Introduction
Numerous studies have shown that heart development
is controlled by an evolutionarily-conserved network of
transcription factors (NK2, MEF2, GATA, Tbx and Hand) that connect
signaling pathways with genes for muscle growth, patterning and
contractility (1–5). In this network, GATA4, MEF2C and Tbx5
are the most significant members which play critical roles in
cardiac cell growth and heart development (6). The disturbance of temporal-spatial
expression or mutations in these cardiac transcription factors may
cause congenital heart disease (1,7).
However, the upstream regulation of these genes remains
unclear.
Previous studies have revealed that bone
morphogenetic protein (BMP) signaling is required for cardiogenesis
(8–10). It has been reported that several
BMP subtypes, including BMP2, 4, 6 and 7, are expressed in the
developing heart (9) and that BMP2
is a major transcription factor among several other members of the
BMP family. BMP2 plays a significant role during cushion and valve
morphogenesis and has a persistent expression in the cushion
mesenchyme from 13.5 to 16 days post coitum (dpc). The
expression of BMP2 is also observed in the valve tissues of adult
mice (11). BMP2 knockout mice die
at 7.5–10.5 dpc and have defects in the heart during development
(12). Clinically, patients with a
heterozygous deletion of BMP2 may present tachycardia as a result
of an abnormal connection between the atria and ventricles
(13). Several studies have
demonstrated that BMP2 is able to elicit ectopic expression of the
early cardiac markers, including Nkx2.5 and GATA4, but not of Tbx5
(9,14). Schlange et al(15) reported that BMP2 was expressed
dynamically during cardiac morphogenesis and regulated the
expression of other cardiac transcription factors, including Nkx2.5
and GATA4, at varying time periods. However, the molecular
mechanisms by which BMP2 regulates the cardiac transcription
factors remain largely unknown.
In recent years, histone acetylation/deacetylation
has been at the center of attention with regard to the control of
gene expression. Acetylation of the conserved lysine residues in
histone tails by histone acetylases (HATs) stimulates gene
expression by neutralizing positive charges, resulting in the
‘open’ state of chromatin, while histone deacetylases (HDACs)
promote chromatin condensation, causing a repression of gene
expression (16–18). It has been reported that histone
acetylation plays a significant role during the development of the
heart by acting as a switch for the regulation of gene expression
(18). In our previous study,
sodium valproate (NaVP), an inhibitor of HDACs, was identified as
being able to increase the expression of CHF1, Tbx5 and MEF2C,
causing cardiac abnormalities in fetal mice (19). Alcohol-induced overexpression of
heart development-related genes was also identified as associated
with the upregulation of histone H3 lysine 9 acetylation (20). One study in rat bone marrow
mesenchymal stem cells also showed that suberoylanilide hydroxamic
acid (SAHA), a HDAC inhibitor, upregulated the expression of
Nkx2.5, GATA4 and MEF2C in a dose-dependent manner (21). These studies indicate that histone
acetylation regulates the expression of cardiac-specific genes and
is essential for heart development.
In the present study, the histone H3 acetylation
levels in the promoter regions of the cardiac-specific genes and
the HAT activities in the cultured H9c2 rat embryonic cardiac
myocytes overexpressing BMP2 were determined. The results indicate
that BMP2 is able to enhance the expression of the cardiac
transcription factors GATA4 and MEF2C, in part by increasing
histone H3 acetylation in the promoter regions of these genes. The
HAT p300 subtype may play an essential role in BMP2-induced histone
hyperacetylation.
Materials and methods
Reagents
The recombinant adenoviruses expressing human BMP2
(AdBMP2), the control adenoviruses expressing green fluorescent
protein (AdGFP), the human embryonic kidney 293 cells and the H9c2
cells were kind gifts from the Molecular Oncology Laboratory at the
University of Chicago Medical Center.
Preparation of adenoviruses in the 293
cells
AdBMP2 and AdGFP were transfected in the 293 cells
to amplify the viruses. Propagations of the viruses were visualized
by GFP expression under a fluorescence microscope. The viral
supernatant was purified in phosphate-buffered saline (PBS) by
ultracentrifugation and the titration of the viruses was measured
by end point dilution assays. The prepared viruses were stored at
−80°C for use.
