Introduction
Osteoporosis is the most common and serious skeletal
disorder among the elderly. Symptomatic osteoporosis occurs due to
a decreased bone mineral density (BMD) leading to reduced bone
strength and an increased risk of fractures (1). Low bone mass in the elderly is highly
dependent on their peak bone mass (PBM) as young adults (2). Therefore, it is necessary to
understand and identify the risk factors for impaired PBM in young
and middle-aged adults.
Osteopenia may result from an imbalance between
increased bone resorption and decreased bone formation (3,4).
Bone resorption involves the dissolution of bone mineral and
degradation of the organic bone matrix. These two functions are
performed by osteoclasts. Osteoclasts are members of the
monocyte/macrophage lineage and are formed by multiple instances of
cellular fusion of their mononuclear precursors (5). Monocytes differentiate into
osteoclasts in the presence of various molecular signals (6). RANKL, one of the most frequently
studied, is a ligand for the receptor activator of nuclear
factor-κB (NF-κB; RANK) on osteoclast precursor cells (7). RANKL/RANK signaling activates four
pathways that mediate osteoclast formation; NF-κB, c-fos and
calcineurin/NFATc1 and three pathways that mediate osteoclast
activation; Src and MKK6/p38/MITF and survival; Src and
extracellular signal-regulated kinase (8). Osteoblasts produce and secrete
osteoprotegerin, a decoy receptor that binds to RANKL and blocks
RANKL/RANK interactions and hence suppresses the ability of RANK to
increase bone resorption (9).
Previous studies have shown that blood monocytes also produce a
wide variety of inflammatory factors and transcription factors
involved in bone metabolism, including interleukin-1 (10), tumor necrosis factor-α (TNF-α)
(11), interleukin-6 (12), platelet-derived growth factor
(13), transforming growth
factor-β (14), resolvinE1
(15), runt-related transcription
factor 2 (Runx2; 16), guanylate binding protein 1 (GBP1), signal
transducer and activator of transcription 1 (STAT1), CXC chemokine
ligand 10 (CXCL10) (17),
chemokine receptor 3, histidine decarboxylase and glucocorticoid
receptor genes (18).
However, it is unknown whether other mechanisms
regulating these factors are significant in the ability of
monocytes to affect bone metabolism. Since biological processes are
mediated by multiple, co-regulated genes working in synchrony,
certain unknown genes may be assigned potential biological
functions when studied in gene sets with known genes and ontology
groups (19). Thus, the objective
of this study was to screen the differential gene expression in
monocytes using a high-throughput microarray platform and to
explore gene expression profiles comprehensively by grouping
individual differentially expressed genes (DEGs) into gene sets and
Gene Ontology (GO) terms. The DEGs between high and low PBM samples
were grouped into nine gene sets using the graph-clustering
approach. GO term enrichment analysis was applied to identify the
relevant molecular functions in response to an impaired PBM. The
current study revealed that the DEGs, as precursors of osteoclasts,
are functionally involved in the immune response. The stimulus
response may contribute to differential osteoclastogenesis, leading
to differential PBM levels.
Materials and methods
Affymetrix microarray data
Circulating monocyte affymetrix microarray datasets
were accessible from the National Center for Biotechnology
Information Gene Expression Omnibus (GEO) data repository
(http://www.ncbi.nlm.nih.gov/geo/) using
the series accession number GSE7158. Fourteen subjects with
extremely high PBM levels and 12 subjects with extremely low PBM
levels were selected for DNA microarray experiments. All the
recruited volunteers signed an informed consent form prior to
entering this project.
Statistical analysis
The limma method (20) was used to identify DEGs. The raw
expression datasets from all conditions were normalized using the
Robust Multiarray Average (RMA) method with the default settings
implemented in Bioconductor and then the linear model was
constructed. DEGs with a fold change >1.5 and P<0.05 were
selected.
The Pearson correlation coefficient (r) was used to
compare the potential correlations between DEGs. Statistical
significance was set at r>0.95 and P<0.05. All statistical
tests were performed using R language (21).
Network analyses and graph
clustering
To identify co-expressed groups, DPClus, a graph
clustering algorithm that extracts densely connected nodes as a
cluster, was used (22). DPClus is
based on the density and periphery tracking of clusters and is
freely available from http://kanaya.naist.jp/DPClus/. In the current study,
the overlapping mode with the DPClus settings were used. The
parameter settings of cluster properties were set; density values
were set to 0.5 (23) and minimum
cluster size was set to 2.
GO term enrichment analysis
The GO (24)
project is a major bioinformatics initiative with the aim of
standardizing the representation of genes and gene product
attributes across species and databases. The project provides a
controlled vocabulary of terms for describing gene product
characteristics and gene product annotation data from GO Consortium
members, as well as tools to access and process this data.
The DAVID tool (25) was used to identify overrepresented
GO terms in biological process. P<0.05 and counts of >2 were
set as the threshold for the analysis using the hypergeometric
distribution.
