Introduction
Medulloblastoma (MB) is the most common type of
malignant brain tumor in children and is characterized by frequent
proliferation and metastasis (1).
Successful treatments, including surgery, radiation and
chemotherapy, have improved in recent years; however, these
therapies result in side effects, including endocrinopathies,
impaired cognition and vasculopathies (2,3), and
survival rates remain to be improved. Thus, the elucidation of the
molecular mechanisms that drive cell proliferation and invasion may
provide more effective therapeutic strategies.
MicroRNAs (miRNAs) are an abundant group of
endogenous, small, non-coding RNAs that regulate gene expression at
the post-transcriptional level; miRNAs undergo base pairing with
target mRNAs in the 3′ untranslated region (3′UTR), leading to
translational inhibition and/or mRNA degradation (4–7).
Numerous studies have demonstrated significantly altered miRNA
expression patterns in types of human cancer, deregulation of miRNA
expression and the contribution of miRNAs to the multistep
processes of carcinogenesis either as oncogenes or as
tumor-suppressor genes (8,9). A number of miRNA expression profiling
studies have demonstrated that miRNA expression is dysregulated, by
the comparison of MBs with normal cerebellum (10–12).
A previous study determined that the expression of miR-17~92 was
upregulated in MBs and promoted cell proliferation (13). Elevated miR-21 expression has been
causally linked with cellular mobility and migration of MB in
vitro due to the suppression of the PDCD4 tumor suppressor
(14). The expression of miR-218
was identified to be significantly decreased in MB compared with
that of normal cerebellum, and miR-218 negatively regulated the
CDK6, Rictor and CTSB proto-oncogenes (15). However, the molecular mechanism of
miR-218 in MB remains unclear.
The present study aimed to observe the expression of
miR-218 in MB cell lines and in vitro experiments were
performed to determine the effect of miR-218 on cell growth and
invasion. Using reporter assays, miR-218 was identified to directly
target the 3′UTR of SH3GL1, which may aid in the elucidation of a
potential molecular therapeutic target for human MB cell lines.
Materials and methods
Cell culture
Daoy, D458 and PFSK human MB cell lines were
purchased from the American Type Culture Collection (Manassas, VA,
USA). UW228 and HEK-293T cells were provided by Professor Wu
(University of Fudan, Shanghai, China). All cell lines were
cultured in Dulbecco’s Modifed Eagle’s Medium (Gibco-BRL, Grand
Island, NY, USA) supplemented with 10% fetal bovine serum
(Gibco-BRL), penicillin (10 IU/ml) and streptomycin (10 μg/ml). All
cells were maintained in humidified air at 37°C and 5%
CO2. Cells were passaged a maximum of eight times.
RNA extraction and qPCR analyses
Total RNA was isolated from cells with TRIzol
reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) and
treated with DNA-free kit (Invitrogen Life Technologies) to remove
any remaining DNA, according to the manufacturer’s instructions.
qPCR assays were performed in triplicate to detect the miR-218 and
SH3GL1 expression using the PrimeScript RT reagent kit (Takara Bio,
Inc., Shiga, Japan) and the SYBR Green PCR Master Mix kit (Qiagen,
Hilden, Germany) according to the manufacturer’s instructions, with
the ABI PRISM® 7900 HT Sequence Detection system
(Invitrogen Life Technologies).
For miRNA detection, gene specific primers were
utilized. The U6 small nuclear RNA was used as a control to
determine the relative miRNA expression. The RT primers were
designed as follows: 5′-GTCGTATCCAGTGCAGGGTCCGAGG
TATTCGCACTGGATACGACACATGG-3′ for miR-218;
5′-AACGCTTCACGAATTTGCGT-3′ for U6. The PCR primers for mature
miR-218 and U6 were designed as follows: Sense:
5′-CGGGCTTGTGCTTGATCTA-3′ and antisense: 5′-GTGCAGGGTCCGAGGT-3′ for
miR-218; sense: 5′-CTCGCTTCGGCAGCACA-3′ and antisense:
5′-AACGCTTCACGAATTTGCGT-3′ for U6.
For the analysis of the SH3GL1 mRNA expression, 500
ng aliquots of total RNA were used to synthesize cDNA at a final
volume of 10 μl, and glyceraldehyde 3-phosphate dehydrogenase
(GAPDH) was used as the internal control. The primer sequences were
as follows: Sense: 5′-ATCGTCTTTCCGATCTTCCG-3′ and antisense:
5′-TGCCCTCGTACCAGTTCTCAT-3′ for SH3GL1; sense:
5′-GGGAGCCAAAAGGGTCAT-3′ and antisense: 5′-GAGTCCTTCCACGATACCAA-3′
for GADPH. The PCR cycle settings were as recommended by Qiagen:
95°C for 10 min; 40 amplification cycles at 95°C for 10 sec and
60°C for 1 min.
