Introduction
CD151, a tetraspanin superfamily protein, contains
two extracellular loops, four hydrophobic transmembrane domains and
two short cytoplasmic tails (1,2).
This tetraspanin is expressed broadly in various tissues and is
particularly abundant in endothelia, epithelia, smooth muscle and
megakaryocytes (3,4). At the cellular level, CD151 is
characteristically localized in intracellular vesicles and at
cell-cell junctions in endothelial cells (ECs) (4). Moreover, the association of CD151
with integrins stands out as a prominent feature (3,5,6). A
‘CD151-integrin’ complex model has been proposed and this model is
functionally linked to CD151-induced biological processes (6–9).
Previous studies have shown that CD151 is involved
in regulating cell motility, cell-cell adhesion and contact, tumor
metastasis and invasion. At present, accumulating evidence has
revealed a role of CD151 in angiogenesis (4–6).
Furthermore, the regulatory role of CD151 in angiogenesis was
supported by CD151 knockout studies (10,11).
Our group focuses on the role of CD151 in angiogenesis: we
previously demonstrated that delivery of the CD151 gene was capable
of increasing angiogenesis in a pig myocardial ischemia model and a
rat ischemic hindlimb model (12,13).
CD151 transfection enhanced ECs proliferation, migration and
capillary network formation on Matrigel (14,15).
Therefore, CD151 may promote angiogenesis in vivo and in
vitro. Although the activation of PI3K and ERK signaling
pathways was demonstrated to be involved in CD151-induced
angiogenesis (15–17), the mechanisms involved remain
unclear.
As previously shown, vesicle trafficking is a
fundamental membrane trafficking event in cell migration, and
endocytic trafficking pathways have become established in recent
years as being important in the regulation of intracellular
signaling pathways during processes such as cell division,
migration and angiogenesis (18–21).
Recently, several studies noted that vesicle trafficking was also
one characteristic feature of CD151 and observed that the YRSL
sequence of CD151 was necessary for internalization and vesicle
trafficking of CD151 (22). It was
revealed that the C-terminal cytoplasmic domain of CD151 contains a
YRSL sequence or YXXϕ sorting motif, in which Y is tyrosine, X is
any amino acid, and ϕ represents the amino acid residue with a
bulky hydrophobic side chain (23,24).
Mutation of this CD151 YRSL motif markedly attenuated
internalization and vesicle trafficking of CD151 (22). The YRSL motif-mediated
internalization of CD151 was thought to promote cell migration by
modulating the endocytosis and/or vesicular trafficking of CD151
(22). Hence, we hypothesized that
the YRSL motif of CD151 may be responsible for the regulation of
CD151-related human umbilical vein endothelial cell (HUVEC)
migration and capillary network formation in vitro, and
CD151 may affect angiogenesis via vesicle trafficking.
In this study, we mutated CD151 YRSL→ARSA, which was
capable of impairing the vesicle trafficking of CD151 (22). We then examined the roles of CD151
and the CD151-ARSA mutant in the cell proliferation, migration and
capillary network formation of HUVECs, with a recombinant
adeno-associated virus (rAAV) construct encoding CD151 and the
CD151-ARSA mutant. The purpose of the present study was to
investigate the critical role of the YRSL sequence of CD151 during
angiogenesis in vitro and the mechanism(s) involved.
Materials and methods
Materials and reagents
All cell culture reagents were obtained from Gibco
BRL Life Technologies, Inc. (Grand Island, NY, USA), including
Dulbecco’s modified Eagle’s medium (DMEM/F12), trypsin and fetal
bovine serum (FBS). Apigenin was supplied by Calbiochem-Novabiochem
(Darmstadt, Germany). Rabbit anti-ERK1 and anti-phospho-ERK1
antibodies were purchased from New England Biolabs (Beverly, MA,
USA). The restriction enzymes were purchased from Takara (Dalian,
China). Antibodies against PI3K (P110), Akt, phospho-Akt (ser473),
CD151 and β-actin were purchased from Santa Cruz Biotechnology Inc.
(Santa Cruz, CA, USA). LY294002 and Hybrisol solution were
purchased from Intergen (Purchase, NY, USA). Tween-20, PMSF and
aprotinin were purchased from B&D Biosciences (Heidelberg,
Germany). All other chemicals and reagents were purchased from
Sigma-Aldrich Inc. (Shanghai, China) unless otherwise specified.
This study was approved by the ethics committee of HuaZhong
University of Science and Technology.
Construction of pAAV-CD151,
pAAV-CD151-ARSA and pAAV-GFP
The PzeoSV-CD151 plasmid was described in an earlier
study (6). The construction of
pAAV-CD151, pAAV-anti-CD151 and pAAV-green fluorescent protein
(GFP) has been described previously (12–15).
The CD151-ARSA mutant was generated by recombinant PCR. The
pAAV-CD151 vector contained the full-length wild-type human CD151,
and CD151 was used as the template. For the CD151-ARSA mutant, the
following primers were used: ATGATCTTCACGTGCT
GCCTGGCTAGGAGTGCCAAGCTGGAGCACTACGCCT ACCCC (internal sense primer
to amplify the 3′-region) and GGGGTAGGCGTAGTGCTCCAGCTTGGCACTCCTAGC
CAGGCAGCACGTGAGATCAT (internal antisense primer to amplify the
5′-region) of the CD151 template. We then used either external
sense GCTTAGATCTGCCACCATGGGTGA GTTCAACGAG or external antisense
GACGCGGCCGCT CAGGCGTAGTCGGG primers. The final recombinant PCR was
performed using purified PCR products and external sense and
antisense primers. The final PCR products were incised by the
BglII and Not restriction enzymes, and then the
incised products were purified and ligated into the
adeno-associated virus (AAV) vector at the BamHI and
NotI restriction sites. Proper ligation was confirmed by
sequencing.