Results
Differential gene expression profiling
and co-expression network construction
GSE7158 microarray datasets were publicly available
from the GEO database. Following microarray analysis, a total of 49
genes were selected as DEGs with a fold change >1.5 and
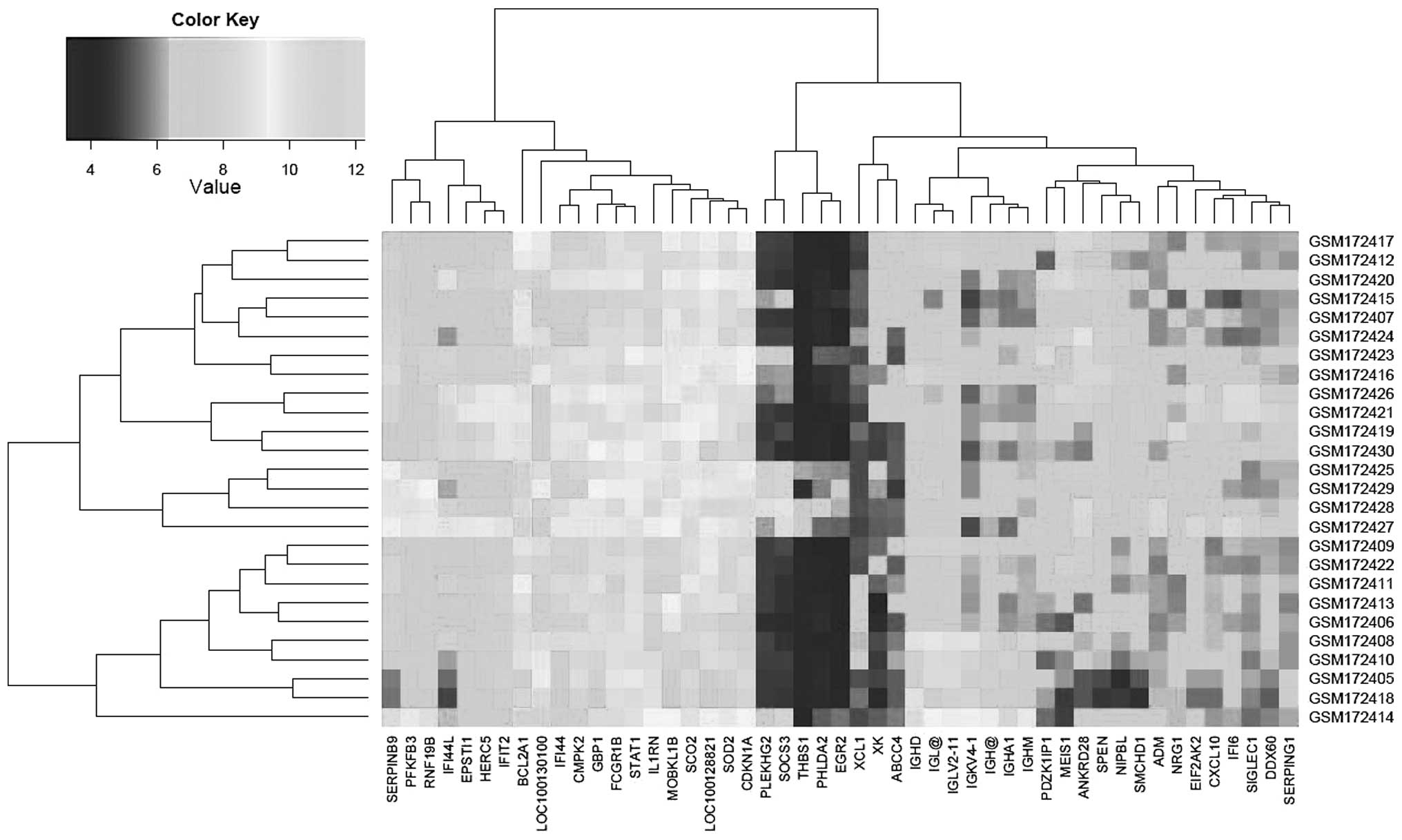
P<0.05. The expression profiling of these 49 DEGs is presented
in Fig. 1.
To form the correlations between DEGs, r>0.7 and
P<0.05 were selected as the cut-off points. A correlation
network was constructed with a total of 159 correlations among 49
DEGs (Fig. 2).
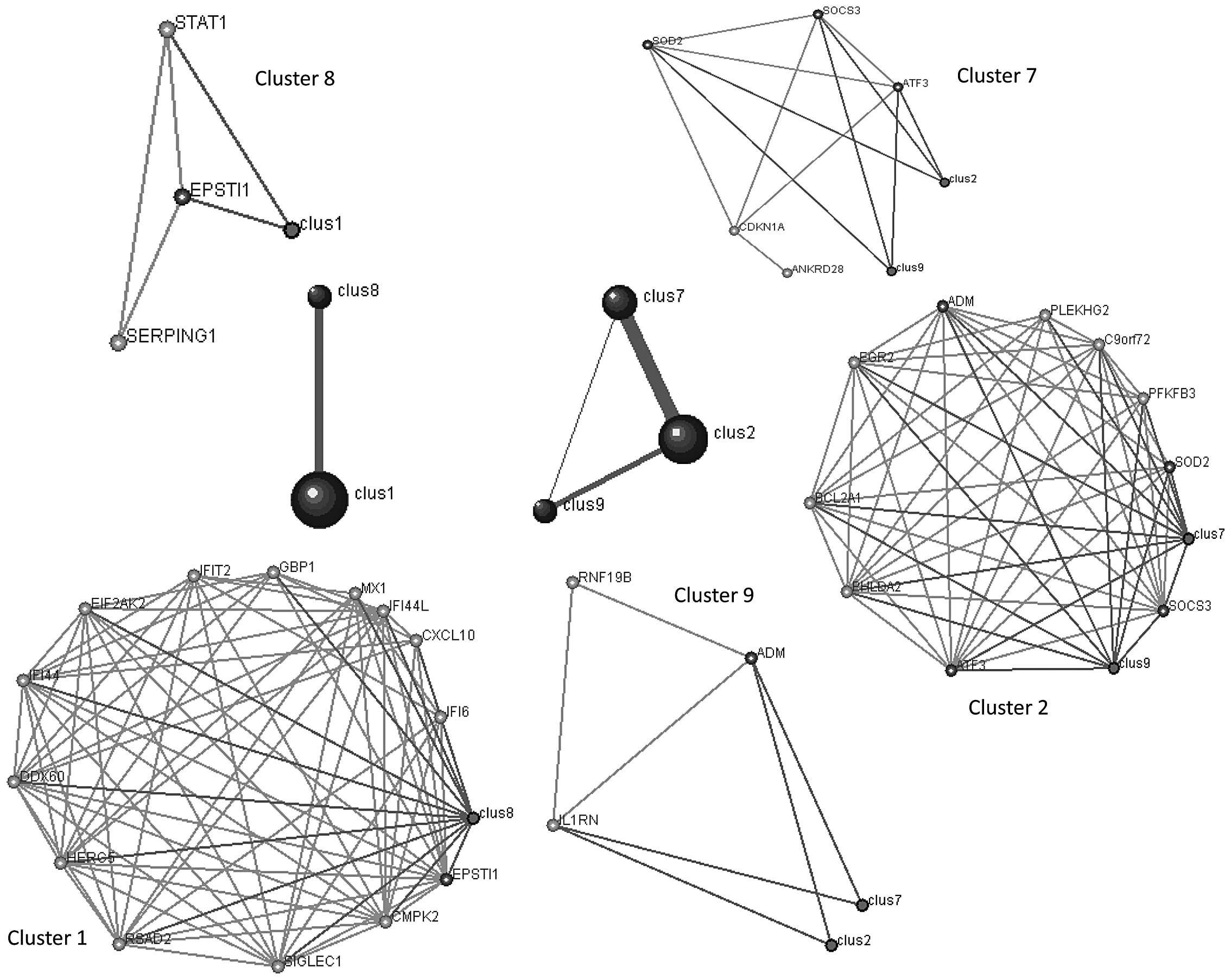
Graph clustering identifies modules
significantly enriched for DEGs contained in GO term pathways
At r>0.7, DPClus (22) identified 9 clusters in the
correlation network for osteoporosis, ranging in size from 3–14
genes. Clusters 1, 2, 7, 8 and 9 were connected as they shared the
same genes. For example, one gene (epithelial stromal interaction
1, EPSTI1) was shared between clusters 1 and 8; three genes
(suppressor of cytokine signaling, SOCS3; superoxide dismutase,
SOD2 and activating transcription factor 3, ATF3) were shared
between cluster 2 and 7 and one gene (adrenomedullin, ADM) was
shared between clusters 2 and 9. The higher the number of genes
shared, the more connectivity among them (corresponding to the
thicker lines; Fig. 3).
To assess the significance of the obtained clusters,
the overrepresented GO terms were used. Enrichment analysis was
performed using the hypergeometrical distribution to find the
significant GO term enrichment pathways. In accordance with the
graph clustering results, the genes in clusters 1 and 8 were
enriched in similar pathways, including immune responses and
circulatory system processes. The genes in clusters 2, 7 and 9 were
enriched in similar pathways regulating apoptosis and responding to
various stimuli, including insulin, hypoxia, nutrients, drugs,
radiation and hormones (Table I).
Clusters 2 and 7 had the most similar GO term enrichment pathways.
These GO biological processes may be relevant to the
differentiation of monocytes into osteoclasts.
 | Table IList of enriched GO terms in clusters
1, 2, 7, 8 and 9 detected by DPClus. |
Table I
List of enriched GO terms in clusters
1, 2, 7, 8 and 9 detected by DPClus.
| Category | Term | Description | Count | P-value | FDR |
|---|
| Cluster 1 | GO:0009615 | Response to
virus | 4 |
5.86e−5 | 0.006833 |
| GO:0006955 | Immune
response | 5 | 0.00110057 | 0.062388 |
| GO:0006952 | Defense
response | 4 | 0.00882795 | 0.292359 |
| Cluster 2 | GO:0032868 | Response to insulin
stimulus | 3 | 0.00147149 | 0.378515 |
| GO:0007568 | Aging | 3 | 0.00177687 | 0.249653 |
| GO:0001666 | Response to
hypoxia | 3 | 0.00262243 | 0.246268 |
| GO:0070482 | Response to oxygen
levels | 3 | 0.00289863 | 0.208958 |
| GO:0043434 | Response to peptide