Plasmid construction and
transfection
To construct a luciferase reporter vector, a cDNA
fragment encoding the SH3GL 3′UTR from UW228 cells was amplified by
PCR and cloned downstream of the Renilla luciferase gene in
psicheck2 (Promega Corporation, Madison, WI, USA). The primer
sequences used were as follows: Sense:
5′-AAAGTTTAAACATCGTCTTTCCGATCTTCCG-3′ and antisense:
5′-AAAGCGGCCGCTGCCCTCGTACCAGTTCTCAT-3′ for SH3GL1. Psicheck2
luciferase constructs containing mutant 3′UTR (AAGCACAA to
AACGAGAA) were also generated using the QuikChange site-directed
mutagenesis kit (Stratagene, La Jolla, CA, USA) and the vectors
were termed SH3GL1-UTR-WT and SH3GL1-UTR-MUT, respectively.
Positive clones were identified by PCR screening and DNA
sequencing.
Knockdown experiments were performed by the
transient transfection of SH3GL1 siRNA using Lipofectamine™ 2000
(Invitrogen Life Technologies) according to the manufacturer’s
instructions. The cell lysates were harvested 48 h following
transfection for western blot analysis. The sequence of the SH3GL1
siRNA was obtained from Origene (SR304356; Rockville, MD, USA).
Briefly, for the reporter assays, cells were transiently
cotransfected with wild-type or mutant reporter plasmids and
miR-218. Renilla and Firefly luciferase activities were measured 36
h following transfection using the Dual-Luciferase assay (Promega
Corporation) and the results were normalized with Firefly
luciferase. Each reporter plasmid was transfected at least three
times (on different days) and samples were assayed in
triplicate.
Lentivirus packaging, infection and
stable cell generation
The lentiviral vector (pCDH-Vector) was obtained
from System Biosciences (Mountain View, CA, USA). A total of 16 μg
of the plasmids, including pCDH-miR-218 or pCDH-Vector, TAT, GAG,
REC and VSVG, were cotransfected with 40 μl Lipofectamine 2000
(Invitrogen Life Technologies) into HEK-293T cells in a 100-mm
diameter culture dish. The supernatant was collected 60 h following
infection, filtered through a 0.45-μm pore filter and used as the
source of the virus. PFSK and UW228 cells were infected with either
pCDH-Vector or pCDH-Vector with 8 μg/ml polybrene (Sigma-Aldrich,
St. Louis, MO, USA) for 24 h, and the medium was replaced. The
efficiency of infection was measured under a fluorescent microscope
72 h following infection.
Cell proliferation
Cells were plated at a density of 4,000 cells/well,
in 4 wells of 96-well plates containing complete medium. Cell
proliferation was detected using CellTiter 96 AQueous One Solution
(Promega Corporation) at 0, 24, 48 and 72 h following plating. For
measurement of cell proliferation, 20 μl methanethiosulfonate
reagent was added to the medium and incubated at 37°C in a
humidified 5% CO2 atmosphere for 1 h. The absorbance was
read at 490 nm using a 96-well microplate reader (ELx800 Absorbance
Microplate Reader; BioTek, Winooski, VT, USA).
Invasion assays
The invasive ability of the cells was determined
using 24-well Matrigel Invasion Chambers according to the
manufacturer’s instructions (Corning Life Sciences, Corning, NY,
USA). Media (600 μl) containing 10% FBS was added to the lower
chamber and a cell suspension of 2×105 cells in 100 μl
DMEM medium was added into each well of the top chamber. Subsequent
to this, the cells were incubated for 48 h at 37°C in a humidified
incubator with 5% CO2. Cells that had passed through the
membrane were stained with methanol and 0.1% crystal violet, imaged
and counted using an IX71 inverted microscope (Olympus, Tokyo,
Japan) from three random microscope fields per filter. Experiments
were repeated three times.
Western blot analysis
Cells were washed with phosphate-buffered saline and
lysed with RIPA lysis buffer (CST) supplied with protease and
phosphatase inhibitor cocktails (1%; Sigma-Aldrich). Total
denatured proteins (50 μg) were subjected to sodium dodecyl sulfate
(SDS)-polyacrylamide gel electrophoresis (10% SDS-acrylamide gel)
and transferred onto polyvinylidene fluoride membranes (Bio-Rad,
Hercules, CA, USA). The membrane was incubated with SH3GL1
(TA309473, Origene), Phospho-c-Jun (Ser73; #9164; Cell Signaling
Technology, Inc., Beverly, MA, USA), Phospho-p44/42 MAPK
[extracellular signal-regulated kinases (Erk)1/2; Thr202/Tyr204;
#9101; Cell Signaling Technology, Inc.] and GAPDH antibodies
(10494-1-AP, Proteintech™, Chicago, IL, USA). Primary antibodies
were detected with horseradish peroxidase-conjugated secondary
antibodies (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA)
and the membranes were subjected to a chemiluminescence detection
assay.