Culture and treatment of the H9c2
cells
The H9c2 cells were grown in Dulbecco’s modified
Eagle’s medium (DMEM)/high glucose (Thermo Scientific, Rochester,
NY, USA) containing 10% fetal bovine serum (FBS; Hyclone, Logan,
UT, USA) in humidified air (5% CO2) at 37°C. The cells
were transfected with AdBMP2 or AdGFP at a varied multiplicity of
infection (MOI). The transfection efficiency was measured by flow
cytometry.
Real-time RT-PCR
The cultured H9c2 cells were collected 24, 48 or 72
h after transfection with AdBMP2 or AdGFP. The total RNA was
extracted using a RNA extraction kit (Bioteck, Beijing, China).
Single-stranded cDNA was reverse transcribed from 500–1,000 ng RNA
using oligo(dT)-adaptor primers and AMV reverse transcriptase
(Takara, Otsu, Japan) according to the manufacturer’s instructions.
The cDNA was then amplified using gene-specific primers (Takara
Biotechnology, Dalian, China) and a SYBR-Green Dye kit (Tiangen,
Beijing, China). The mRNA expression levels of BMP2, GATA4, MEF2C,
Tbx5, p300, GCN5 and β-actin were quantified by real-time RT-PCR
using an IQcycler kit (Bio-Rad, Hercules, CA, USA). The annealing
temperatures were 54°C for BMP2, 57°C for GATA4 and Tbx5 and 65°C
for MEF2C, p300, GCN5 and β-actin. The gene-specific primers were
designed using the Primer-3 software as follows: BMP2 forward,
5′-gacatccactccacaaacgaga-3′ and reverse,
5′-gtcattccaccccacatcact-3′; GATA4 forward,
5′-caactgccagactaccaccac-3′ and reverse, 5′-ccatggagcttc
atgtagagg-3′; MEF2C forward, 5′-gcgaaagttcggattgatgaaga-3′ and
reverse, 5′-gtggatgtcagtgctggcgta-3′; Tbx5 forward,
5′-cctgggtccgtaggtggaatag-3′ and reverse, 5′-ctttgatgctct
gtctcgggtag-3′; p300 forward, 5′-agattcagagggcagcagagac-3′ and
reverse, 5′-gccataggaggtgggttcatac-3′; GCN5 forward,
5′-ggaaaggagaagggcaaggag-3′ and reverse, 5′-gtcaatggggaa
gcggataac-3′; β-actin forward, 5′-ggagattactgccctggctccta-3′ and
reverse, 5′-gactcatcgtactcctgcttgctg-3′. The analyses of relative
mRNA expression were carried out using the 2−ΔΔCt method
as described previously (22). The
values were normalized using β-actin as an endogenous housekeeping
control gene.
Western blot analysis
The cultured H9c2 cells were collected 24 h after
transfection with AdBMP2 or AdGFP. Nuclear proteins were extracted
using a Nuclear and Cytoplasmic Protein Extraction kit (KeyGen,
Nanjing, China) according to the manufacturer’s instructions. The
nuclear proteins were separated and electrophoresed on 15% Bis-Tris
polyacrylamide gels and then electrophoretically transferred to
polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica,
MA, USA). The PVDF blots were then blocked with 5% nonfat milk in
PBST (PBS plus 0.05% Tween-20) for 1 h. Subsequently, the blots
were incubated with rabbit monoclonal antibodies for acetylated
histone H3 (Ac-H3; Millipore, Temecula, CA, USA; 1:500 dilution) or
for histone H3 (Millipore, Charlottesville, VA, USA; 1:500
dilution) in PBST containing 5% nonfat milk at 4°C overnight.
HRP-conjugated goat anti-rabbit antibodies (Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA) were used as the
secondary antibodies. The immunoreactive protein bands were
detected with an Enhanced Chemiluminescence Luminal reagent
(KeyGen), then scanned with a chemiluminescence kit (Gene, Hong
Kong, China) and analyzed with Quantity One version 4.4 software
(Bio-Rad). All western blotting experiments were repeated a minimum
of 3 times.
HAT activity assay
The nuclear proteins were extracted as mentioned
previously. The HAT activities of the nuclear protein extraction
were determined using a HAT Activity Colorimetric Assay kit
(BioVision, Mountain View, CA, USA) according to the manufacturer’s
instructions. Nuclear proteins (40 μg) were prepared for each assay
in 96-well plates at a concentration of 1 μg/μl, with 40 μl water
prepared in one plate for the background reading. The assay mix was
then added to each well and incubated at 37°C for 4 h. The samples
were read by an enzyme micro-plate reader (Thermo Scientific) at a
wavelength of 440 nm. Background readings were subtracted from the
reading of all the samples. The HAT activities were expressed as
relative OD values.