hormone stimulus | 3 | 0.00344654 | 0.19991 |
| GO:0006915 | Apoptosis | 4 | 0.00415134 | 0.200641 |
| GO:0012501 | Programmed cell
death | 4 | 0.00432956 | 0.181443 |
| GO:0031667 | Response to
nutrient levels | 3 | 0.00557639 | 0.202104 |
| GO:0006916 | Anti-apoptosis | 3 | 0.00608264 | 0.196651 |
| GO:0008219 | Cell death | 4 | 0.00684652 | 0.199007 |
| GO:0009991 | Response to
extracellular stimulus | 3 | 0.00691087 | 0.184238 |
| GO:0016265 | Death | 4 | 0.00698058 | 0.171843 |
| GO:0042981 | Regulation of
apoptosis | 4 | 0.00934738 | 0.208115 |
| GO:0043067 | Regulation of
programmed cell death | 4 | 0.00960751 | 0.19967 |
| GO:0010941 | Regulation of cell
death | 4 | 0.00970618 | 0.189438 |
| GO:0010332 | Response to gamma
radiation | 2 | 0.01352424 | 0.240339 |
| GO:0031100 | Organ
regeneration | 2 | 0.01527641 | 0.253599 |
| GO:0048666 | Neuron
development | 3 | 0.01586432 | 0.249457 |
| GO:0043066 | Negative regulation
of apoptosis | 3 | 0.01722442 | 0.255741 |
| GO:0043069 | Negative regulation
of programmed cell death | 3 | 0.01768878 | 0.250411 |
| GO:0060548 | Negative regulation
of cell death | 3 | 0.01778231 | 0.241164 |
| GO:0009725 | Response to hormone
stimulus | 3 | 0.01844308 | 0.23914 |
| GO:0009628 | Response to abiotic
stimulus | 3 | 0.01853835 | 0.231094 |
| GO:0006873 | Cellular ion
homeostasis | 3 | 0.01911445 | 0.228747 |
| GO:0055082 | Cellular chemical
homeostasis | 3 | 0.01969829 | 0.226664 |
| GO:0009719 | Response to
endogenous stimulus | 3 | 0.0222131 | 0.24351 |
| GO:0050801 | Ion
homeostasis | 3 | 0.02262762 | 0.239518 |
| GO:0030182 | Neuron
differentiation | 3 | 0.02573112 | 0.259709 |
| GO:0019725 | Cellular
homeostasis | 3 | 0.02888801 | 0.27855 |
| GO:0048878 | Chemical
homeostasis | 3 | 0.03440423 | 0.314043 |
| GO:0010212 | Response to
ionizing radiation | 2 | 0.03494492 | 0.309692 |
| GO:0031099 | Regeneration | 2 | 0.0400934 | 0.338357 |
| GO:0032496 | Response to
lipopolysaccharide | 2 | 0.04464963 | 0.360509 |
| GO:0009266 | Response to
temperature stimulus | 2 | 0.04805437 | 0.373652 |
| GO:0002237 | Response to
molecule of bacterial origin | 2 | 0.04975276 | 0.375599 |
| Cluster 7 | GO:0070482 | Response to oxygen
levels | 3 |
3.21e−4 | 0.079885 |
| GO:0009314 | Response to
radiation | 3 |
6.46e−4 | 0.080292 |
| GO:0042493 | Response to
drug | 3 |
7.53e−4 | 0.062989 |
| GO:0009991 | Response to
extracellular stimulus | 3 |
7.81e−4 | 0.049354 |
| GO:0055093 | Response to
hyperoxia | 2 | 0.00177318 | 0.087833 |
| GO:0043066 | Negative regulation
of apoptosis | 3 | 0.00201309 | 0.08331 |
| GO:0043069 | Negative regulation
of programmed cell death | 3 | 0.00206992 | 0.073801 |
| GO:0060548 | Negative regulation
of cell death | 3 | 0.00208138 | 0.06523 |
| GO:0009628 | Response to abiotic
stimulus | 3 | 0.00217417 | 0.060715 |