Statistical analysis
All experiments were performed at least in
triplicate. Values are presented as the mean ± standard deviation.
Significance was examined by Student’s t-test (two-tailed) or
one-way analysis of variance. P<0.05 was considered to indicate
a statistically significant difference.
Results
miR-218 is downregulated in MB and its
expression is inversely correlated with that of SH3GL1
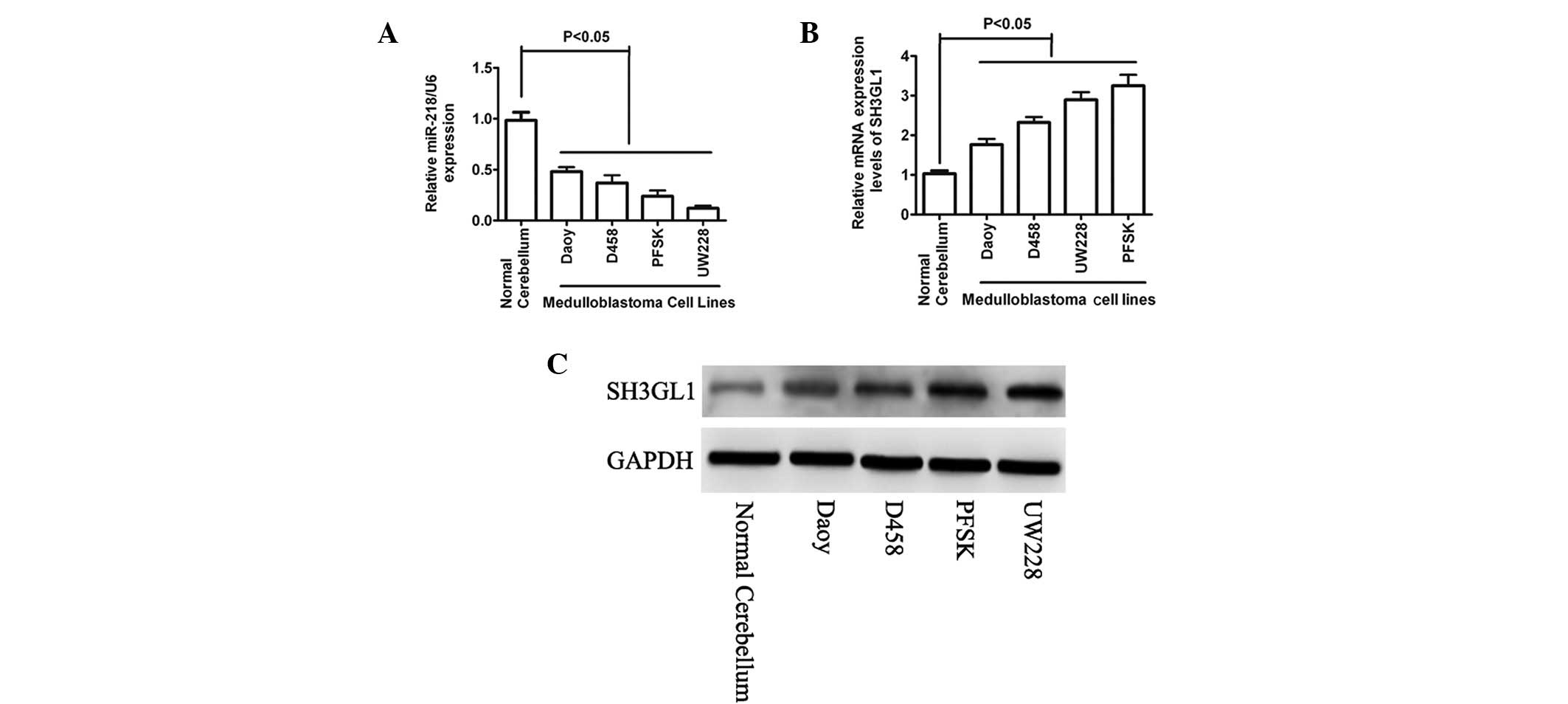
To investigate the involvement of miR-218 in MB, the
miR-218 expression level was detected using qPCR in a number of
selected cancer cell lines as well as in normal cerebellum
(Fig. 1A). The expression level of
miR-218 was significantly lower or undetectable in all tested
cancer cell lines compared with that of the normal cerebellum. To
identify the proteins that miR-218 affected, the mRNA targets were
predicted by bioinformatics. Among the list of potential targets,
the seed sequence of miR-218 was complementary to the 3′-UTR of
SH3GL1. To investigate the correlation between the expression of
miR-218 and SH3GL1, the expression of SH3GL1 at the mRNA and
protein levels were determined in the same panel of cell lines. As
expected, the mRNA and protein expression levels of SH3GL1 were
higher in the cancer cell lines when compared with that in the
normal cerebellum (Fig. 1B and C).