Chromatin immunoprecipitation (ChIP)
assay
The cultured H9c2 cells were washed with PBS 48 h
after treatment with AdBMP2 or AdGFP, then the cells were fixed
with formaldehyde to cross link the proteins and DNA. ChIP
experiments were performed using a ChIP assay kit (Millipore,
Billerica, MA, USA). Subsequent to cross-linking, the DNA was cut
into small fragments by sonication. The conditions used for the
sonication to shear the DNA were 10 sec/time, with an interval of
80 sec for cooling. These steps were repeated 270 times. The
protein-DNA complex was then recruited and precipitated by rabbit
monoclonal antibodies for Ac-H3 (ChIP grade; Millipore). Anti-RNA
polymerase was used as a positive control and normal mouse IgG was
used as a negative control. The DNA from the samples was then
obtained by phenol/chloroform extraction and ethanol precipitation.
The promoter regions of GATA4, MEF2C and Tbx5 were assayed by
real-time PCR of the total DNA using specific primers. The
sequences of these primers were as follows: GATA4 forward,
5′-actgac gccgactccaaactaag-3′ and reverse,
5′-gtgtccctgttctccctgtagc-3′; MEF2C forward,
5′-ctttccaggttggctcttactcc-3′ and reverse,
5′-gcctcctcctaacaaagtgggta-3′; Tbx5 forward, 5′-actgac
gccgactccaaactaag-3′ and reverse, 5′-gtgtccctgttctccctgtagc-3′. The
annealing temperatures were 57°C for Tbx5 and 65°C for GATA4 and
MEF2C. The analyses of the relative promoter precipitation levels
were carried out using the 2−ΔΔCt method as described
previously (22). The values were
normalized using an input sample as the internal standard.
Statistical analysis
All experiments were repeated independently at least
3 times. All data are reported as mean ± SD and statistical
analyses was performed using a one-way ANOVA. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of BMP2 in the H9c2 cells
To identify the expression of BMP2 in the H9c2 cells
following transfection with AdBMP2, BMP2 mRNA levels were measured
using real-time RT-PCR assays. The expression of the β-actin gene
was used as an endogenous control. BMP2 was highly expressed in the
AdBMP2-transfected cells, reaching a peak 48 h after the
transfection (1,432±313-fold, AdBMP2 group vs. blank group,
P<0.05). Extremely low expression levels of BMP2 were detected
in the AdGFP and blank groups (Fig.
1A).
Expression of GATA4 and MEF2C in H9c2
cells overexpressing BMP2
The results of real-time RT-PCR assays showed that
the mRNA expression levels of GATA4 and MEF2C were significantly
enhanced in H9c2 cells overexpressing BMP2. As shown in Fig. 1B and C, GATA4 and MEF2C expression
levels were enhanced by BMP2 and reached a peak 48 h after AdBMP2
transfection (1.94±0.23-fold for GATA4 and 4.84±0.43-fold for
MEF2C, AdBMP2 vs. blank group, P<0.05). The expression levels of
GATA4 and MEF2C in the AdGFP group were not altered (AdGFP vs.
blank group, P>0.05). In addition, there was no significant
change observed in the expression of Tbx5 in the cells following
AdBMP2 transfection (AdBMP2 vs. blank group, P>0.05; Fig. 1D).
Increased histone H3 acetylation in the
whole chromatin of H9c2 cells overexpressing BMP2
Using western blotting assays, the effect of BMP2 on
histone H3 acetylation in H9c2 cells overexpressing BMP2 was
analyzed. The ratio of Ac-H3 to H3 was determined in order to
normalize the sample recovery and loading. As shown in Fig. 2, the histone H3 acetylation levels
were significantly increased in H9c2 cells overexpressing BMP2
(3.07±0.16-fold, AdBMP2 vs. blank group, P<0.05). There was no
significant difference in histone H3 acetylation levels between the
AdGFP and blank groups (P>0.05).
