Introduction
Nogo-A, a myelin-associated endoplasmic reticulum
protein, is encoded by the reticulon-4 (RTN4) gene located
at chromosome 2p13-14 (1). The
human RTN4 gene encodes three alternatively spliced
variants, Nogo-A, Nogo-B and Nogo-C. Nogo-A is the largest Nogo
isoform, which exhibits a unique N-terminal extension and is mainly
expressed by oligodendrocytes in the brain. By contrast, Nogo-B
demonstrates a ubiquitous pattern of expression and Nogo-C is
highly expressed in skeletal muscles (2,3). The
three Nogo isoforms share a common C-terminal domain of 188 amino
acids, which contains two putative transmembrane domains and an
endoplasmic reticulum retention motif (4). In vitro studies have reported
that Nogo-A contributes to the axonal growth inhibitory function of
central myelin; however, there have been contradictory results
regarding this growth inhibitory effect in Nogo-A knockout mice
(5,6). Whilst the role of Nogo-A in the
nervous system is well established, including activation of growth
cone collapse, axonal outgrowth inhibition and synaptic plasticity
(7–9), the role of endogenous Nogo-A in
non-neural systems is yet to be elucidated.
Hepatocellular carcinoma (HCC) is the most common
primary malignant tumor and accounts for ~5.6% of all tumors
(10). As a result of the poor
prognosis associated with HCC, a high mortality rate is observed
among patients with the tumor. Therefore, exploring the molecular
mechanisms associated with HCC and developing more effective
targeted therapies are of utmost importance. The Arg114Gly mutation
in the Nogo-C gene, which affects the dimensional structure of the
Nogo-66 domain, was observed to promote apoptosis in a liver cancer
cell line (11). The Nogo-66
domain has been identified as a conserved functional group among
the three Nogo isoforms; therefore, it is hypothesized that Nogo-A
may have a role in HCC.
The present study aimed to explore the role of the
neural-associated Nogo-A gene in liver cancer cells. Nogo-A was
observed to exhibit high expression in the HCC SMMC-7721 cell line.
Furthermore, a novel lentivirus vector that mediated RNA
interference (RNAi) targeting of Nogo-A (LV-Nogo-A-siRNA) was
employed to study the effect of Nogo-A knockdown on SMMC-7721 cell
growth in vitro.
Materials and methods
Cell culture
The human liver cancer cell lines HepG2, Huh-7,
BEL-7402 and SMMC-7721 (Shanghai GeneChem Co., Shanghai, China)
were maintained in Dulbecco’s modified Eagle medium (DMEM;
Gibco-BRL, Grand Island, NY, USA) supplemented with 10% (v/v)
heat-inactivated fetal bovine serum (FBS; Gibco-BRL) at 37°C with
5% CO2.
Construction of lentiviral vectors, and
preparation and transduction of the lentivirus
The complementary DNA sequence of Nogo-A was
designed from full-length Nogo-A by Shanghai GeneChem Co., Ltd. The
sequence of the small interfering RNA (siRNA) targeting Nogo-A was
as follows: ccggGC TATATCTGAGGAGTTGGTTTTCAAGAGAACCAACTC
CTCAGATATAGCTTTTTgaatt. Following the testing of knockdown
efficiencies, stem-loop oligonucleotides were synthesized and
cloned into the lentivirus-based vector, pGCSIL-enhanced green
fluorescence protein (eGFP). A non-targeting stem-loop DNA was
generated as a negative control. An improved lentivirus system of
pGC-LV was developed by Shanghai GeneChem Co, Ltd. This system
consisted of the following plasmids: pHELPER1.0, a packaging
plasmid in which the accessory genes, vif, vpr, vpu and nef, and a
regulatory gene, tat, were deleted; pHELPER2.0, an envelope plasmid
for vesicular stomatitis virus G glycoprotein (VSV-G); and
pGCSIL-eGFP, a transferring plasmid with a multiple cloning site
and the gene encoding eGFP (Fig.