The results indicated that a potential tumor suppressor role of
miR-218 is downregulated in MB, which may have directly targeted
SH3GL1.
miR-218 directly targets the 3′UTR of
SH3GL1
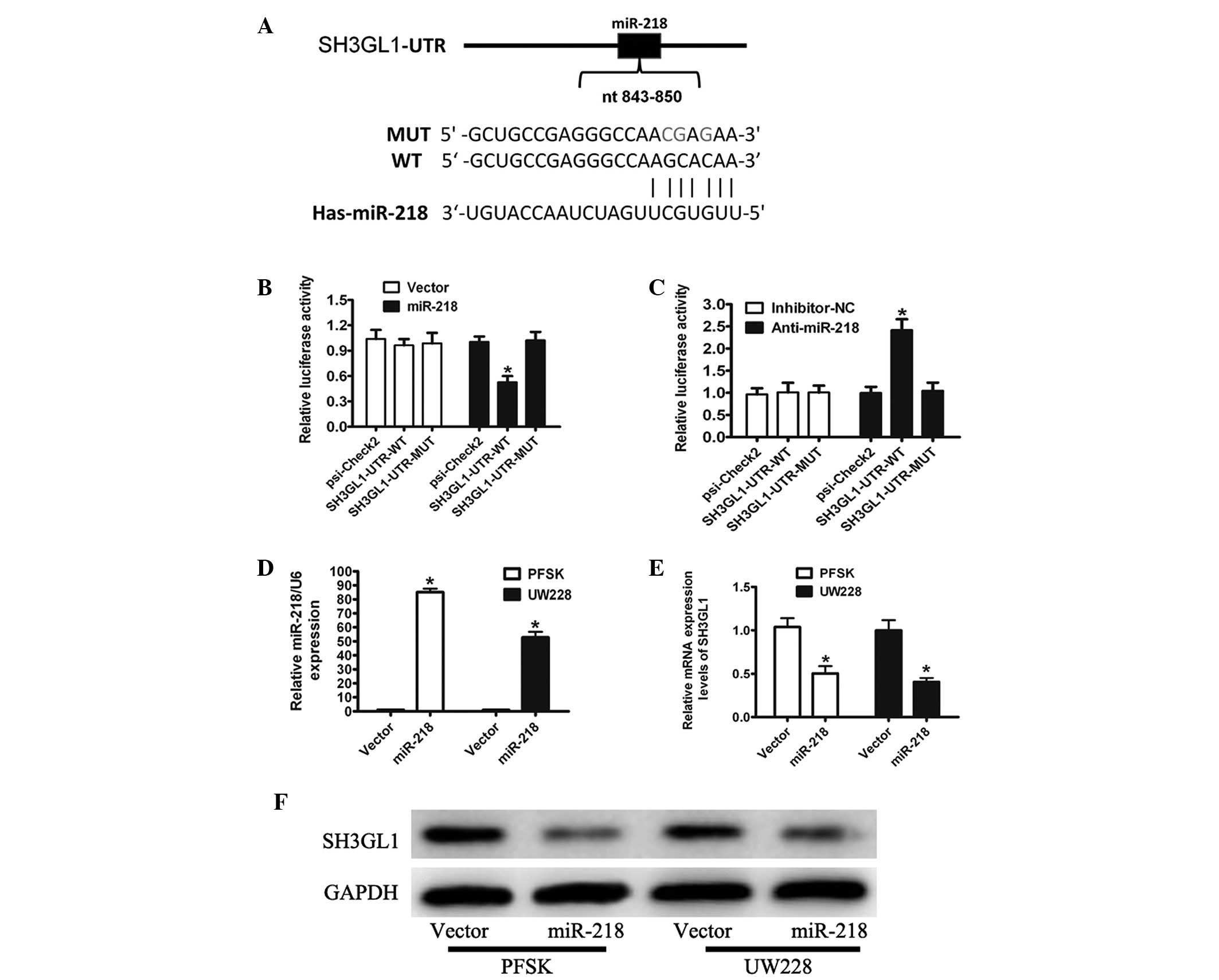
To determine whether miR-218 regulates SH3GL1, a
reporter plasmid (in which the full-length 3′-UTR of SH3GL1 was
inserted downstream of the Renilla luciferase gene) was generated
for reporter gene assays (Fig.
2A). The transient transfection of HEK293T cells with the
SH3GL1-UTR-WT or SH3GL1-UTR-MUT construct and miR-218 resulted in a
significant reduction in reporter gene expression when compared
with that of the control vector (Fig.