Preparation of recombinant
adeno-associated viruses (rAAVs)
The rAAV vector pXXUF1, packaging plasmid pXX2,
adenovirus helper plasmid pHelper, and a rAAV plasmid containing
GFP cDNA was obtained from Dr Xiao Xiao (University of North
Carolina, Chapel Hill, NC, USA). The packing and production of
rAAV-GFP, rAAV-CD151 and rAAV-CD151-ARSA were carried out using a
triple-plasmid cotransfection method in human embryonic kidney
cells [293 cells, American Type Culture Collection (ATCC),
Manassas, VA, USA] (12,13). For purification, a single-step
gravity-flow column was applied (25). The titers of vector particles were
determined.
HUVEC culture and transfection
HUVECs were obtained from ATCC and grown in DMEM/F12
medium supplemented with 10% FBS (Gibco), streptomycin 100 μg/ml
and penicillin 100 U/ml (all obtained from Sigma) at 37°C under 5%
CO2 and 95% air. Only cells passaged less than five
times were used for experiments. Cells were grown to 50–60%
confluence and transfected with rAAV-GFP, rAAV-CD151 and
rAAV-CD151-ARSA, as described previously (14). For the control group, PBS or
HD-Fugene was added. Cells were incubated in the conditions above
for 3 days and then subjected to the following assays.
Cell proliferation assay
Assessment of cell viability was performed using the
Cell Counting Kit-8 (CCK-8) assay. At 12, 24 and 48 h after rAAV
transfection, HUVECs were incubated in 10% CCK-8 (Beyotime
Institute of Biotechnology, Nantong, China) diluted in normal
culture medium at 37°C until visual color conversion occurred in
96-well culture plates. The number of viable cells was assessed by
measurement of absorbance at 450 nm using a microplate reader.
Cell migration assay
Assessment of cell migration was performed using a
cell wound-healing assay. Briefly, transfectant cells were cultured
in 60-mm diameter dishes and synchronized in 0.5% FBS. After HUVECs
were grown to confluence, wounds were generated by scraping the
monolayers with sterile pipette tips. After 0, 12, 24 and 48 h
culture at 37°C, respectively, images of the wound in each well
were captured using an inverted microscope (Nikon TE 2000; Nikon,
Tokyo, Japan).
Capillary network formation assay
Assessment of cell migration was performed using
capillary network formation on Matrigel. Briefly, Matrigel (0.5 ml)
was polymerized on 24-well plates, and 5×104
transfectant cells were then plated in full-growth medium for 1 h.
Once the cells were seeded, the medium was replaced with medium
containing 0.5% serum. After incubation at 37°C for 12, 24 and 48
h, the capillary network formation was visualized using an inverted
microscope (Nikon TE 2000) equipped with digital imaging. For each
treatment, 10 field images were captured, and the area containing
endothelial tubes and networks that had formed was quantified using
the Scion Image Analysis System (Windows version of Scion Image,
NIH) with background subtraction.
Protein extraction and western
blotting
HUVEC proteins were extracted and used for western
blot analysis (12,14), with specific primary antibodies
against CD151, PI3K, Akt, phospho-Akt, ERK1, phospho-ERK1 and
β-actin. The HRP-conjugated secondary antibodies were used
respectively to reveal the specific protein bands with ECL
detection reagents. The intensities of protein bands were
quantified by densitometry. In addition, inhibitors of MAPK
(apigenin) and PI3-kinase (LY294002) were added to cultured HUVECs.
The protein levels of PI3K, Akt, phospho-Akt, ERK1 and phospho-ERK1
were observed.
Statistical analysis
Data were analyzed using SPSS 18.0 statistical
software (Chicago, IL, USA). Data were presented as the means ±
standard deviation (SD) unless otherwise specified. Statistical
comparisons between two groups were carried out using the Student’s
t-test or one-way ANOVA. P<0.05 was considered to indicate a
statistically significant result.
