosome 9q34. Cytogenet Cell Genet. 48:48–50.
1988. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kopecková M, Paclt I, Petrásek J, Pacltová
D, Malíková M and Zagatová V: Some ADHD polymorphisms (in genes
DAT1, DRD2, DRD3, DBH, 5-HTT) in case-control study of 100 subjects
6–10 age. Neuro Endocrinol Lett. 29:246–251. 2008.PubMed/NCBI
|
|
17
|
Kieling C, Genro JP, Hutz MH and Rohde LA:
The -1021 C/T DBH polymorphism is associated with
neuropsychological performance among children and adolescents with
ADHD. Am J Med Genet B Neuropsychiatr Genet. 147B:485–490. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Misener VL, Luca P, Azeke O, et al:
Linkage of the dopamine receptor D1 gene to
attention-deficit/hyperactivity disorder. Mol Psychiatry.
9:500–509. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bobb AJ, Addington AM, Sidransky E, et al:
Support for association between ADHD and two candidate genes: NET1
and DRD1. Am J Med Genet B Neuropsychiatr Genet. 134B:67–72. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Balk JH, Picetti R, Saiardi A, et al:
Parkinsonian-like locomotor impairment in mice lacking dopamine D2
receptors. Nature. 377:424–428. 1995. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Grandy DK, Litt M, Allen L, et al: The
human dopamine D2 receptor gene is located on chromosome 11 at q22-
q23 and identifies a TaqI RFLP. Am J Hum Genet. 45:778–785.
1989.PubMed/NCBI
|
|
22
|
Paclt I, Drtilkova I, Kopeckova M, Theiner
P, Serý O and Cermakova N: The association between TaqI A
polymorphism of ANKK1 (DRD2) gene and ADHD in the Czech boys aged
between 6 and 13 years. Neuro Endocrinol Lett. 31:131–136.
2010.PubMed/NCBI
|
|
23
|
Serý O, Drtílková I, Theiner P, et al:
Polymorphism of DRD2 gene and ADHD. Neuro Endocrinol Lett.
27:236–240. 2006.PubMed/NCBI
|
|
24
|
Todd RD and Lobos EA: Mutation screening
of the dopamine D2 receptor gene in attention-deficit hyperactivity
disorder subtypes: preliminary report of a research strategy. Am J
Med Genet. 114:34–41. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pálmason H, Moser D, Sigmund J, et al:
Attention-deficit/hyperactivity disorder phenotype is influenced by
a functional catechol-O-methyltransferase variant. J Neural Transm.
117:259–267. 2010.PubMed/NCBI
|
|
26
|
Comings DE: Clinical and molecular
genetics of ADHD and Tourette syndrome. Two related polygenic
disorders. Ann N Y Acad Sci. 931:50–83. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sagvolden T, Johansen EB, Aase H and
Russell VA: A dynamic developmental theory of
attention-deficit/hyperactivity disorder (ADHD) predominantly
hyperactive/impulsive and combined subtypes. Behav Brain Sci.
28:397–468. 2005. View Article : Google Scholar
|
|
28
|
Amantea D and Bowery NG: Reduced
inhibitory action of a GABAB receptor agonist on [3H]-dopamine
release from rat ventral tegmental area in vitro after chronic
nicotine administration. BMC Pharmacol. 4:242004.
|
|
29
|
Hossain MA and Weiner N: Interactions of
dopaminergic and GABAergic neurotransmission: impact of
6-hydroxydopamine lesions into the substantia nigra of rats. J
Pharmacol Exp Ther. 275:237–244. 1995.PubMed/NCBI
|