| GO:0010332 | Response to gamma
radiation | 2 | 0.00509224 | 0.123857 |
| GO:0031100 | Organ
regeneration | 2 | 0.00575517 | 0.12707 |
| GO:0048145 | Regulation of
fibroblast proliferation | 2 | 0.00774219 | 0.154437 |
| GO:0042127 | Regulation of cell
proliferation | 3 | 0.0097487 | 0.177311 |
| GO:0042981 | Regulation of
apoptosis | 3 | 0.01016581 | 0.172238 |
| GO:0043067 | Regulation of
programmed cell death | 3 | 0.01036499 | 0.164649 |
| GO:0010941 | Regulation of cell
death | 3 | 0.01044016 | 0.156241 |
| GO:0010212 | Response to
ionizing radiation | 2 | 0.01324778 | 0.183871 |
| GO:0031099 | Regeneration | 2 | 0.0152248 | 0.198085 |
| GO:0007568 | Aging | 2 | 0.02419781 | 0.283882 |
| GO:0014070 | Response to organic
cyclic substance | 2 | 0.02659589 | 0.294663 |
| GO:0001666 | Response to
hypoxia | 2 | 0.02942492 | 0.308128 |
| GO:0048545 | Response to steroid
hormone stimulus | 2 | 0.04197997 | 0.396429 |
| GO:0031667 | Response to
nutrient levels | 2 | 0.0430572 | 0.390801 |
| GO:0010035 | Response to
inorganic substance | 2 | 0.0447791 | 0.39006 |
| GO:0006916 | Anti-apoptosis | 2 | 0.04499419 | 0.379328 |
| Cluster 8 | GO:0008015 | Blood
circulation | 2 | 0.01374926 | 0.839184 |
| GO:0003013 | Circulatory system
process | 2 | 0.01374926 | 0.839184 |
| Cluster 9 | GO:0051384 | Response to
glucocorticoid stimulus | 2 | 0.01149882 | 0.709892 |
| GO:0031960 | Response to
corticosteroid stimulus | 2 | 0.01252751 | 0.490566 |
| GO:0048545 | Response to steroid
hormone stimulus | 2 | 0.02818517 | 0.6393 |
Discussion
In the current study, differential expression
profiling was systematically investigated and its possible role in
the differentiation of osteoclasts was explored. A total of 49 DEGs
were identified and correlated to produce 159 network connections.
These DEGs were assigned into nine clusters using the graph
clustering method in response to different PBM levels. A total of
14 genes were included in cluster 1 [GBP1; interferon-induced
protein with tetratricopeptide repeats 2, IFIT2; eukaryotic
translation initiation factor 2-α kinase 2, EIF2AK2;
interferon-induced protein 44, IFI44; IFI44L; DEAD
(Asp-Glu-Ala-Asp) box polypeptide 60, DDX60; HECT and RLD domain
containing E3 ubiquitin protein ligase 5, HERC5; radical S-adenosyl
methionine domain containing 2, RSAD2; sialic acid binding Ig-like
lectin 1, sialoadhesin, SIGLEC1; cytidine monophosphate kinase 2,
CMPK2; EPSTI1; interferon, α-inducible protein 6, IFI6; CXCL10; and
myxovirus resistance 1, interferon-inducible protein p78, MX1] and
3 genes were involved in cluster 8 (STAT1; EPSTI1; and serpin
peptidase inhibitor, clade G, SERPING1). Notably, cluster 8 was
connected with all the genes of cluster 1 by STAT1 and EPSTI1 in
order to be involved in immune responses and circulatory system
processes, as demonstrated in previous studies.