Results
Expression of CD151 protein after
transfection
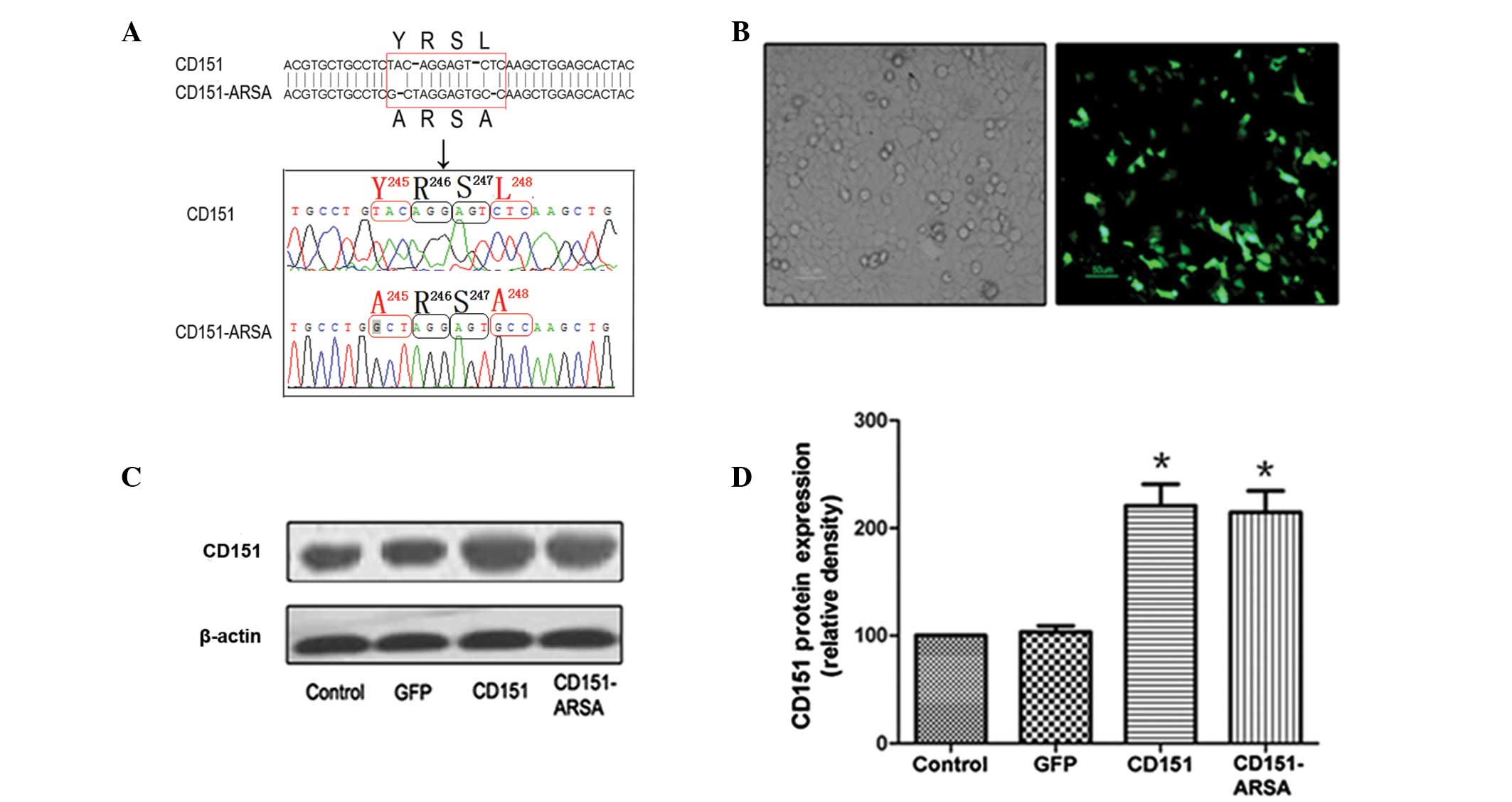
In the present study, we mutated the YRSL motif in
the human CD151 molecule. Gene sequencing analysis showed that the
Y and L residues in the CD151 YRSL motif were replaced with alanine
(A) residues simultaneously, and the resulting
YRSL→ARSA mutant was designated as the
CD151-ARSA mutant (Fig. 1A). As
shown in Fig. 1B, HUVECs
transfected with rAAV-GFP were observed using an inverted
fluorescence microscope.
Compared with the control group and the GFP group,
the expression of CD151 protein was increased significantly in the
CD151 and CD151-ARSA groups (Fig. 1C
and D). However, there was no significant difference in CD151
protein expression between the CD151 group and the CD151-ARSA
mutant transfectant group (Fig. 1C and
D). The experiment suggests that the CD151-ARSA mutant does not
affect the expression of CD151 protein.
Effects of CD151 and CD151-ARSA
transfection on the proliferation of HUVECs
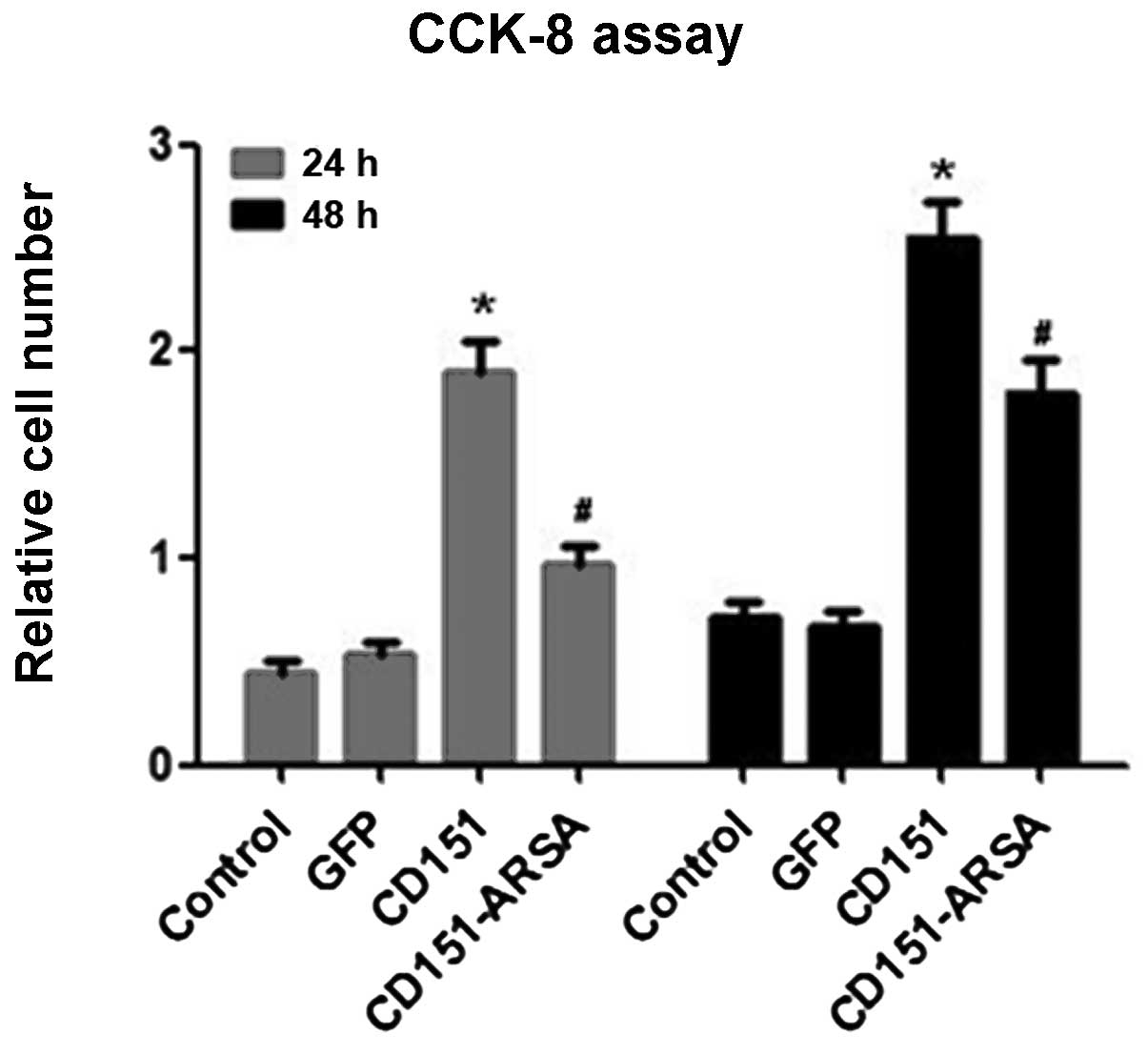
To determine the proliferative effects of CD151 and
CD151-ARSA, we performed the CCK-8 assay at 12, 24 and 48 h after
delivery. As shown in Fig. 2, the
CD151 group showed increased proliferation ability, compared with
the control group and the GFP group at 24 and 48 h after rAAV
transfection. No significant difference was observed in the groups