Increased histone H3 acetylation in the
promoter regions of GATA4 and MEF2C in H9c2 cells overexpressing
BMP2
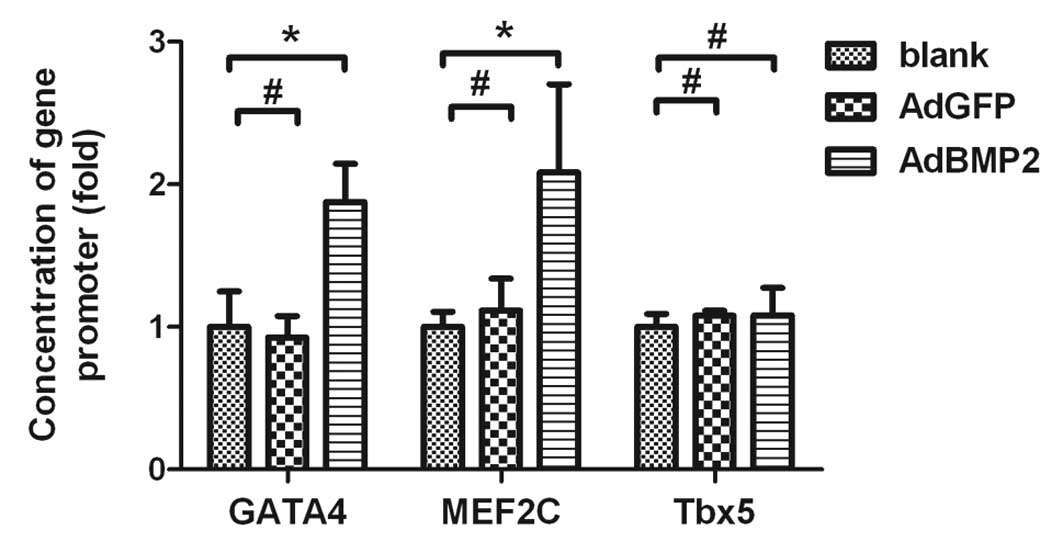
ChIP and real-time PCR assays were conducted to
further investigate whether the upregulation of GATA4 and MEF2C by
BMP2 was associated with changes in the histone H3 acetylation of
these genes. The real-time PCR results showed that the quantities
of promoter DNA for GATA4 and MEF2C were increased in H9c2 cells
overexpressing BMP2 (1.88±0.27-fold for GATA4, 2.08±0.62-fold for
MEF2C, AdBMP2 vs. blank group, P<0.05). There was no significant
difference in these values between the AdGFP and blank groups
(P>0.05; Fig. 3). Accordingly,
the ChIP data indicated that the histone H3 acetylation levels in
the promoter regions of GATA4 and MEF2C were significantly
increased in H9c2 cells overexpressing BMP2. However, the histone
H3 acetylation in the promoter region of Tbx5 was not significantly
changed in the same cells overexpressing BMP2 (P>0.05). These
data indicate that increased expression of GATA4 and MEF2C are
associated with increased histone H3 acetylation in the promoter
regions of these genes in H9c2 cells overexpressing BMP2.
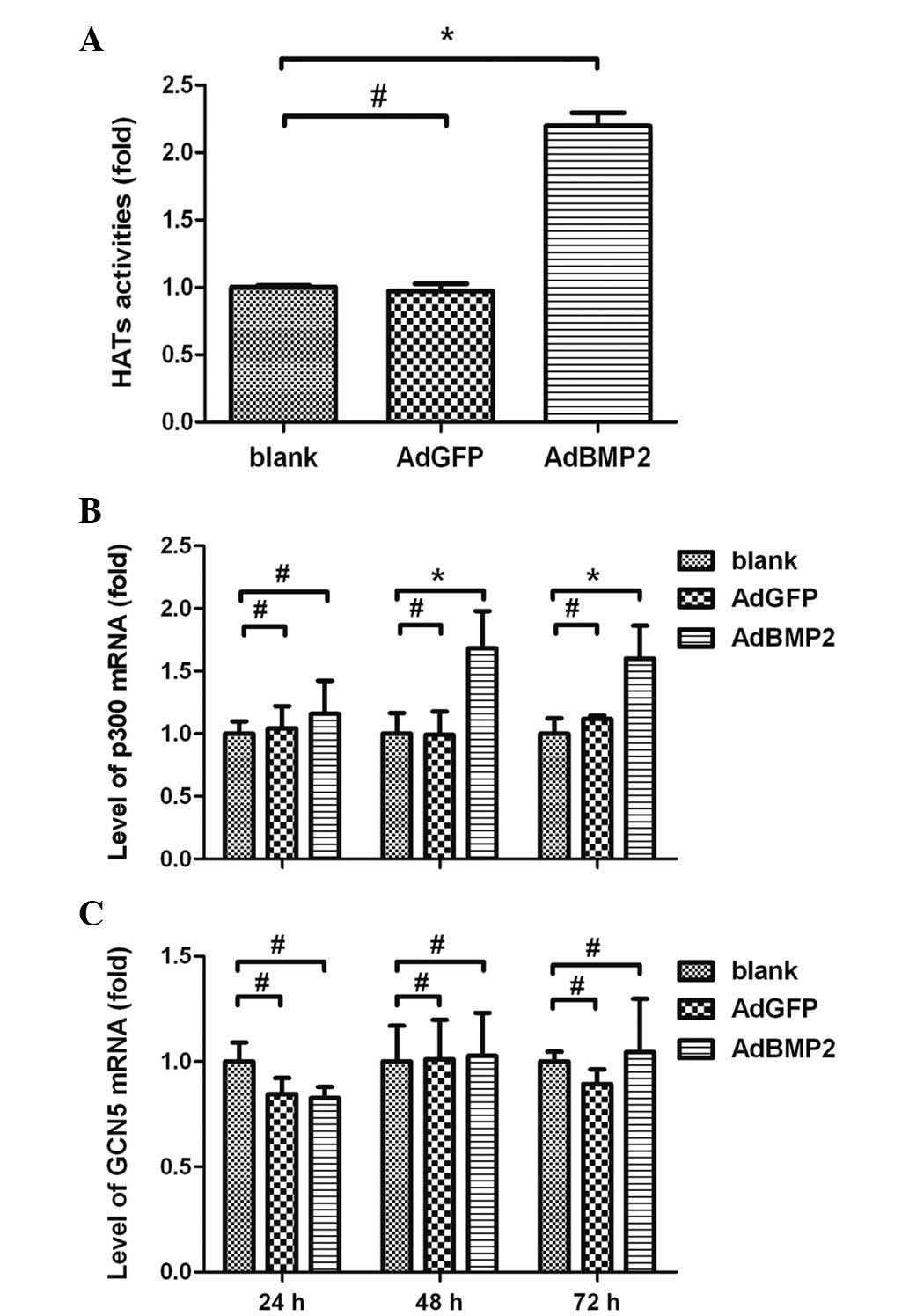
Enhanced HAT activities and increased
expression of HAT subtype p300 in H9c2 cells overexpressing
BMP2
To further investigate whether the BMP2-induced
histone H3 hyperacetylation was associated with HATs, experiments
were performed using colorimetric assays to measure the HAT