1A).
 | Figure 1Packaging system of pGC-LV and
identification of pGCSIL-eGFP using PCR. (A) Recombinant
self-inactivating lentivirus vectors were produced by
co-transfection of the envelope plasmid pHELPER2.0, packaging
plasmid pHELPER1.0 and transfer vector plasmid. (B) Detection of
the positive clone-containing pGCSIL-eGFP using PCR. Lane 1,
negative control with H2O (NC1); lane 2, empty vector of
306 bp (NC2); lane 3, DNA ladder (M) from top to bottom, 5 kb, 3
kb, 2 kb, 1.5 kb, 1 kb, 750 bp, 500 bp, 250 bp and 100 bp; lanes
4–7, recombinant vector of 343 bp (V1–V4). LTR, long terminal
repeat; RRE, Rev response element; hU6, human U6 promoter; MCS,
multiple cloning sites; CMV, cytomegalovirus promoter; eGFP,
enhanced green fluorescent protein marker gene; WPRE, woodchuck
post-transcriptional regulatory element; cPPT, central polypurine
tract; Env, envelope protein; VSV-G, vesicular stomatitis virus G
protein envelope; GAG, group-specific antigen; POL, reverse
transcriptase; PRO, protease; PCR, polymerase chain reaction. |
The recombinant Nogo-A-siRNA lentivirus vector was
generated using co-transfection of 293T cells with 20 μg
pGCSIL-Nogo-A-siRNA-eGFP, 15 μg pHELPER1.0, 10 μg pHELPER2.0 and
Opti-MEM® (Invitrogen Life Technologies, Carlsbad, CA,
USA) of the same volume in 15-cm dishes with
Lipofectamine® 2000 (Invitrogen Life Technologies). The
293T cells were cultured in DMEM containing 10% FBS. Culture
supernatants were collected after two days, filtered through a
0.45-μm pore size filter and concentrated using
Centricon® Plus-20 (Millipore Corporation, Hayward, CA,
USA). The viral filtrate was collected and stored at −80°C.
Cells were incubated with the lentivirus in a small
volume of serum-free DMEM at 37°C for 4 h. Medium was then replaced
with DMEM containing 10% FBS, and cells were cultured further as
indicated for the following experiments. eGFP revealed that the
infection efficiency in SMMC-7721 cells was ~90% and at a
multiplicity of infection (MOI) of 30. No viral toxicity was
observed at this concentration in the SMMC-7721 cells; therefore,
the following experiments were performed using viruses at such
MOIs, unless indicated. The lentivirus package with eGFP was used
for the cell proliferation assay, and that without eGFP was used
for the cell apoptosis assay.
Western blot analysis of exogenous
flag-Nogo-A expression
To assess the gene knockdown efficiency of
Nogo-A-siRNA, an exogenous 3flag-Nogo-A fusion protein was
constructed into a GV143 vector (Shanghai GeneChem Co.). The
constructed 3flag-Nogo-A vector was co-transfected with
pGCSIL-Nogo-A-siRNA-eGFP or pGCSIL-scr-siRNA-eGFP, respectively,
into 293T cells. The transfection ratio was estimated using
fluorescence microscopy. Total proteins were extracted 48 h
following transfection and the protein was quantified using the
Coomassie brilliant blue assay. Subsequently, 20 μg protein was
boiled in loading buffer, separated on 10% SDS-polyacrylamide gels,
electro-transferred to polyvinylidene fluoride (PVDF) membranes and
probed with mouse anti-flag (Sigma Aldrich, St. Louis, MO, USA) and
mouse anti-GAPDH (Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA) antibodies overnight at dilutions of 1:3,000 and 1:5,000
respectively. Membranes were then incubated with a goat anti-mouse
immunoglobulin G (IgG) peroxidase-conjugated secondary antibody
(Santa-Cruz Biotechnology, Inc., 1:5,000), prior to development
using the Amersham enhanced chemiluminescence (ECL) plus western
blotting detection system (Amersham Pharmacia Biotech, Piscataway,
NJ, USA).
RNA isolation and quantitative polymerase
chain reaction (qPCR) analysis
Total RNA was extracted using TRIzol®
reagent (Invitrogen Life Technologies) in accordance with the
manufacturer’s instructions. A total of 2 μg RNA was subjected to
reverse transcription. The PCR primer sequences were as follows:
GAPDH, 5′-TGACTTCAACAGCGACAC CCA-3′ (forward) and
5′-CACCCTGTTGCTGTAGCCAAA-3′ (reverse); Nogo-A,
5′-AGGAGCAGCCAGGTAACAC-3′ (forward) and
5′-GAGACAGAGAAGGAAGAGAAGC-3′ (reverse), as described previously
(11). PCR products were separated
on a 1% agarose gel, and visualized and photographed under
ultraviolet light for semi-quantitation.