2B). Mutations in the targeting sites of miR-218 were
unaffected by the simultaneous transfection with miR-218 (Fig. 2B). To further confirm that
miR-218-mediated reduction of the luciferase activity of SH3GL1 was
due to the direct interaction between miR-218 and its putative
binding site, cotransfection of SH3GL1-UTR-WT or SH3GL1-UTR-MUT
with anti-miR-218 in HEK293T cells was conducted. As expected,
anti-miR-218 markedly increased the luciferase activity of
SH3GL1-UTR-WT compared with that of anti-miR-normal cerebellum
(NC); however, the effect was completely abolished in this mutant
construct (Fig. 2C).
Significance of SH3GL1 suppression
To further address the significance of the
suppression of SH3GL1 by miR-218 in MB cells, PSFK and UW228 cells
stably expressing miR-218 were generated and the overexpression of
miR-218 was confirmed by qPCR analysis (Fig. 2D). The mRNA and protein levels of
SH3GL1 by qPCR and western blot analysis demonstrated that the
overexpression of miR-218 downregulated the endogenous SH3GL1
levels (Fig. 2E and F). These
results suggested that miR-218 downregulated the endogenous SH3GL1
expression by directly binding to the 3′-UTR sequence of
SH3GL1.
Stable overexpression of miR-218 inhibits
cell proliferation and invasion
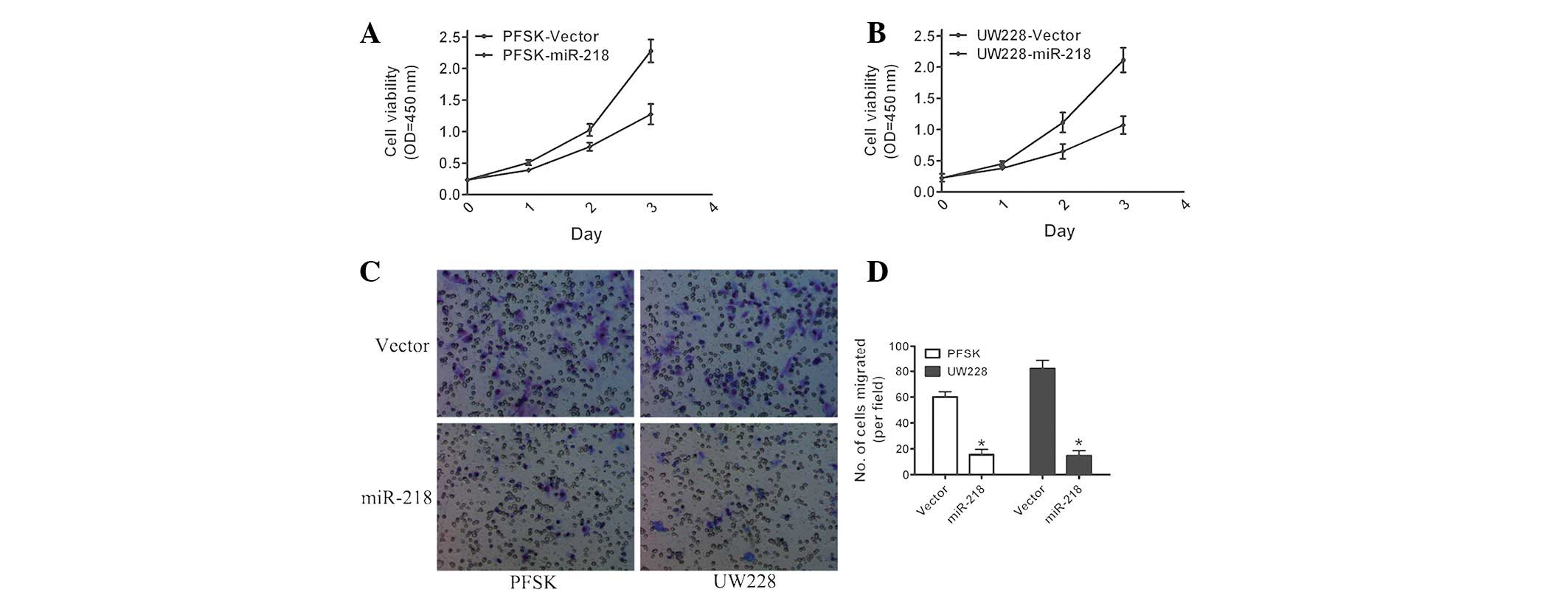
To assess the biological role of miR-218, the effect
of the overexpression of miR-218 on cell proliferation and invasion
was determined. MTT assays demonstrated that the overexpression of
miR-218 markedly impaired the growth rate of MB cells as compared
with that of the control vector (Fig.