The immune system has been correlated with bone
resorption through a complex interaction involving T and B
lymphocytes, dendritic cells (DCs), cytokines and cell-cell
interactions (26). There is
strong evidence that STAT1 is significant in bone metabolism as
STAT1 has been reported to be upregulated in the femur tissue of
osteoporotic mice (27) and humans
(18). STAT1 may serve as a
primary mediator of interferon (IFN) signaling pathways involving
osteoclast differentiation. Through the p38 MAPK pathway, RANKL
stimulates the serine phosphorylation of STAT1, resulting in the
migration and adhesion of osteoclast precursors (28). STAT1 interacts with Runx2, an
essential transcription factor for osteoblast differentiation, in
its latent form in the cytoplasm, thereby inhibiting the nuclear
localization of Runx2. This function of STAT1 does not require the
Tyr 701 that is phosphorylated when STAT1 becomes a transcriptional
activator (29).
The GBP1 gene is also predicted to be involved in
bone metabolism or osteoclast differentiation (30) in a STAT1-dependent manner (31). The sumoylation-defective STAT1
mutant exhibits increased induction of GBP1 and transporters
associated with antigen presentation 1 (TAP1) transcription
(32). The mutation in the STAT1
gene dramatically reduces the inducibility of the GBP1 and TAP1
genes by IFN (33). In this study,
STAT1 and GBP1 directly interacted with each other (Figs. 2 and 3).
Chemokines have a potential role in the regulation
of osteoclast functions. For example, IFN-γ-inducible protein-10
(CXCL10) is expressed in human osteoclasts with changing expression
levels during osteoclast differentiation (34). CXCL10 has been suggested to
contribute to osteoclastogenesis by increasing RANKL expression in
CD4+ T cells in an animal model of rheumatoid arthritis
(35). Notably, previous studies
have shown that osteoblasts secrete IFN-β in response to viral
infections and that endogenous IFN-β induces CXCL10 and IFI44L
production via an IFN-α/β receptor-STAT1 pathway (36,37).
EIF2AK2 is also reported to interact with STAT1 and
increase its degradation. Reduction of EIF2AK2 activity also
reduces RUNX2 activity and murine osteoblast differentiation
(38,39). Therefore, it appears illogical that
EIF2AK2 is upregulated in human osteoblasts following IFN-β
treatment which results in an inhibition of mineralization
(40).
Ten genes were included in cluster 2 [ADM; early
growth response 2, EGR2; BCL2-related protein A1, BCL2A1;
chromosome 9 open reading frame 72, C9orf72; pleckstrin
homology-like domain, family A, member 2, PHLDA2; ATF3; SOCS3;
SOD2; 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3,
PFKFB3; and pleckstrin homology domain containing, family G (with
RhoGef domain) member 2, PLEKHG2], five genes were included in
cluster 7 (SOCS3; SOD2; ATF3; cyclin-dependent kinase inhibitor 1A,
CDKN1A; and ankyrin repeat domain 28, ANKRD28) and three genes were
included in cluster 9 (ADM; interleukin 1 receptor antagonist,
IL1RN; and ring finger protein 19B, RNF19B). Cluster 7 was
connected with all the genes of cluster 2 and 9 by SOCS3, SOD2 and
ATF3. Cluster 9 was connected with all the genes of clusters 2 and
7 by ADM and IL1RN. These findings indicate that SOCS3, SOD2, ATF3
and ADM are significant genes for responding to various stimuli,
including insulin, hypoxia, nutrients, drugs, radiation and
hormones regulating apoptosis.
The SOCS3 family are cytoplasmic adaptor proteins
that negatively regulate various cytokine responses in leukocytes.
SOCS3 overexpression augments TGF-β, TNF-α and RANKL-induced
osteoclast formation, priming precursors to the osteoclast lineage
by suppressing specific anti-osteoclastic JAK/STAT signals
(41). Zhang et al
demonstrated that a higher SOCS3 expression level is associated
with RANKL-mediated alveolar bone loss and enhances
CD11c+ DC-derived osteoclastogenesis in vivo and
in vitro. The reduced expression of functional SOCS3 in
CD11c+ DCs results in significantly lower
osteoclastogenesis and dendritic cell-derived osteoclasts
development during immune interactions with T cells, based on TRAP
expression and bone resorptive activity (42). In SOCS3-deficient bone
marrow-derived monocytes, the expression levels of
TNF-receptor-associated factor-6 and IκB are drastically reduced.
The receptor activation of NF-κB ligand-induced IκB phosphorylation
is severely impaired, indicating that SOCS3 regulates
osteoclastogenesis by blocking the inhibitory effect of
inflammatory cytokines on receptor activation of the NF-κB
ligand-mediated osteoclast differentiation signals (43).
ADM is a 52-amino acid peptide first described in a
human phaeochromocytoma but has since been identified in numerous
tissues, including the bone (44).
Systemic administration of ADM stimulates the proliferation of
osteoblasts and promotes bone growth (45). Treatment with ADM significantly
blunts the apoptosis of serum-deprived osteoblastic cells,
evaluated by caspase-3 activity, DNA fragmentation quantification
and Annexin V-FITC labeling. This effect is eliminated by
calcitonin-related polypeptide α (CGRP1) and insulin-like growth
factor-I (46). The selective
inhibitor of MAPK kinase (MEK), PD98059, also eliminates the
protective effect of ADM on apoptosis and prevents ADM activation
of ERK1/2. These data show that ADM acts as a survival factor in
osteoblastic cells via a CGRP1 receptor-MEK-ERK pathway, which
provides further understanding on the physiological function of ADM
in osteoblasts (47).