Cell growth assay
The cell growth rate was determined using the
Cellomics® ArrayScan® VTI machine (Thermo
Fisher Scientific, San Jose, CA, USA). Cells were transduced with
lentivirus vectors for 72 h, prior to being seeded into flat-bottom
96-well plates at 2,000 cells per well. Cells were observed at one,
two, three, four and five days using the Cellomics ArrayScan VTI
machine. In the Cellomics ArrayScan VTI system (Thermo Fisher
Scientific), an automated inverted epifluorescence microscope was
used to record images from multiple fields in each individual well.
Fluorescence images were acquired using a high-resolution
charge-coupled device (CCD) camera. Cells were identified based on
the presence of valid nuclei and cell body measurements, determined
by analyzing size, shape and eGFP fluorescence intensity. Images
were then analyzed using the Cellomics software and cells were
counted (12). Cell growth curves
were generated following three experimental repeats.
Colony formation assay
The Cellomics ArrayScan VTI system was used to
perform colony formation assays to assess the anchorage-independent
growth ability of cells as a characteristic of in vitro
tumorigenicity. SMMC-7721 cells were infected with the virus for 24
h, then detached using Trypsin (Chemreagent, Shanghai, China) and
plated in 96-well plates at 500 cells/well. The number of foci
(>100 μm) was counted after 14 days. Each experiment was
performed in triplicate.
Flow cytometric analysis
Flow cytometric analysis was performed to determine
the distribution of cells throughout the cell cycle and those
undergoing apoptosis, and was performed as described previously
(13). SMMC-7721 cells were seeded
and transduced with lentivirus vectors and then cultured for 96 h
in complete medium. Following one wash with Hanks’ balanced salt
solution (Sigma Aldrich) adherent cells were detached using
trypsin, and cells were washed once with ice cooled
phosphate-buffered saline (PBS). Prior to analysis, the
transduction efficiency and cell death rate were estimated using a
fluorescence microscope. Cells with >90% transduction efficiency
and <1% death rate were used for the subsequent
fluorescence-activated cell sorting (FACS) analysis.
For analysis of apoptosis, cells were washed with 1X
binding buffer before 1×106–1×107 cells were
pelleted and resuspended in 1 ml 1X staining buffer. A total of 100
μl of the cell suspension was added to 5 μl Annexin
V-allophycocyanin (APC) and incubated in the dark at room
temperature for 10–15 min. Data from ≥10,000 cells were collected
and analyzed using CellQuest software (Becton Dickinson, San Diego,
CA, USA). The apoptosis ratio was calculated as the number of
apoptotic cells/total number of cells.
For cell cycle analysis, cells were fixed using 0.5
ml 70% iced alcohol and incubated at 4°C for 1 h, prior to being
washed once with ice cooled PBS. The cell pellet was resuspended
with staining buffer at 4°C for 30 min [40× 2 mg/ml propidium
iodide (PI): 100× 10 mg/ml RNase: 1X PBS=25:10:1,000]. The
suspension was filtered through a 50-μm nylon mesh, and flow
cytometry was used to analyze the DNA content of the stained nuclei
with a BD FACSCalibur machine (Becton Dickinson). Cell cycle
distribution was assessed using Multicycle cell cycle analysis
software (Phoenix Flow Systems, San Diego, CA, USA).
Statistical analysis
Each experiment was repeated at least three times.
Bands from western blot analysis or qPCR were quantified using
Quantity One® software (Bio-Rad Laboratories Inc.,
Berkley, CA, USA). Relative protein or mRNA levels were calculated
using GAPDH as a normalization control. T-test analysis was
performed to determine the significance of the differences between
means. All statistical analyses were performed using Excel 2003
software (Microsoft, Redmond, WA, USA). A value of P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of Nogo-A in liver cancer cell
lines
To determine the expression of Nogo-A in different
liver cancer cell lines, qPCR analysis was performed. As shown in
Fig. 2, Nogo-A expression was
observed in four different liver cancer cell lines.