3A and B). Similarly, the results of the invasion assays
revealed that invasive ability was decreased following the
overexpression of miR-218 in PSFK and UW228 cells (Fig. 3C and D). In conclusion, these
results demonstrated that miR-218 inhibited MB cell proliferation
and invasion.
miR-218 suppresses SH3GL1 and negatively
regulates the ERK pathway in MB cell lines
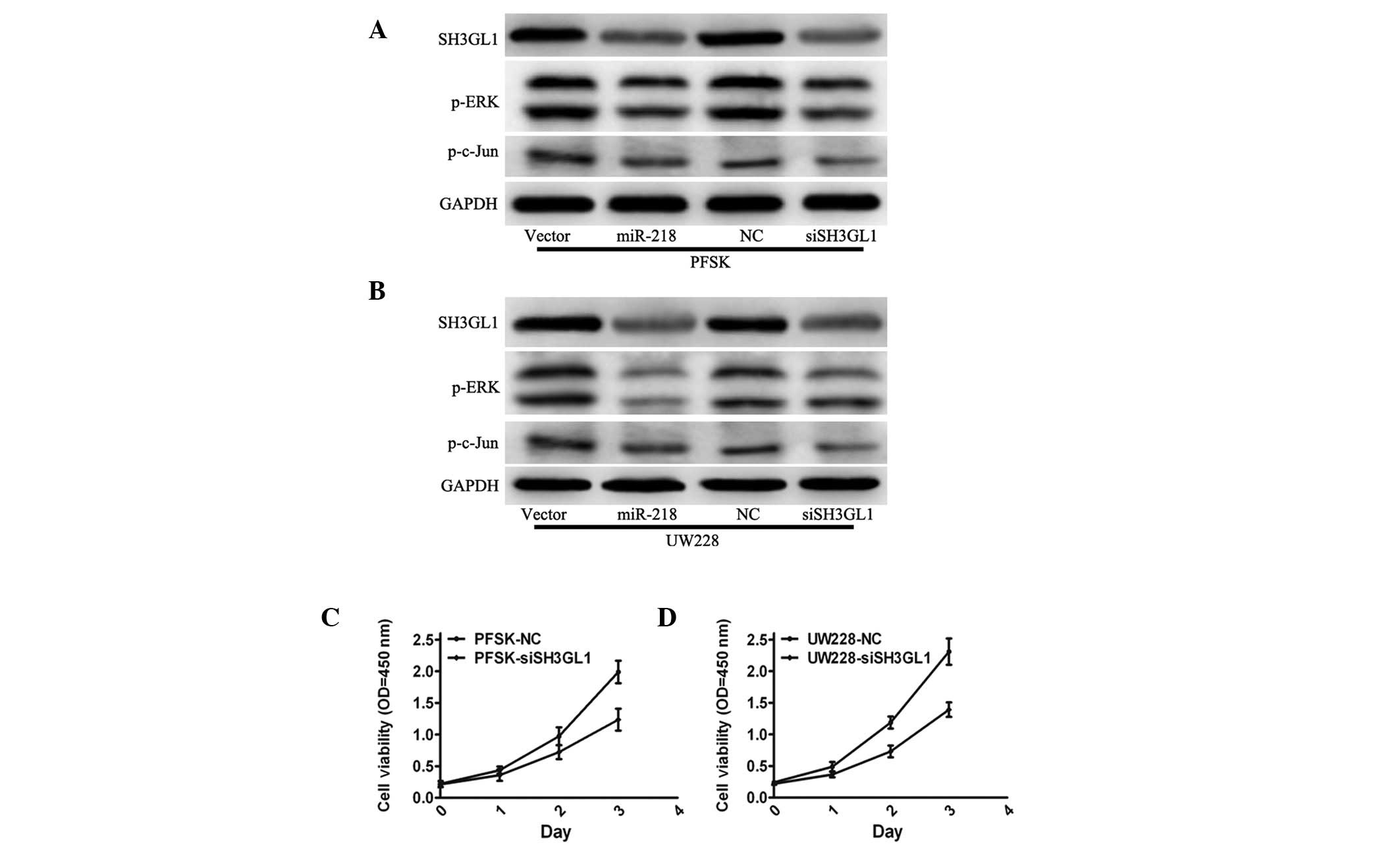
To confirm the involvement of SH3GL1 in the
miR-218-mediated inhibition of cell growth, siRNA targeting SH3GL1
was used to downregulate SH3GL1 expression and analyze its effect
on cell growth. The protein expression levels of SH3GL1,
phosphorylated (p)-ERK and p-Jun were significantly downregulated
in PSFK and UW228 cells with siSH3GL1 compared with those of the
normal cerebellum (Fig. 4A and B).