at 12 h after transfection (data not shown). However, the results
showed that the CD151-induced proliferation of HUVECs was decreased
significantly by the CD151-ARSA transfection, at 24 or 48 h after
delivery (Fig. 2). These data
suggest that the CD151-ARSA mutant was capable of impairing the
cell proliferation ability of CD151.
Effects of CD151 and CD151-ARSA
transfection on the migration of HUVECs
Regulation of cell motility is a prominent feature
of CD151 (4,22). Cell migration was assessed by a
cell wound-healing assay and observed at 0, 12, 24 and 48 h after
CD151 transfection. As shown in Fig.
3A and B, the expression of CD151 significantly enhanced cell
migration at 24 and 48 h, compared with the control and GFP groups.
However, at 24 h after delivery, the migration ability of
CD151-ARSA mutant cells was reduced compared with the CD151 group,
and the most notable effect was at 48 h (Fig. 3).
Thus, these results suggest that CD151 transfection
promotes wound healing while the CD151-ARSA mutant transfection
delays wound healing.
Effects of CD151 and CD151-ARSA
transfection on the capillary network formation of HUVECs
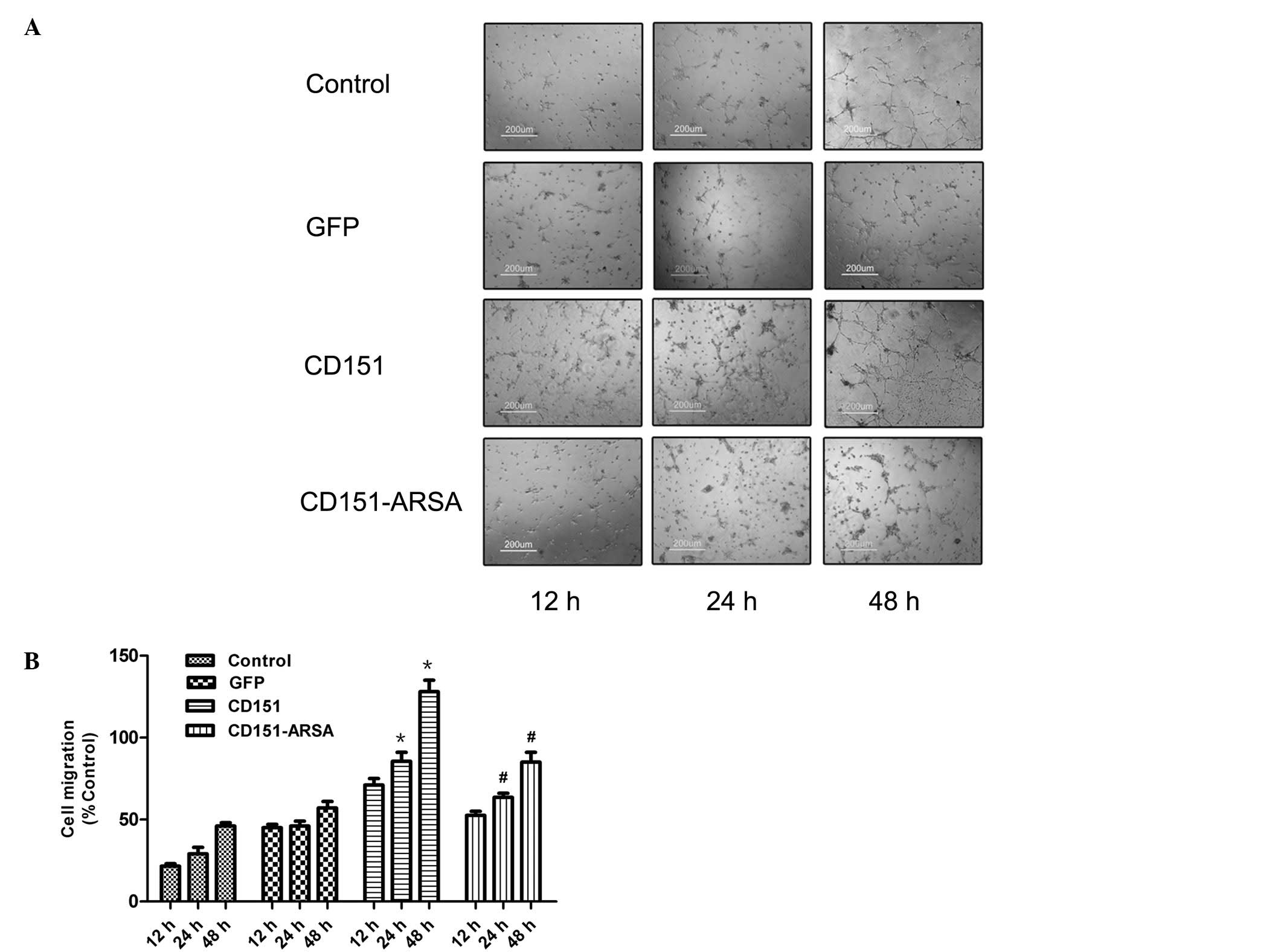
Upon plating on Matrigel basement membrane, HUVECs
assembled into capillary network formation structures, which were
observed at 12, 24 and 48 h after the rAAV transfection. As shown