Effect of RNAi targeting Nogo-A on Nogo-A
expression using the LV-Nogo-A-siRNA lentivirus vector
To examine the interrelation between Nogo-A and
liver cancer cells, a lentivirus-delivered Nogo-A-specific siRNA
vector (LV-Nogo-A-siRNA) and a negative control scramble-siRNA
vector (LV-scr-siRNA) were constructed. The two vectors were then
respectively transduced into SMMC-7721 cells for three days. qPCR
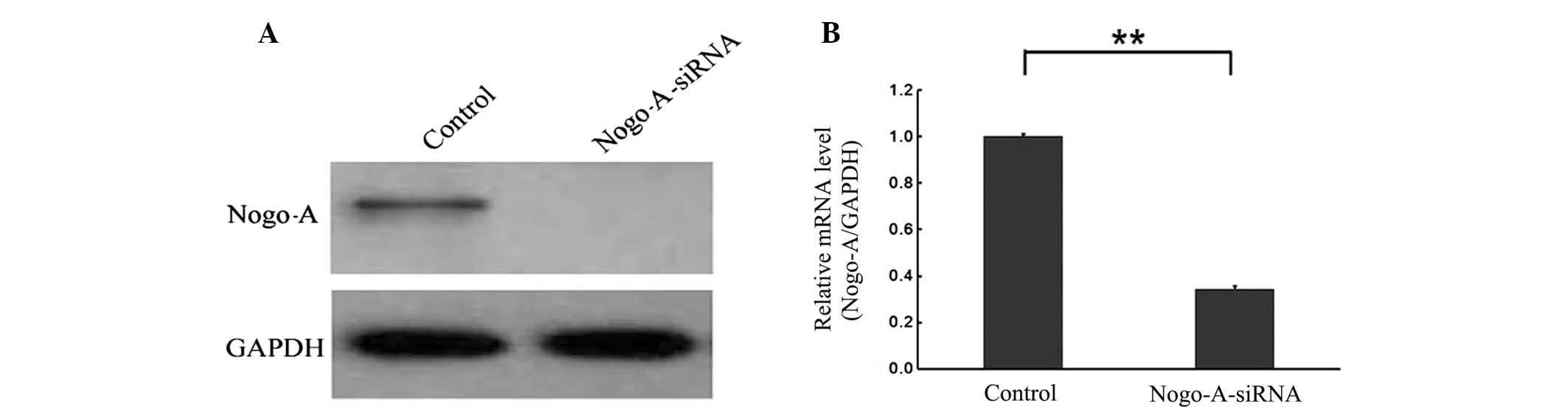
and western blot analyses demonstrated that the level of Nogo-A
expression was lower in SMMC-7721/Nogo-A-siRNA cells than in
SMMC-7721/scr-siRNA cells (P<0.001). Furthermore, Nogo-A-siRNA
was also observed to knockdown expression of exogenous 3flag-Nogo-A
(Fig. 3). These data show that the
constructed recombinant lentiviral vector of Nogo-A-siRNA is
capable of efficiently downregulating endogenous and exogenous
Nogo-A expression in SMMC-7721 cells.