PSFK and UW228 cells in which SH3GL1 had been knocked down
demonstrated a decrease in growth rate in the MTT assay (Fig. 4C and D), which was similar to the
phenotype observed following miR-218 overexpression in PSFK and
UW228 cells. The expression of p-ERK following ectopic expression
of miR-218 in MB cells was also observed. Consistent with the
effect of SH3GL1, miR-218 overexpression significantly reduced the
expression of p-ERK and p-Jun in PSFK and UW228 cells and induced
the maintained overexpression of miR-218 (Fig. 4A and B). In conclusion, the
tumor-suppressive role of miR-218 may mediate SH3GL1 via the
mitogen-activated protein kinase pathway.
Discussion
Understanding of the biological mechanisms of
malignant brain tumors, such as MB, is essential for advances in
therapy (13,16–18).
Numerous studies observed the aberrant expression of microRNAs in
MB and suggested that the determination of key microRNAs may lead
to the identification of novel targets for more effective therapies
with fewer side effects (14,15).
The reduced expression of miR-218 was also demonstrated in gastric,
colon, prostate, pancreatic, glioma cell and cervical cancer
(19–21). In the present study, low expression
levels of miR-218 were identified in MB cell lines compared with in
normal cerebellum; the expression of miR-218 was significantly
downregulated in all examined MB cell lines, including Daoy, D458,
PFSK and UW228, compared with in normal cerebellum. This was
consistent with the previous studies, which demonstrated that
miR-218 functions as a tumor suppressor in MB (15).
Identification of putative miRNA targets is
important for the understanding of the function of miRNAs. In this
study, we identified SH3GL1 as a direct target of miR-218 with the
aid of bioinformatics, and demonstrated that the upregulation of
miR-218 markedly reduced the endogenous SH3GL1 expression at the
transcriptional and protein levels in MB cells. SH3GL1 has been
observed to be involved in endocytosis and signal transduction
(22,23). The epidermal growth factor receptor
(EGFR) signaling pathway is an important pathway that regulates
cellular proliferation, differentiation and invasion (24). SH3GL1 binds to BPGAP1, which is
involved in the activation of EGFR endocytosis and ERK1/2
signaling. In addition, overexpression of SH3GL1 has been
demonstrated to promote cell growth (25,26).
The reduced expression of miR-218 in MB suggested
that miR-218 acted as a tumor suppressor. The overexpression of
miR-218 suppressed cell growth and invasion in vitro. It has
been demonstrated that miR-218 inhibited the growth of oral cancer
by targeting the mTOR component Rictor with the inhibition of Akt
phosphorylation (27). Exogenous
expression of miR-218 suppressed the growth of nasopharyngeal
carcinoma by BIRC5 and suppressed cell invasion via the SLIT-ROBO
pathway (28). Previously, miR-218
was observed to be downregulated in MB and the restoration of
miR-218 resulted in a marked decrease in MB cell growth, colony
formation, migration and invasion, as well as tumor sphere size, by
directly targeting CDK6, RICTOR and CTSB (15). However, miR-218 has been determined
to stimulate the Wnt pathway by downregulating SOST, DKK2 and SFRP2
during the process of osteogenesis (29), which is inconsistent with our
findings. This inconsistency may be due to the use of different
cancer cell lines (gastric and lung), and therefore the
organ-specific targets and cellular context may differ.
To further determine the effect of miR-218 on MB,
SH3GL1 was knocked down in PFSK and UW228 cells, and the
proliferation was decreased, which was similar to the phenotype
observed following miR-218 overexpression in PFSK and UW228 cells.
Previous studies implicated that SH3GL1 activated P-ERK activity,
which regulates cancer cell growth and migration. In the current
study, overexpression of miR-218 decreased the expression of P-ERK
and P-Jun, which was consistent with the results of the knockdown
of SH3GL1. Thus, this study demonstrated that miR-218 exerted tumor
suppressor effects in MB cells by downregulating SH3GL1. The
mechanisms underlying SH3GL1 in carcinogenesis require further
investigation.
In conclusion, the present study confirmed that
miR-218 is a tumor suppressor miRNA involved in the initiation and
progression of human MB, by affecting multiple signal pathways.
These results also suggested that miR-218 may provide therapeutic
potential for the treatment of MB.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (grant no. 81201765).
References
|
1
|
Gilbertson RJ and Ellison DW: The origins
of medulloblastoma subtypes. Annu Rev Pathol. 3:341–365. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ellison DW, Kocak M, Dalton J, et al:
Definition of disease-risk stratification groups in childhood
medulloblastoma using combined clinical, pathologic, and molecular
variables. J Clin Oncol. 29:1400–1407. 2011. View Article : Google Scholar
|
|
3
|
No authors listed. 43rd Congress of the
International Society of Paediatric Oncology (SIOP) 2011. In:
Auckland, New Zealand. 28th–30th October, 2011; SIOP Abstracts
Pediatr Blood Cancer. 57. pp. 705–897. 2011, PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu L, Fan J and Belasco JG: MicroRNAs
direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA.