in Fig. 4A and B, CD151
transfection promoted the capillary network formation at 24 and 48
h after gene delivery, compared with the control and GFP groups
(Fig. 4). By contrast, network
formation was inhibited by transfection with the CD151-ARSA mutant
at 24 and 48 h after gene delivery, compared with the CD151 group,
and there was a marked difference at 48 h. These findings indicate
that CD151-ARSA mutant delivery impairs the tube formation improved
by CD151 gene transfer.
Effects of CD151 and CD151-ARSA mutant on
the PI3K/Akt and ERK signaling pathways
A number of signaling pathways are involved in
angiogenesis, such as Akt, eNOS, ERK and p38 MAPK (26,27).
We analyzed the levels of PI3K, Akt and ERK proteins, and applied
inhibitors of MAPK (apigenin) and PI3K (LY294002) to HUVECs
following transfection with CD151 and CD151-ARSA.
Consistent with earlier data, the present study
showed the activation of PI3K/Akt and ERK signaling pathways in the
CD151 group (15,16). However, in the CD151-ARSA group,
the protein expression levels of PI3K, p-Akt and p-ERK were all
reduced compared with the CD151 group (Fig. 5). The CD151-ARSA mutant resulted in
reduced activation of PI3K/Akt and ERK signaling pathways (Fig. 5). In addition, MAPK inhibitor
(apigenin) and PI3K inhibitor (LY294002) significantly reduced the
activation of ERK and PI3K induced by CD151, respectively (Fig. 5).