The SOD2 gene encodes a free radical-scavenging
enzyme that removes superoxidate and catalyzes the production of
hydrogen peroxide. Oxidative stress is significant in the
pathogenesis of osteoporosis (48). Previous studies have revealed that
SOD2 is significantly upregulated in circulating monocytes at the
mRNA and protein level in vivo in Chinese patients with low
versus high hip BMD levels (49).
Women with postmenopausal osteoporosis have significantly higher
plasma SOD enzyme activity levels than those in controls (50). This indicates that SOD2 is
significant in the pathogenesis of osteoporosis, promoting
osteoclast differentiation, formation and activity (51).
EGR2 is a highly conserved transcription factor
involved in bone remodeling. The upregulation of EGR2 is involved
in the biological affinity of titanium for osteogenic cells and in
the promotion of osteoblast differentiation (52). Macrophage colony-stimulating factor
activates MEK/ERK and induces the MEK-dependent expression of the
immediate early gene EGR2. Inhibition of either MEK1/2 or EGR2
increases osteoclast apoptosis (53). Previous studies have revealed a
novel role for EGR2 in postnatal skeletal metabolism. EGR2+/− mice
reveal a low bone mass phenotype. EGR2 silencing in pre-osteoclasts
increases the expression of cFms and the response to macrophage
colony-stimulating factor, leading to a cell-autonomous stimulation
of cell-cycle progression. Thus, the anti-mitogenic role of EGR2 in
pre-osteoclasts is the predominant mechanism underlying the low
bone mass phenotype of EGR2-deficient mice (54).
The osteoporotic state increases ATF3 expression in
dorsal root ganglia neurons innervating L3 vertebrae (55). BCL2A1, an anti-apoptotic activated
macrophage protein, is also heavily overexpressed in osteolysis
patients, providing a possible mechanism for the persistence of the
particle-laden cells expressing macrophage phenotype activation
markers (56).
In conclusion, the present findings shed new light
on the biology of osteoporosis and have implications for future
research. The changes in the immune system (GBP1, STAT1, CXCL10 and
EIF2AK2) and stimulus response (SOCS3, SOD2, ATF3, ADM EGR2 and
BCL2A1) may be associated with osteoclast differentiation. This
study provides a number of candidate genes that warrant further
investigation, including DDX60, HERC5, RSAD2, SIGLEC1, CMPK2, MX1,
SERPING1, EPSTI1, C9orf72, PHLDA2, PFKFB3, PLEKHG2, ANKRD28, IL1RN
and RNF19B.
References
|
1
|
Chao TH, Yu HN, Huang CC, Liu WS, Tsai YW
and Wu WT: Association of interleukin-1β (-511C/T) polymorphisms
with osteoporosis in postmenopausal women. Ann Saudi Med.
30:437–441. 2010.
|
|
2
|
Lapauw BM, Taes Y, Bogaert V, Vanbillemont
G, Goemaere S, Zmierczak HG, De Bacquer D and Kaufman JM: Serum
estradiol is associated with volumetric BMD and modulates the
impact of physical activity on bone size at the age of peak bone
mass: a study in healthy male siblings. J Bone Miner Res.
24:1075–1085. 2009.PubMed/NCBI
|
|
3
|
Andersen TL, Sondergaard TE, Skorzynska
KE, Dagnaes-Hansen F, Plesner TL, Hauge EM, Plesner T and Delaisse
JM: A physical mechanism for coupling bone resorption and formation
in adult human bone. Am J Pathol. 174:239–247. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Marie PJ: The calcium-sensing receptor in
bone cells: a potential therapeutic target in osteoporosis. Bone.
46:571–576. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Quinn JM, Neale S, Fujikawa Y, McGee JO
and Athanasou NA: Human osteoclast formation from blood monocytes,
peritoneal macrophages and bone marrow cells. Calcif Tissue Int.
62:527–531. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim HJ, Minashima T, McCarthy EF, Winkles
JA and Kirsch T: Progressive ankylosis protein (ANK) in osteoblasts
and osteoclasts controls bone formation and bone remodeling. J Bone
Miner Res. 25:1771–1783. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wada T, Nakashima T, Hiroshi N and
Penninger JM: RANKL-RANK signaling in osteoclastogenesis and bone
disease. Trends Mol Med. 12:17–25. 2006. View Article : Google Scholar
|
|
8
|
Kobayashi Y, Udagawa N and Takahashi N:
Action of RANKL and OPG for osteoclastogenesis. Crit Rev Eukaryot
Gene Expr. 19:61–72. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Boyce BF and Xing L: Functions of
RANKL/RANK/OPG in bone modeling and remodeling. Arch Biochem
Biophys. 473:139–146. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yao Z, Xing L, Qin C, Schwarz EM and Boyce
BF: Osteoclast precursor interaction with bone matrix induces
osteoclast formation directly by an interleukin-1-mediated
autocrine mechanism. J Biol Chem. 283:9917–9924. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Goto H, Hozumi A, Osaki M, Fukushima T,
Sakamoto K, Yonekura A, Tomita M, Furukawa K, Shindo H and Baba H:
Primary human bone marrow adipocytes support TNFα-induced
osteoclast differentiation and function through RANKL expression.
Cytokine. 56:662–668. 2011.
|
|
12
|
Axmann R, Böhm C, Krönke G, Zwerina J,
Smolen J and Schett G: Inhibition of interleukin-6 receptor
directly blocks osteoclast formation in vitro and in
vivo. Arthritis Rheum. 60:2747–2756. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
McCarthy HS, Williams JH, Davie MW and
Marshall MJ: Platelet-derived growth factor stimulates
osteoprotegerin production in osteoblastic cells. J Cell Physiol.