activities and to determine the expression levels of the HAT p300
subtype in H9c2 cells overexpressing BMP2. The results showed that
HAT activities were significantly increased in H9c2 cells
overexpressing BMP2 (2.20±0.17-fold, AdBMP2 vs. blank group,
P<0.05). There were no significant differences in the HAT
activities between the AdGFP and blank groups (P>0.05; Fig. 4A). The real-time RT-PCR data showed
that the expression levels of p300 were significantly increased and
reached a peak in the H9c2 cells 48 h after AdBMP2 transfection
(1.68±0.30-fold, AdBMP2 group vs. blank group, P<0.05). There
was no significant difference in the p300 expression levels between
the AdGFP and blank groups (P>0.05; Fig. 4B). Moreover, the expression of GCN5
was not significantly altered in H9c2 cells overexpressing BMP2
(P>0.05; Fig. 4C).
Discussion
GATA4, MEF2C and Tbx5 are the most significant
transcription factors during the early stage of heart development
(6). In the present study, the
expression levels of GATA4 and MEF2C, but not Tbx5, were identified
as significantly enhanced in H9c2 cells overexpressing BMP2. These
data are in agreement with the previous reports that BMP2 is able
to induce GATA4 and MEF2C expression ectopically, resulting in an
ectopic cardiac mesoderm specification in chicken embryos (9). These studies also observed that BMP2
was not able to affect the expression of Tbx5 in chicken embryos
(9,14). Consistent with these studies, the
results of the present study confirm that the expression of Tbx5 is
not altered in H9c2 cells overexpressing BMP2.