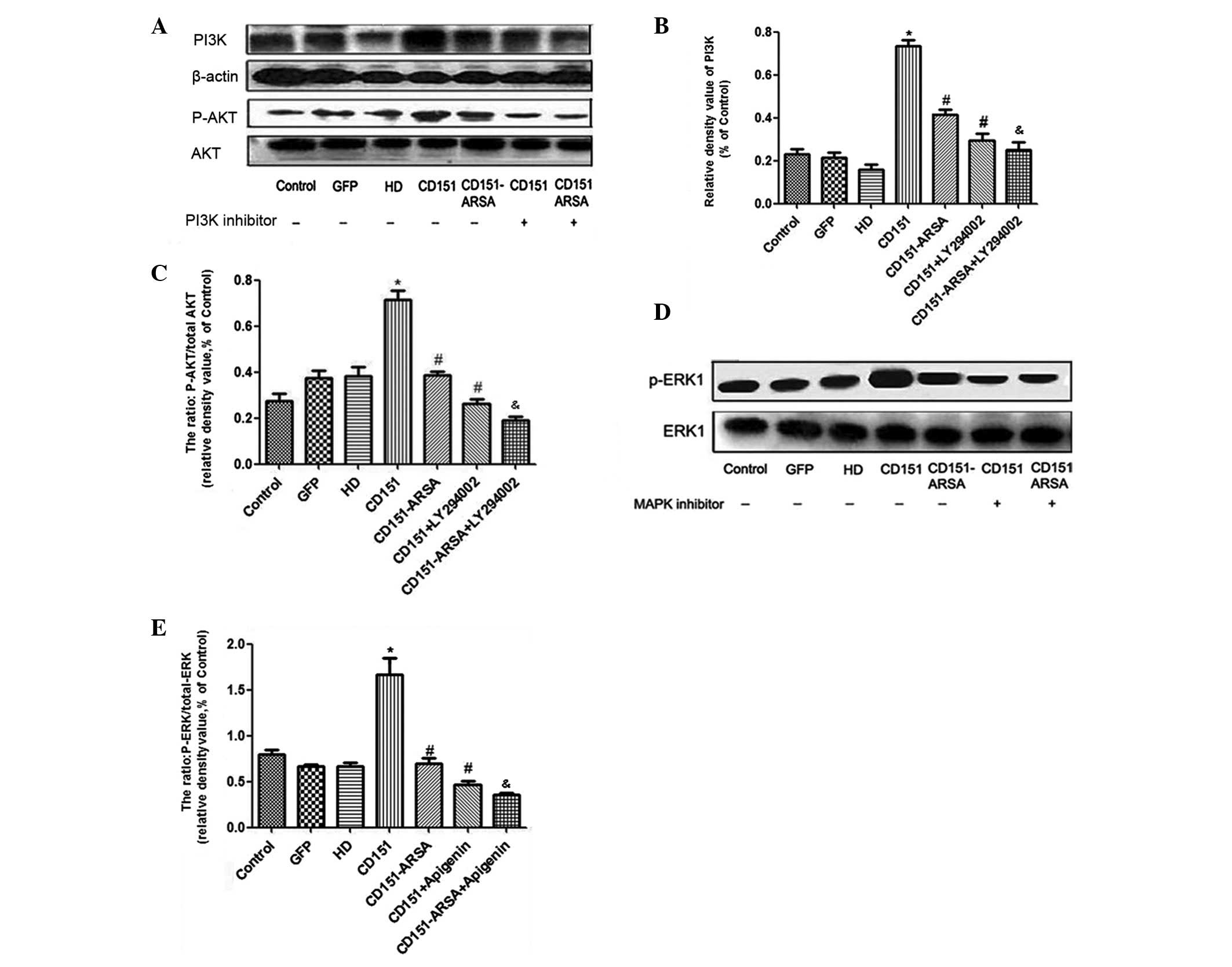 | Figure 5Effects of CD151 and CD151-ARSA
transfection on PI3K/Akt and ERK signaling pathways. Western blot
analysis for PI3K, phosphorylated Akt, total Akt, phosphorylated
ERK and total ERK. Inhibitors of MAPK (apigenin) and PI3K
(LY294002) were applied to HUVECs following transfection with CD151
or CD151-ARSA. (A-C) The protein levels of PI3K, Akt, phospho-Akt
and quantitative analysis. (D and E) Levels of ERK1, phospho-ERK1
and quantitative analysis. Inhibitor of PI3K (LY294002, 15 μM);
inhibitor of MAPK (apigenin, 25 μM). HD group (HD-Fugene 6
transfection reagent) was used as a control group. Each experiment
was performed at least in triplicate. *p<0.05 vs.
control, GFP and HD groups. #p<0.05 vs. CD151 group.
&p<0.05 vs. CD151-ARSA group (no signaling
pathway inhibitor group). HUVECs, human umbilical vein endothelial
cells; GFP, green fluorescent protein. |
As a whole, these results suggest that the gene
transfer of CD151-ARSA mutant attenuates the activation of the
PI3K/Akt and ERK signaling pathways.
Discussion
The present study was designed to investigate the
molecular mechanisms that govern the effects of CD151 in
angiogenesis. In this in vitro study, our results showed
that the delivery of the CD151-ARSA mutant (YRSL→ARSA) into HUVECs
decreased cell migration and capillary network formation on
Matrigel, contrary to the effects of CD151 gene delivery.
Furthermore, we demonstrated that mutation of the CD151 YRSL motif
resulted in diminished activation of PI3K/Akt and ERK signaling
pathways in HUVECs. These data provide evidence that the CD151 YRSL
motif was indeed a key region of CD151 for regulating cell
migration, capillary network formation and angiogenesis.
Multiple lines of evidence indicate that the YRSL
sequence of the CD151 C-terminal cytoplasmic tail is important and
it is thought to determine its intracellular trafficking and
function (24). These motifs in
the cytoplasmic domain of CD151 could be recognized by adaptor
protein (AP)-2 complex, a core component of clathrin endocytic
machinery (28). It was found that
the YRSL motif was required for CD151 endocytic processes,
indicating that the YRSL sequence in the CD151 cytoplasmic domain
determines its trafficking (22).
When the YRSL→ARSA mutant human CD151 molecule was created and
transfected into NIH3T3 mouse fibroblast cells, the vesicle
trafficking was completely impaired and CD151-promoted NIH3T3
migration was diminished (22).
Therefore, the YRSL sequence of CD151 is necessary for vesicle
trafficking of CD151 and this sequence is also a key region for
CD151-related cell migration.