218:350–354. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Futakuchi M, Nannuru KC, Varney ML,
Sadanandam A, Nakao K, Asai K, Shirai T, Sato S and Singh RK:
Transforming growth factor-β signaling at the tumor-bone interface
promotes mammary tumor growth and osteoclast activation. Cancer
Sci. 100:71–81. 2009.
|
|
15
|
Herrera BS, Ohira T, Gao L, Omori K, Yang
R, Zhu M, Muscara MN, Serhan CN, Van Dyke TE and Gyurko R: An
endogenous regulator of inflammation, resolvin E1, modulates
osteoclast differentiation and bone resorption. Br J Pharmacol.
155:1214–1223. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nakashima K, Zhou X, Kunkel G, Zhang Z,
Deng JM, Behringer RR and de Crombrugghe B: The novel zinc
finger-containing transcription factor osterix is required for
osteoblast differentiation and bone formation. Cell. 108:17–29.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lei SF, Wu S, Li LM, Deng FY, Xiao SM,
Jiang C, Chen Y, Jiang H, Yang F, Tan LJ, et al: An in vivo
genome wide gene expression study of circulating monocytes
suggested GBP1, STAT1 and CXCL10 as novel risk genes for the
differentiation of peak bone mass. Bone. 44:1010–1014. 2009.
|
|
18
|
Chen XD, Xiao P, Lei SF, Liu YZ, Guo YF,
Deng FY, Tan LJ, Zhu XZ, Chen FR, Recker RR and Deng HW: Gene
expression profiling in monocytes and SNP association suggest the
importance of the STAT1 gene for osteoporosis in both Chinese and
Caucasians. J Bone Miner Res. 25:339–355. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Horan K, Jang C, Bailey-Serres J, Mittler
R, Shelton C, Harper JF, Zhu JK, Cushman JC, Gollery M and Girke T:
Annotating genes of known and unknown function by large-scale
coexpression analysis. Plant Physiol. 147:41–57. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Smyth GK: Linear models and empirical
bayes methods for assessing differential expression in microarray
experiments. Stat Appl Genet Mol Biol. 3:Article 3. 2004.PubMed/NCBI
|
|
21
|
Gentleman RC, Carey VJ, Bates DM, Bolstad
B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al:
Bioconductor: open software development for computational biology
and bioinformatics. Genome Biol. 5:R802004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Altaf-Ul-Amin M, Shinbo Y, Mihara K,
Kurokawa K and Kanaya S: Development and implementation of an
algorithm for detection of protein complexes in large interaction
networks. BMC Bioinformatics. 7:2072006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fukushima A, Kusano M, Redestig H, Arita M
and Saito K: Metabolomic correlation-network modules in
Arabidopsis based on a graph-clustering approach. BMC Syst
Biol. 5:12011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
26
|
Clowes JA, Riggs BL and Khosla S: The role
of the immune system in the pathophysiology of osteoporosis.
Immunol Rev. 208:207–227. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Orlić I, Borovecki F, Simić P and
Vukicević S: Gene expression profiling in bone tissue of
osteoporotic mice. J Ind Hyg Toxicol. 58:3–11. 2007.PubMed/NCBI
|
|
28
|
Kwak HB, Lee SW, Jin HM, Ha H, Lee SH,
Takeshita S, Tanaka S, Kim HM, Kim HH and Lee ZH: Monokine induced
by interferon-γ is induced by receptor activator of nuclear factor
κB ligand and is involved in osteoclast adhesion and migration.
Blood. 105:2963–2969. 2005.
|
|
29
|
Kim S, Koga T, Isobe M, Kern BE, Yokochi
T, Chin YE, Karsenty G, Taniguchi T and Takayanagi H: Stat1
functions as a cytoplasmic attenuator of Runx2 in the
transcriptional program of osteoblast differentiation. Genes Dev.
17:1979–1991. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lei SF, Wu S, Li LM, Deng FY, Xiao SM,
Jiang C, Chen Y, Jiang H, Yang F and Tan LJ: An in vivo
genome wide gene expression study of circulating monocytes
suggested GBP1, STAT1 and CXCL10 as novel risk genes for the
differentiation of peak bone mass. Bone. 44:1010–1014. 2009.
|
|
31
|
Cheng YS, Patterson CE and Staeheli P:
Interferon-induced guanylate-binding proteins lack an N (T) KXD
consensus motif and bind GMP in addition to GDP and GTP. Mol Cell
Biol. 11:4717–4725. 1991.PubMed/NCBI
|
|
32
|
Ungureanu D, Vanhatupa S, Grönholm J,
Palvimo JJ and Silvennoinen O: SUMO-1 conjugation selectively
modulates STAT1-mediated gene responses. Blood. 106:224–226. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kovarik P, Mangold M, Ramsauer K, Heidari
H, Steinborn R, Zotter A, Levy DE, Müller M and Decker T:
Specificity of signaling by STAT1 depends on SH2 and C-terminal
domains that regulate Ser727 phosphorylation, differentially
affecting specific target gene expression. EMBO J. 20:91–100. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Grassi F, Piacentini A, Cristino S,
Toneguzzi S, Cavallo C, Facchini A and Lisignoli G: Human
osteoclasts express different CXC chemokines depending on cell
culture substrate: molecular and immunocytochemical evidence of
high levels of CXCL10 and CXCL12. Histochem Cell Biol. 120:391–400.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee EY, Seo M, Juhnn YS, Kim JY, Hong YJ,
Lee YJ, Lee EB and Song YW: Potential role and mechanism of
IFN-γ-inducible protein-10 on receptor activator of nuclear factor
kappa-B ligand (RANKL) expression in rheumatoid arthritis.
Arthritis Res Ther. 13:R1042011.
|
|
36
|
Nakamura K, Deyama Y, Yoshimura Y, Suzuki
K and Morita M: Toll-like receptor 3 ligand-induced antiviral
response in mouse osteoblastic cells. Int J Mol Med. 19:771–775.