Numerous studies have reported that BMP2 is required
for heart development by inducing expression in various cardiac
genes. However, the underlying mechanisms are largely unknown. One
previous study in bovine granulosa cells suggested that BMP4 was
able to suppress the expression of StAR by inhibiting histone H3
acetylation in the promoter regions of the gene (23). Pan et al(24) suggested that the activation of Sox9
gene transcription by BMP2 was associated with chromatin remodeling
and histone modification in primary mouse embryo fibroblasts. In
the present study, western blotting assays were performed to
determine whether BMP2 was able to affect the histone H3
acetylation in H9c2 cells overexpressing BMP2. First an increase in
the total amount of Ac-H3 in the whole chromatin extracted from
H9c2 cells overexpressing BMP2 was identified. Further analysis of
histone H3 acetylation in the promoter regions of the specific
cardiac genes was then conducted. The results demonstrate that BMP2
is able to enhance histone H3 acetylation levels in the promoter
regions of GATA4 and MEF2C. The increase of histone H3 acetylation
levels in these genes is accompanied by an increase in the
expression levels of these genes, suggesting that histone H3
acetylation may be one of the molecular mechanisms by which BMP2
upregulates the expression of GATA4 and MEF2C in H9c2 cells
overexpressing BMP2. By contrast, no change in Tbx5 expression or
in histone H3 acetylation in the Tbx5 gene promoter was observed in
the same cells overexpressing BMP2.
The status of histone acetylation is determined by
the balanced actions between the HATs and HDACs. In the present
study, the colorimetric assay data show that BMP2 is able to
enhance the HAT activities in H9c2 cells overexpressing BMP2.
However, it is unclear which HAT subtype(s) participates in the
regulation of histone H3 acetylation in response to BMP2 in the
H9c2 cells. In our previous studies, p300 was identified with a
higher expression level than other HAT subtype in crescent-shaped
cardiogenic plates (25), and
inhibition of p300 HAT activity resulted in reduced histone
acetylation and decreased expression of GATA4 and MEF2C in mouse
cardiac myocytes (26). Other
studies also indicated a critical role for p300 in cardiac gene
expression and heart development (27). In the present study, the data
indicate that the expression level of p300 was significantly
increased in H9c2 cells overexpressing BMP2, while another HAT
subtype, GCN5, had no significant changes in the same cells
overexpressing BMP2. This suggests that p300 is more important than
GCN5, in BMP2-induced histone H3 hyperacetylation in the H9c2
cells.
In conclusion, the data demonstrate that BMP2
upregulates histone H3 acetylation levels in the promoter regions
of specific cardiac genes, GATA4 and MEF2C. BMP2 also increases HAT
activities and the expression of the HAT p300 subtype. The
upregulatory effects of BMP2 on GATA4 and MEF2C are due, at least
in part, to the increased acetylation levels of histone H3 in the
promoter regions of these genes, which is associated with increased
HAT activities and the expression of the HAT p300 subtype in the
H9c2 cells.
Acknowledgements
The authors thank Professor Xupei Huang from Florida
Atlantic University and Mr. Geoffrey Gatts from Ohio State
University, USA, for their critical reading and editing of the
manuscript. This study was supported by research grants from the
Natural Science Foundation of China (Grant no. 81070132) and from
the Natural Science Foundation of Chongqing (Grant no.
CSTC2009BA5084).
References
|
1
|
Olson EN: Gene regulatory networks in the
evolution and development of the heart. Science. 313:1922–1927.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bryantsev AL and Cripps RM: Cardiac gene
regulatory networks in Drosophila. Biochim Biophys Acta.
1789:343–353. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Black BL: Transcriptional pathways in
second heart field development. Semin Cell Dev Biol. 18:67–76.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Satou Y and Satoh N: Gene regulatory
networks for the development and evolution of the chordate heart.
Genes Dev. 20:2634–2638. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cripps RM and Olson EN: Control of cardiac
development by an evolutionarily conserved transcriptional network.
Dev Biol. 246:14–28. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Qian L, Huang Y, Spencer CI, Foley A,
Vedantham V, Liu L, Conway SJ, Fu JD and Srivastava D: In vivo
reprogramming of murine cardiac fibroblasts into induced
cardiomyocytes. Nature. 485:593–598. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brand T: Heart development: molecular
insights into cardiac specification and early morphogenesis. Dev
Biol. 258:1–19. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Song L, Yan W, Chen X, Deng CX, Wang Q and
Jiao K: Myocardial smad4 is essential for cardiogenesis in mouse
embryos. Circ Res. 101:277–285. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van Wijk B, Moorman AF and van den Hoff
MJ: Role of bone morphogenetic proteins in cardiac differentiation.