103:4034–4039. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI
|
|
7
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shenouda SK and Alahari SK: MicroRNA
function in cancer: oncogene or a tumor suppressor? Cancer
Metastasis Rev. 28:369–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar
|
|
10
|
Uziel T, Karginov FV, Xie S, et al: The
miR-17~92 cluster collaborates with the Sonic Hedgehog pathway in
medulloblastoma. Proc Natl Acad Sci USA. 106:2812–2817. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ferretti E, De Smaele E, Po A, et al:
MicroRNA profiling in human medulloblastoma. Int J Cancer.
124:568–577. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Genovesi LA, Anderson D, Carter KW, et al:
Identification of suitable endogenous control genes for microRNA
expression profiling of childhood medulloblastoma and human neural
stem cells. BMC Res Notes. 5:5072012. View Article : Google Scholar
|
|
13
|
Northcott PA, Fernandez-L A, Hagan JP, et
al: The miR-17/92 polycistron is up-regulated in sonic
hedgehog-driven medulloblastomas and induced by N-myc in sonic
hedgehog-treated cerebellar neural precursors. Cancer Res.
69:3249–3255. 2009. View Article : Google Scholar
|
|
14
|
Grunder E, D’Ambrosio R, Fiaschetti G, et
al: MicroRNA-21 suppression impedes medulloblastoma cell migration.
Eur J Cancer. 47:2479–2490. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Venkataraman S, Birks DK, Balakrishnan I,
et al: MicroRNA 218 acts as a tumor suppressor by targeting
multiple cancer phenotype-associated genes in medulloblastoma. J
Biol Chem. 288:1918–1928. 2013. View Article : Google Scholar
|
|
16
|
Shapiro RH and Chang AL: Urgent
radiotherapy is effective in the treatment of metastatic
medulloblastoma causing symptomatic brainstem edema. Pediatr Blood
Cancer. 57:1077–1080. 2011. View Article : Google Scholar
|
|
17
|
Gajjar A and Pizer B: Role of high-dose
chemotherapy for recurrent medulloblastoma and other CNS primitive
neuroectodermal tumors. Pediatr Blood Cancer. 54:649–651. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Taniguchi E, Cho MJ, Arenkiel BR, et al:
Bortezomib reverses a post-translational mechanism of tumorigenesis
for patched1 haploinsufficiency in medulloblastoma. Pediatr Blood
Cancer. 53:136–144. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Volinia S, Calin GA, Liu CG, et al: A
microRNA expression signature of human solid tumors defines cancer
gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Song L, Huang Q, Chen K, et al: miR-218
inhibits the invasive ability of glioma cells by direct
downregulation of IKK-β. Biochem Biophys Res Commun. 402:135–140.
2010.PubMed/NCBI
|
|
21
|
Volinia S, Calin GA, Liu CG, et al: A
microRNA expression signature of human solid tumors defines cancer
gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ringstad N, Nemoto Y and De Camilli P: The
SH3p4/Sh3p8/SH3p13 protein family: binding partners for
synaptojanin and dynamin via a Grb2-like Src homology 3 domain.
Proc Natl Acad Sci USA. 94:8569–8574. 1997. View Article : Google Scholar
|
|
23
|
So CW, So CK, Cheung N, et al: The
interaction between EEN and Abi-1, two MLL fusion partners, and
synaptojanin and dynamin: implications for leukaemogenesis.
Leukemia. 14:594–601. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tanaka K, Babic I, Nathanson D, et al:
Oncogenic EGFR signaling activates an mTORC2-NF-kappaB pathway that
promotes chemotherapy resistance. Cancer Discov. 1:524–538. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lua BL and Low BC: Activation of EGF
receptor endocytosis and ERK1/2 signaling by BPGAP1 requires direct
interaction with EEN/endophilin II and a functional RhoGAP domain.
J Cell Sci. 118:2707–2721. 2005. View Article : Google Scholar
|
|
26
|
Ma LH, Liu H, Xiong H, et al: Aberrant
transcriptional regulation of the MLL fusion partner EEN by
AML1-ETO and its implication in leukemogenesis. Blood. 109:769–777.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Uesugi A, Kozaki K, Tsuruta T, et al: The
tumor suppressive microRNA miR-218 targets the mTOR component
Rictor and inhibits AKT phosphorylation in oral cancer. Cancer Res.
71:5765–5778. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fish JE, Wythe JD, Xiao T, et al: A
Slit/miR-218/Robo regulatory loop is required during heart tube
formation in zebrafish. Development. 138:1409–1419. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hassan MQ, Maeda Y, Taipaleenmaki H, et
al: miR-218 directs a Wnt signaling circuit to promote
differentiation of osteoblasts and osteomimicry of metastatic
cancer cells. J Biol Chem. 287:42084–42092. 2012. View Article : Google Scholar : PubMed/NCBI
|