2007.PubMed/NCBI
|
|
37
|
Woeckel V, Eijken M, van de Peppel J,
Chiba H, van der Eerden B and van Leeuwen J: IFNβ impairs
extracellular matrix formation leading to inhibition of
mineralization by effects in the early stage of human osteoblast
differentiation. J Cell Physiol. 227:2668–2676. 2011.
|
|
38
|
Yoshida K, Okamura H, Amorim BR, Hinode D,
Yoshida H and Haneji T: PKR-mediated degradation of STAT1 regulates
osteoblast differentiation. Exp Cell Res. 315:2105–2114. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yoshida K, Okamura H, Amorim BR, Ozaki A,
Tanaka H, Morimoto H and Haneji T: Double-stranded RNA-dependent
protein kinase is required for bone calcification in MC3T3-E1 cells
in vitro. Exp Cell Res. 311:117–125. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Woeckel VS, Koedam M, van de Peppel J,
Chiba H, van der Eerden BC and van Leeuwen JP: Evidence of Vitamin
D and interferon-β cross-talk in human osteoblasts with 1α,
25-dihydroxyvitamin D3 being dominant over interferon-β in
stimulating mineralization. J Cell Physiol. 227:3258–3266.
2011.
|
|
41
|
Lovibond AC, Haque SJ, Chambers TJ and Fox
SW: TGFβ-induced SOCS3 expression augments TNFα-induced osteoclast
formation. Biochem Biophys Res Commun. 309:762–767. 2003.
|
|
42
|
Zhang X, Alnaeeli M, Singh B and Teng YT:
Involvement of SOCS3 in regulation of CD11c+ dendritic
cell-derived osteoclastogenesis and severe alveolar bone loss.
Infect Immun. 77:2000–2009. 2009.PubMed/NCBI
|
|
43
|
Yoshimura A, Sanada T, Takaki H and Ohishi
M: Suppressor of cytokine signalling-1 (SOCS1) and SOCS3 regulate
osteoclastogenesis by blocking inflammatory cytokine signals. Nihon
Riumachi Gakkai, Gakujutsu Shukai, Kokusai Riumachi Simpojiumu
Puroguramu, Shorokushu. 50:152006.PubMed/NCBI
|
|
44
|
Cornish J, Callon KE, Bava U, Coy DH,
Mulvey TB, Murray MA, Cooper GJ and Reid IR: Systemic
administration of adrenomedullin (27–52) increases bone volume and
strength in male mice. J Endocrinol. 170:251–257. 2001.
|
|
45
|
Cornish J, Callon KE, Coy DH, Jiang NY,
Xiao L, Cooper GJ and Reid IR: Adrenomedullin is a potent
stimulator of osteoblastic activity in vitro and in
vivo. Am J Physiol. 273:E1113–1120. 1997.PubMed/NCBI
|
|
46
|
Cornish J, Grey A, Callon KE, Naot D, Hill
BL, Lin CQ, Balchin LM and Reid IR: Shared pathways of osteoblast
mitogenesis induced by amylin, adrenomedullin and IGF-1. Biochem
Biophys Res Commun. 318:240–246. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Uzan B, Villemin A, Garel JM and Cressent
M: Adrenomedullin is anti-apoptotic in osteoblasts through CGRP1
receptors and MEK-ERK pathway. J Cell Physiol. 215:122–128. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Abdollahi M, Larijani B, Rahimi R and
Salari P: Role of oxidative stress in osteoporosis. Therapy.
2:787–796. 2005. View Article : Google Scholar
|
|
49
|
Deng FY, Lei SF, Chen XD, Tan LJ, Zhu XZ
and Deng HW: An integrative study ascertained SOD2 as a
susceptibility gene for osteoporosis in Chinese. J Bone Miner Res.
26:2695–2701. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ozgocmen S, Kaya H, Fadillioglu E, Aydogan
R and Yilmaz Z: Role of antioxidant systems, lipid peroxidation and
nitric oxide in postmenopausal osteoporosis. Mol Cell Biochem.
295:45–52. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Deng FY, Liu YZ, Li LM, Jiang C, Wu S,
Chen Y, Jiang H, Yang F, Xiong JX and Xiao P: Proteomic analysis of
circulating monocytes in Chinese premenopausal females with
extremely discordant bone mineral density. Proteomics. 8:4259–4272.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Levi G, Topilko P, Schneider-Maunoury S,
Lasagna M, Mantero S, Pesce B, Ghersi G, Cancedda R and Charnay P:
Role of Krox-20 in endochondral bone formationa. Ann NY Acad Sci.
785:288–291. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Bradley EW, Ruan MM and Oursler MJ: Novel
pro-survival functions of the Kruppel-like transcription factor
EGR2 in promotion of M-CSF-mediated osteoclast survival downstream
of the MEK/ERK pathway. J Biol Chem. 283:8055–8064. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Gabet Y, Baniwal SK, Leclerc N, Shi Y,
Kohn-Gabet AE, Cogan J, Dixon A, Bachar M, Guo L, Turman JE Jr and
Frenkel B: Krox20/EGR2 deficiency accelerates cell growth and
differentiation in the monocytic lineage and decreases bone mass.
Blood. 116:3964–3971. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Suzuki M, Ohtori S, Inoue G, Orita S,
Ishikawa T, Miyagi M, Kamoda H, Eguchi Y, Arai G, et al: ATF3 and
GAP43 immunoreactive DRG neurons innervate the vertebral body in a
rat model of osteoporosis: GP18. Spine. 2011.
|
|
56
|
Purdue PE: Alternative macrophage
activation in periprosthetic osteolysis. Autoimmunity. 41:212–217.
2008. View Article : Google Scholar : PubMed/NCBI
|