Cardiovasc Res. 74:244–255. 2007.PubMed/NCBI
|
|
10
|
Wang J, Greene SB and Martin JF: BMP
signaling in congenital heart disease: new developments and future
directions. Birth Defects Res A Clin Mol Teratol. 91:441–448. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sugi Y, Yamamura H, Okagawa H and Markwald
RR: Bone morphogenetic protein-2 can mediate myocardial regulation
of atrioventricular cushion mesenchymal cell formation in mice. Dev
Biol. 269:505–518. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang H and Bradley A: Mice deficient for
BMP2 are nonviable and have defects in amnion/chorion and cardiac
development. Development. 122:2977–2986. 1996.PubMed/NCBI
|
|
13
|
Lalani SR, Thakuria JV, Cox GF, Wang X, Bi
W, Bray MS, Shaw C, Cheung SW, Chinault AC, Boggs BA, Ou Z,
Brundage EK, Lupski JR, Gentile J, Waisbren S, Pursley A, Ma L,
Khajavi M, Zapata G, Friedman R, Kim JJ, Towbin JA, Stankiewicz P,
Schnittger S, Hansmann I, Ai T, Sood S, Wehrens XH, Martin JF,
Belmont JW and Potocki L: 20p12.3 microdeletion predisposes to
Wolff-Parkinson-White syndrome with variable neurocognitive
deficits. J Med Genet. 46:168–175. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamada M, Revelli JP, Eichele G, Barron M
and Schwartz RJ: Expression of chick Tbx-2, Tbx-3, and Tbx-5 genes
during early heart development: evidence for BMP2 induction of
Tbx2. Dev Biol. 228:95–105. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schlange T, Andrée B, Arnold HH and Brand
T: BMP2 is required for early heart development during a distinct
time period. Mech Dev. 91:259–270. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jenuwein T and Allis CD: Translating the
histone code. Science. 293:1074–1080. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Clayton AL, Hazzalin CA and Mahadevan LC:
Enhanced histone acetylation and transcription: a dynamic
perspective. Mol Cell. 23:289–296. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Backs J and Olson EN: Control of cardiac
growth by histone acetylation/deacetylation. Circ Res. 98:15–24.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu G, Nan C, Rollo JC, Huang X and Tian J:
Sodium valproate-induced congenital cardiac abnormalities in mice
are associated with the inhibition of histone deacetylase. J Biomed
Sci. 17:162010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhong L, Zhu J, Lv T, Chen G, Sun H, Yang
X, Huang X and Tian J: Ethanol and its metabolites induce histone
lysine 9 acetylation and an alteration of the expression of heart
development-related genes in cardiac progenitor cells. Cardiovasc
Toxicol. 10:268–274. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Feng C, Zhu J, Zhao L, Lu T, Zhang W, Liu
Z and Tian J: Suberoylanilide hydroxamic acid promotes
cardiomyocyte differentiation of rat mesenchymal stem cells. Exp
Cell Res. 315:3044–3051. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
|
|
23
|
Yamashita H, Murayama C, Takasugi R,
Miyamoto A and Shimizu T: BMP-4 suppresses progesterone production
by inhibiting histone H3 acetylation of StAR in bovine granulosa
cells in vitro. Mol Cell Biochem. 348:183–190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pan Q, Wu Y, Lin T, Yao H, Yang Z, Gao G,
Song E and Shen H: Bone morphogenetic protein-2 induces chromatin
remodeling and modification at the proximal promoter of Sox9 gene.
Biochem Biophys Res Commun. 379:356–361. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen G, Zhu J, Lv T, Wu G, Sun H, Huang X
and Tian J: Spatiotemporal expression of histone
acetyltransferases, p300 and CBP, in developing embryonic hearts. J
Biomed Sci. 16:242009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sun H, Yang X, Zhu J, Lv T, Chen Y, Chen
G, Zhong L, Li Y, Huang X, Huang G and Tian J: Inhibition of
p300-HAT results in a reduced histone acetylation and
down-regulation of gene expression in cardiac myocytes. Life Sci.
87:707–714. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shikama N, Lutz W, Kretzschmar R, Sauter
N, Roth JF, Marino S, Wittwer J, Scheidweiler A and Eckner R:
Essential function of p300 acetyltransferase activity in heart,
lung and small intestine formation. EMBO J. 22:5175–5185. 2003.
View Article : Google Scholar : PubMed/NCBI
|