Depletion of Nogo-A inhibits cell
growth
The present study further investigated the effect of
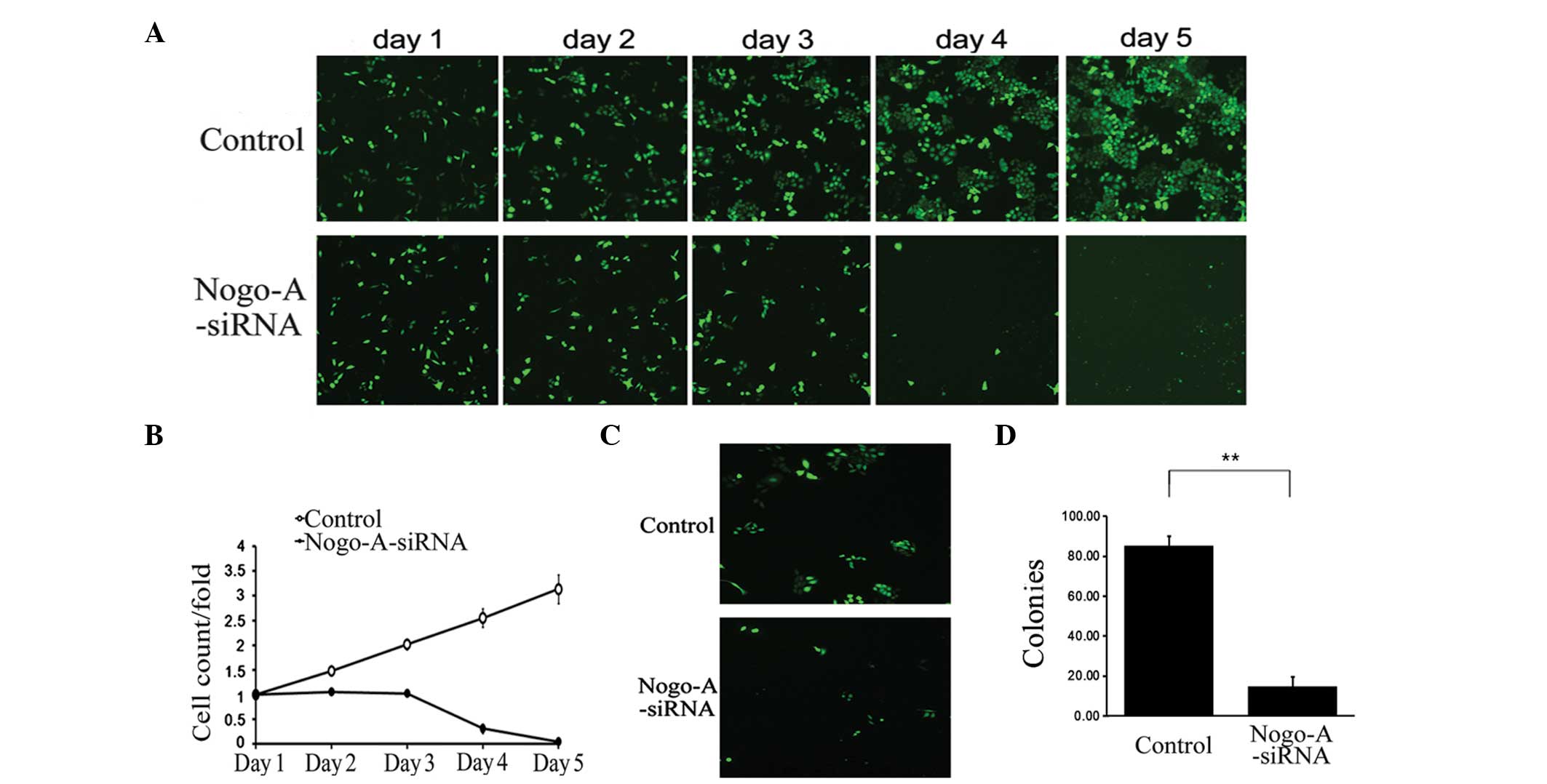
Nogo-A siRNA treatment on SMMC-7721 cell viability. As shown in
Fig. 4, the growth of SMMC-7721
cells treated with Nogo-A siRNA was significantly inhibited
compared with those treated with scr-siRNA (Fig. 4A and B). This growth inhibitory
effect was also confirmed by colony formation assay, which
demonstrated that LV-Nogo-A-siRNA transfection significantly
decreased the number and size of SMMC-7721 cell colonies (Fig. 4C and D). These data indicate that
knockdown of Nogo-A is capable of inhibiting proliferation in the
HCC SMMC-7721 cell line. Furthermore, the effect of Nogo-A
depletion was assessed in three other cancer cell lines: H1299, RKO
and SKOV3. Compared with the scr-siRNA, Nogo-A siRNA was not
observed to significantly inhibit growth in these cancer cell lines
(data not shown). These results suggested that the growth
inhibitory effect of Nogo-A siRNA may be liver cancer-specific.