Angiogenesis is a complex process involving
extracellular matrix degradation, endothelial cell proliferation
and migration, formation of tube structures and morphological
differentiation (29–31). Although our previous data showed
that CD151 was capable of promoting cell proliferation, cell
migration and angiogenesis both in vivo and in vitro,
the mechanisms remain to be elucidated. Based on above data, we
hypothesized that the YRSL sequence of CD151 may also play an
important role in the CD151-mediated process of angiogenesis. To
test this hypothesis, in the present study we mutated the YRSL→ARSA
motif in the human CD151 molecule and transfected it into HUVECs.
As shown in the western blot analysis, CD151 and the CD151-ARSA
mutant were both well expressed at the protein level, and the
mutation did not influence the expression of CD151 protein.
Furthermore, evidence indicates that the CD151-ARSA mutant
transfection abrogates CD151-induced cell proliferation and
migration in vitro. First, CD151-ARSA treatment decreased
the HUVEC proliferation, in contrast to the promoting effects of
CD151. Second, the directional motility of HUVECs (cell-wounding
healing assay) showed that the CD151-ARSA mutant exhibited delayed
cell motility, compared with CD151 transfectant. The YRSL motif was
required for CD151 regulating cell migration, consistent with the
results obtained from Liu et al(22).
Our previous work has shown that CD151 gene delivery
promoted capillary formation in vitro(16). Although the specific region of
CD151 QRD194–196 was emphasized (14,16),
the YRSL motif may be another key region in CD151-induced
angiogenesis. As was previously reported, endocytosis of adhesion
molecules from cell-cell contacts may be regulated according to the
YRSL motif (4,22,32).
In particular, the capillary network formation was related to the
maintenance of cell-cell contacts or junctions, which could be
regulated by the lateral trafficking of cell adhesion molecules
(4,21,22),
and it was questioned whether CD151 may affect the capillary
formation caused by the YRSL motif through vesicular trafficking.
In the present study, it was shown that CD151-ARSA mutant
transfection significantly disrupted the capillary network
formation, while CD151 promoted the capillary network formation on
Matrigel. The capillary network formation was inhibited by
transfection with the CD151-ARSA mutant at 24 and 48 h after gene
delivery. Notably, the difference became more significant over
time, and a marked difference was observed at 48 h. Thus, it was
observed that CD151 is capable of regulating the capillary network
formation through the YRSL motif. Based on these data, it was
accepted that the CD151 YRSL sequence is critical for
CD151-mediated capillary network formation. Therefore, CD151 YRSL
sequence-mediated vesicular trafficking may be another mechanism
for CD151 regulation of angiogenesis in vitro.
The signaling mechanism of CD151 has been explored
in the last decade (9,11,31).
A growing list of signaling pathways that may be involved includes
FAK, ERK and PI3K/Akt (11,13–15,31).
Takeda et al showed that the adhesion-dependent activation
of PKB/c-Akt and e-NOS was diminished in CD151-null mouse lung
endothelial cells (11). In the
present study, the CD151-ARSA mutant transfection resulted in
diminished activation of PI3K/Akt and ERK signaling pathways, which
was opposite to the findings with CD151 transfection. Thus, the
activation of PI3K/Akt and ERK signaling pathways may be involved
in CD151-mediated endocytosis trafficking.
In conclusion, our data demonstrated that the
CD151-ARSA mutant abrogated cell proliferation, migration and
capillary network formation in HUVECs. Furthermore, we observed the
CD151-mediated activation of ERK and PI3K/Akt signaling pathways,
but these effects were all impaired by CD151-ARSA gene delivery.
Our observations emphasize that the specific YRSL region of CD151
is important for vesicle trafficking, which plays a key role in
CD151-induced cell proliferation, migration and capillary network
formation. Our study demonstrates for the first time the effect of
CD151 YRSL in CD151-induced angiogenesis. Further studies should be
performed and may provide indications for a better understanding of
CD151 and CD151-ARSA via integrin-vesicle trafficking.
Acknowledgements
The project was supported by the grant no. 81000047
from the National Natural Science Foundation of China.
References
|
1
|
Fitter S, Tetaz TJ, Berndt MC and Ashman
LK: Molecular cloning of cDNA encoding a novel platelet-endothelial
cell tetra-span antigen, PETA-3. Blood. 86:1348–1355.
1995.PubMed/NCBI
|
|
2
|
Hasegawa H, Utsunomiya Y, Kishimoto K,
Yanagisawa K and Fujita S: SFA-1, a novel cellular gene induced by
human T-cell leukemia virus type 1, is a member of the
transmembrane 4 superfamily. J Virol. 70:3258–3263. 1996.PubMed/NCBI
|
|
3
|
Sincock PM, Mayrhofer G and Ashman LK:
Localization of the transmembrane 4 superfamily (TM4SF) member
PETA-3 (CD151) in normal human tissues: comparison with CD9, CD63,
and alpha5beta1 integrin. J Histochem Cytochem. 45:515–525. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sincock PM, Fitter S, Parton RG, Berndt
MC, Gamble JR and Ashman LK: PETA-3/CD151, a member of the
transmembrane 4 superfamily, is localised to the plasma membrane
and endocytic system of endothelial cells, associates with multiple
integrins and modulates cell function. J Cell Sci. 112:833–844.
1999.PubMed/NCBI
|
|
5
|
Yauch RL, Berditchevski F, Harler MB,
Reichner J and Hemler ME: Highly stoichiometric, stable, and
specific association of integrin alpha3beta1 with CD151 provides a
major link to phosphatidylinositol 4-kinase, and may regulate cell
migration. Mol Biol Cell. 9:2751–2765. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang XA, Kazarov AR, Yang X, Bontrager
AL, Stipp CS and Hemler ME: Function of the tetraspanin
CD151-alpha6beta1 integrin complex during cellular morphogenesis.