Cell cycle arrest in G2/M phase by
LV-Nogo-A siRNA in SMMC-7721 cells
Experiments were also performed to assess the role
of the cell cycle in mediating the anti-proliferative effects of
Nogo-A inhibition in SMMC-7721 cells. Cell cycle distribution was
analyzed at 96 h following siRNA transduction, using flow
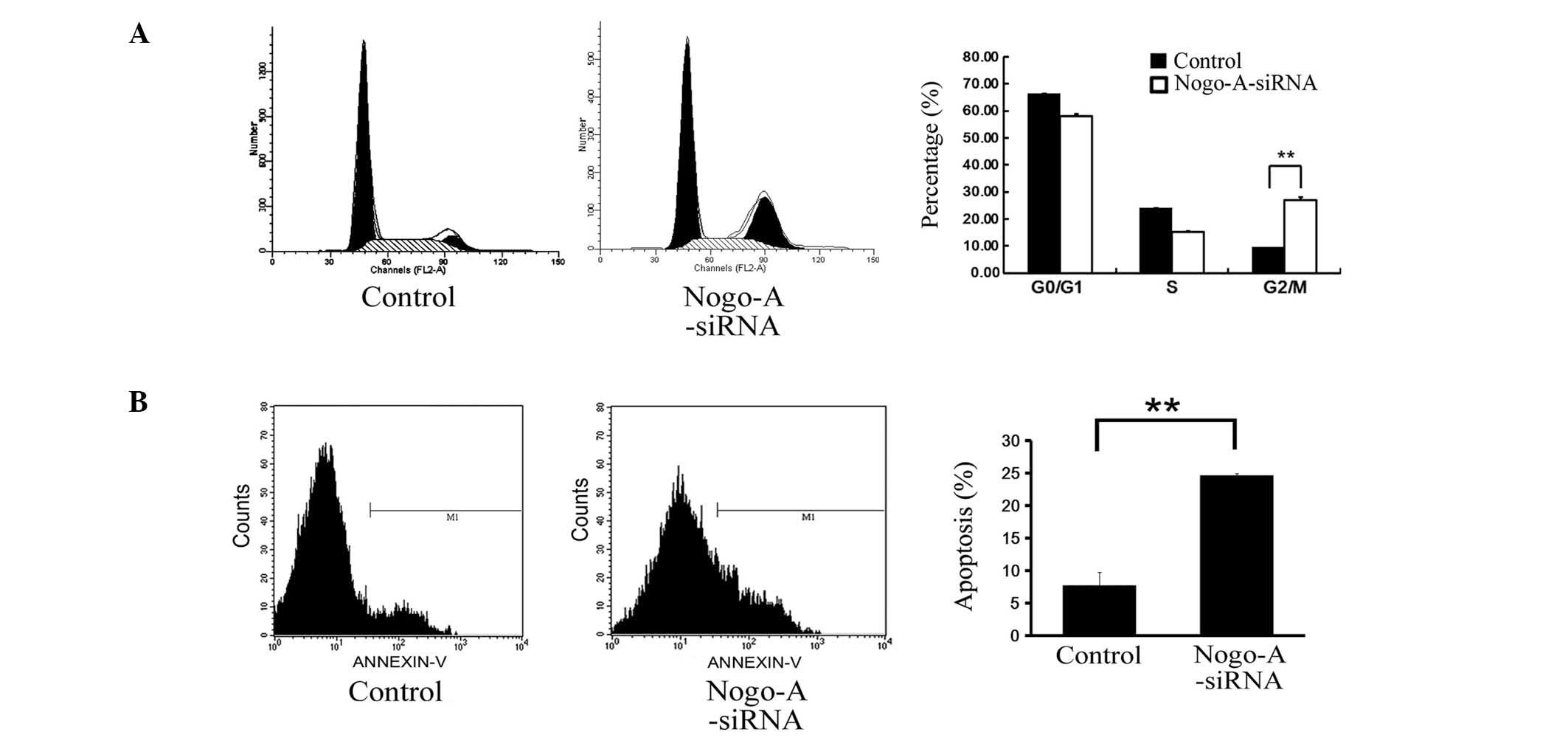
cytometry. Cell cycle analysis revealed that inhibition of Nogo-A
expression using siRNA caused a significant increase in the
proportion of the cell population in G2/M phase compared with that
of the control population transduced with scr-siRNA (Fig. 5A; P=0.0014). These data indicate
that knockdown of Nogo-A expression is capable of inducing G2/M
arrest in SMMC-7721 cells.
Induction of apoptosis by LV-Nogo-A siRNA
in SMMC-7721 cells
To determine whether Nogo-A depletion is capable of
inducing apoptosis in SMMC-7721 cells, flow cytometric analysis was
performed 96 h following siRNA transduction (Fig. 5B). Apoptosis was observed in 24.7%
of the SMMC-7721 cells transduced with Nogo-A-siRNA compared with
7.6% of the control cells transduced with scr-siRNA (P=0.004).
Since the transduction efficiency was >90%, these data suggest
that knockdown of Nogo-A expression promotes an increase in
apoptosis in the HCC SMMC-7721 cell line.
Discussion
Liver cancer is associated with the highest
mortality rate among all types of cancer. However, even when
diagnosed early, few patients represent candidates for surgery, due
the high likelihood of HCC cell relapse and metastasis following
surgery. Therefore, the development of novel therapies for the
treatment of liver cancer is of great importance. It has been
previously reported that Nogo-A has a significant role in the
development of myelin and other central nervous tissues (4,9).
Numerous studies have suggested that the Nogo-A, Nogo-B and Nogo-C
isoforms have a role in apoptosis, particularly in cancer cells
(14–16). This study has presented the Nogo-A
expression profile in four liver cancer cell lines, and has
investigated the inhibitory effect of endogenous Nogo-A on HCC
SMMC-7721 cells using an improved lentivirus packaging system.
Lentivirus vectors, which integrate their
complementary DNA (cDNA) into dividing and non-dividing cells, are
capable of permanently integrating into their target cells
(17). In comparison with the
small hairpin RNA (shRNA) expression vector systems, whose
transduction efficiency and transient shRNA expression are low
(18), lentivirus-delivered siRNAs
are capable of specific, highly stable and functional silencing of
gene expression in a variety of human cells (19). This study employed an improved
lentivirus vector with a deletion of the U3 region, including the
TATA box, of the 3′ long terminal repeat (LTR), which deactivated
the LTR promoter and significantly enhanced the safety level of
transgene expression. Using this lentivirus system, a lentivirus
vector that mediated RNAi targeting of Nogo-A (LV-Nogo-A-siRNA) was
constructed, and was observed to effectively and specifically
downregulate Nogo-A expression in SMMC-7721 cells by ≤90%.
The proliferation of SMMC-7721 cells was observed to
be markedly reduced following infection with LV-Nogo-A-siRNA for 72
h. Furthermore, the growth inhibitory effect associated with
LV-Nogo-A-siRNA infection was identified as being SMMC-7721
cell-specific. Moreover, the present study revealed that Nogo-A
depletion may achieve such growth inhibition by arresting the
SMMC-7721 cells in the G2/M phase of the cell cycle and
consequently promoting apoptosis. These results are in accordance
with the findings of Sutendra et al (20), who suggested that the absence of
Nogo-B may increase the susceptibility of pulmonary arterial smooth
muscle cells to apoptosis.
Contrary to the findings of the present study, Chen
et al (14) observed that
the overexpression of mutant Nogo-C was capable of inducing
apoptosis in HCC SMMC-7721 cells. Among the three Nogo isoforms,
Nogo-C is the shortest, while Nogo-A is the longest. These
conflicting findings suggest that the exogenous mutant Nogo-C may
be capable of antagonizing endogenous Nogo-A, thereby reducing the
activity of Nogo-A and increasing apoptosis. Further evidence is
required to verify this hypothesis.
In conclusion, the present study found that Nogo-A
depletion was capable of inhibiting HCC SMMC-7721 cell
proliferation by promoting G2/M cell cycle arrest and apoptosis. To
the best of our knowledge, this is the first investigation into the
effect of endogenous Nogo-A in a liver cancer cell line. The
present findings suggest that Nogo-A may represent an effective
molecular target for the therapeutic treatment of liver cancer, in
addition to its significant roles in neural systems.
Acknowledgements
The authors would like to thank Professor Krzysztof
Trzciński and Dr. Bing-Jun Qian for their generous assistance and
editing skills. This study was supported by the 973 Program (no.