Mol Biol Cell. 13:1–11. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hemler ME: Integrin associated proteins.
Curr Opin Cell Biol. 10:578–585. 1998. View Article : Google Scholar
|
|
8
|
Serru V, Le Naour F, Billard M, et al:
Selective tetraspan-integrin complexes (CD81/alpha4beta1,
CD151/alpha3beta1, CD151/alpha6beta1) under conditions disrupting
tetraspan interactions. Biochem J. 340:103–111. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang XA, Bontrager AL and Hemler ME:
Transmembrane-4 superfamily proteins associate with activated
protein kinase C (PKC) and link PKC to specific beta(1) integrins.
J Biol Chem. 276:25005–25013. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wright MD, Geary SM, Fitter S, et al:
Characterization of mice lacking the tetraspanin superfamily member
CD151. Mol Cell Biol. 24:5978–5988. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Takeda Y, Kazarov AR, Butterfield CE, et
al: Deletion of tetraspanin Cd151 results in decreased pathologic
angiogenesis in vivo and in vitro. Blood. 109:1524–1532. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lan RF, Liu ZX, Liu XC, Song YE and Wang
DW: CD151 promotes neovascularization and improves blood perfusion
in a rat hind-limb ischemia model. J Endovasc Ther. 12:469–478.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zuo H, Liu Z, Liu X, et al: CD151 gene
delivery after myocardial infarction promotes functional
neovascularization and activates FAK signaling. Mol Med.
15:307–315. 2009.PubMed/NCBI
|
|
14
|
Liu WF, Zuo HJ, Chai BL, et al: Role of
tetraspanin CD151-alpha3/alpha6 integrin complex: Implication in
angiogenesis CD151-integrin complex in angiogenesis. Int J Biochem
Cell Biol. 43:642–650. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zheng ZZ and Liu ZX: Activation of the
phosphatidylinositol 3-kinase/protein kinase Akt pathway mediates
CD151-induced endothelial cell proliferation and cell migration.
Int J Biochem Cell Biol. 39:340–348. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zuo H, Liu Z, Yang J, et al: Activation of
the ERK signaling pathway is involved in CD151-induced angiogenic
effects on the formation of CD151-integrin complexes. Acta
Pharmacol Sin. 31:805–812. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zuo H, Liu Z, Liu X, et al: Assessment of
myocardial blood perfusion improved by CD151 in pig myocardial
infarction model. Acta Pharmacol Sin. 30:70–77. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bretscher MS and Aguado-Velasco C:
Membrane traffic during cell locomotion. Curr Opin Cell Biol.
10:537–541. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kobayashi T, Vischer UM, Rosnoblet C, et
al: The tetraspanin CD63/lamp3 cycles between endocytic and
secretory compartments in human endothelial cells. Mol Biol Cell.
11:1829–1843. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Duffield A, Kamsteeg EJ, Brown AN, Pagel P
and Caplan MJ: The tetraspanin CD63 enhances the internalization of
the H,K-ATPase beta-subunit. Proc Natl Acad Sci USA.
100:15560–15565. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Caswell PT, Vadrevu S and Norman JC:
Integrins: masters and slaves of endocytic transport. Nat Rev Mol
Cell Biol. 10:843–853. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu L, He B, Liu WM, Zhou D, Cox JV and
Zhang XA: Tetraspanin CD151 promotes cell migration by regulating
integrin trafficking. J Biol Chem. 282:31631–31642. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ohno H, Stewart J, Fournier MC, et al:
Interaction of tyrosine-based sorting signals with
clathrin-associated proteins. Science. 269:1872–1875. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bonifacino JS and Traub LM: Signals for
sorting of transmembrane proteins to endosomes and lysosomes. Annu
Rev Biochem. 72:395–447. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Auricchio A, Hildinger M, O’Connor E, Gao
GP and Wilson JM: Isolation of highly infectious and pure
adeno-associated virus type 2 vectors with a single-step
gravity-flow column. Hum Gene Ther. 12:71–76. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Berditchevski F and Odintsova E:
Characterization of integrin-tetraspanin adhesion complexes: role
of tetraspanins in integrin signaling. J Cell Biol. 146:477–492.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fitter S, Sincock PM, Jolliffe CN and
Ashman LK: Transmembrane 4 superfamily protein CD151 (PETA-3)
associates with beta 1 and alpha IIb beta 3 integrins in
haemopoietic cell lines and modulates cell-cell adhesion. Biochem
J. 338:61–70. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Conner SD and Schmid SL: Regulated portals
of entry into the cell. Nature. 422:37–44. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Carmeliet P: Mechanisms of angiogenesis
and arteriogenesis. Nat Med. 6:389–395. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rey S and Semenza GL: Hypoxia-inducible
factor-1-dependent mechanisms of vascularization and vascular
remodelling. Cardiovasc Res. 86:236–242. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ulrich F and Heisenberg CP: Trafficking
and cell migration. Traffic. 10:811–818. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mellman I and Nelson WJ: Coordinated
protein sorting, targeting and distribution in polarized cells. Nat
Rev Mol Cell Biol. 9:833–845. 2008. View Article : Google Scholar : PubMed/NCBI
|