2012CB910100) to CLW, the National Nature Science Foundation of
China (no. 31101015) and the Scientific Research Foundation for the
Returned Overseas Chinese Scholars to LC, State Education Ministry
(no. 12Z102050009).
References
|
1
|
Yang J, Yu L, Bi AD and Zhao SY:
Assignment of the human reticulon 4 gene (RTN4) to chromosome
2p14-->2p13 by radiation hybrid mapping. Cytogenet Cell Genet.
88:101–102. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
GrandPré T, Nakamura F, Vartanian T and
Strittmatter SM: Identification of the Nogo inhibitor of axon
regeneration as a Reticulon protein. Nature. 403:439–444.
2000.PubMed/NCBI
|
|
3
|
Woolf CJ: No Nogo: now where to go?
Neuron. 38:153–156. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen MS, Huber AB, van der Haar ME, et al:
Nogo-A is a myelin-associated neurite outgrowth inhibitor and an
antigen for monoclonal antibody IN-1. Nature. 403:434–439. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zheng B, Ho C, Li S, Keirstead H, Steward
O and Tessier-Lavigne M: Lack of enhanced spinal regeneration in
Nogo-deficient mice. Neuron. 38:213–224. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim JE, Li S, GrandPré T, Qiu D and
Strittmatter SM: Axon regeneration in young adult mice lacking
Nogo-A/B. Neuron. 38:187–199. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fournier AE, GrandPré T and Strittmatter
SM: Identification of a receptor mediating Nogo-66 inhibition of
axonal regeneration. Nature. 409:341–346. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xie F and Zheng B: White matter inhibitors
in CNS axon regeneration failure. Exp Neurol. 209:302–312. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
McGee AW, Yang Y, Fischer QS, Daw NW and
Strittmatter SM: Experience-driven plasticity of visual cortex
limited by myelin and Nogo receptor. Science. 309:2222–2226. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sherman M: Hepatocellular carcinoma:
epidemiology, surveillance, and diagnosis. Semin Liver Dis.
30:3–16. 2010. View Article : Google Scholar
|
|
11
|
Novak G and Tallerico T: Nogo A, B and C
expression in schizophrenia, depression and bipolar frontal cortex,
and correlation of Nogo expression with CAA/TATC polymorphism in
3′-UTR. Brain Res. 1120:161–171. 2006.PubMed/NCBI
|
|
12
|
Lie M, Grover M and Whitlon DS:
Accelerated neurite growth from spiral ganglion neurons exposed to
the Rho kinase inhibitor H-1152. Neuroscience. 169:855–862. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu L, Zhang N, Liu J, et al:
Lentivirus-mediated siRNA interference targeting SGO-1 inhibits
human NSCLC cell growth. Tumour Biol. 33:515–521. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen YC, Lu DD, Cao XR and Zhang XR:
RTN4-C gene expression in hepatocellular carcinoma and its
influence on SMMC7721 cell growth and apoptosis. Yi Chuan Xue Bao.
32:891–897. 2005.PubMed/NCBI
|
|
15
|
Kuang E, Wan Q, Li X, Xu H, Zou T and Qi
Y: ER stress triggers apoptosis induced by Nogo-B/ASY
overexpression. Exp Cell Res. 312:1983–1988. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zheng H, Xue S, Lian F and Wang YY: A
novel promising therapy for vein graft restenosis: overexpressed
Nogo-B induces vascular smooth muscle cell apoptosis by activation
of the JNK/p38 MAPK signaling pathway. Med Hypotheses. 77:278–281.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Naldini L, Blömer U, Gallay P, et al: In
vivo gene delivery and stable transduction of nondividing cells by
a lentiviral vector. Science. 272:263–267. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Brummelkamp TR, Bernards R and Agami R: A
system for stable expression of short interfering RNAs in mammalian
cells. Science. 296:550–553. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rubinson DA, Dillon CP, Kwiatkowski AV, et
al: A lentivirus-based system to functionally silence genes in
primary mammalian cells, stem cells and transgenic mice by RNA
interference. Nat Genet. 33:401–406. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sutendra G, Dromparis P, Wright P, et al:
The role of Nogo and the mitochondria-endoplasmic reticulum unit in
pulmonary hypertension. Sci Transl Med. 3:88ra552011.PubMed/NCBI
|