Introduction
Breast cancer is one of the most common types of
cancers among females globally. The disease has a high fatality
rate, with ~465,000 mortalities from breast cancer annually
worldwide (1). Early diagnosis
reduces the rate of mortality from breast cancer. MicroRNAs
(miRNAs/miR) are a class of small non-coding RNAs that were first
reported in 1993 (2). miRNAs
regulate the post-transcriptional expression of mRNAs (3) and are involved in cell
differentiation, survival and apoptosis (4). Due to their significant regulatory
role in these processes, the misregulation of miRNAs may lead to
oncogenesis by increasing cell proliferation, decreasing apoptosis
and enhancing the metastatic potential of affected cells (5). In breast cancer, miRNAs have been
shown to function as oncogenes and tumor suppressors (6). Several expression profiling studies
have identified miRNAs that exhibit upregulated expression patterns
in breast tumors. These miRNAs, which are classified as oncogenic,
include miR-21, miR-29b, miR-29c, miR-98, miR-122a, miR-128b,
miR-136, miR-149, miR-155, miR-181b, miR-181d, miR-191, miR-202,
miR-203, miR-210, miR-213, miR-365, miR-373 and miR-520c (7–15).
By contrast, let-7a, miR-10b, miR-17-5p, miR-26b, miR-30a-3p,
miR-31, miR-34a, miR-101-1, miR-125a, miR-125b, miR-127, miR-143,
miR-145, miR-200, miR-204, miR-205, miR-206, miR-320, miR-355, and
miR-497 are downregulated in breast cancer (11–18)
and are therefore classified as tumor suppressors.
There is a high degree of correlation between serum
and tissue miRNA expression profiles (19). Several potential pathways have been
hypothesized as the origin of circulating miRNAs in serum: i) Free
miRNAs may be directly secreted by cells; ii) miRNAs may be
selectively packed into microparticles (MPs) or exosomes and
released by cells via shedding of microvesicles (MVs); iii) miRNAs
may be enclosed in apoptotic bodies; and iv) miRNAs may also be
vesicle-free and associated with either argonaute proteins alone or
incorporated into high-density lipoprotein (HDL) particles
(20). Recent studies have
revealed that secreted miRNA levels in the blood and other body
fluids correlate significantly with cancer progression, therapeutic
response and patient survival (21). Thus, serum miRNA testing is viewed
as a potential non-invasive method for detecting the risk of
tumors.
The aim of the present study was to evaluate the
feasibility and clinical utility of serum-derived miRNAs as
biomarkers for the detection and staging of breast cancer. Six
candidate miRNAs (miR-374, miR-666-5p, miR-451, miR-148a, miR-27a
and miR-30b) were tested that were selected from 750 breast
cancer-associated miRNAs initially identified by pedigree screening
conducted in our preliminary experiments. This finding supports the
use of miRNAs as biomarkers that may serve as sensitive and
specific tools for the diagnosis of breast cancer.
Materials and methods
Ethics
All subjects in this study provided written informed
consent, and the study was approved by the Committee on Ethics of
the Chinese PLA General Hospital (Beijing, China).
Patients and specimen collection
Between August and November 2011, 129 serum samples
were obtained from patients at the Chinese PLA General Hospital.
These included 29 normal serum samples from healthy volunteers, 20
from benign breast tumor patients and 60 from patients with
malignant breast cancer. A total of 20 pairs of pre-operative and
post-operative serum samples were collected from patients who had
suffered breast cancer and had undergone surgery. Patients were
categorized into two groups, stages I/II and stages III/IV,
according to the WHO classification of breast cancer (22). Based on this categorization, the
present study included 41 samples of breast tumors (stages I–II)
and 19 samples of metastatic breast cancer with lymph node
metastasis or distant metastasis (stages III–IV). The patient ages
were between 20 and 75 years old (46.81±12.29 years old), and a
mean body surface area of 166.65±11.04 cm2 was recorded.
Specimens were stored at −80°C until further analysis (Table I).
 | Table IClinicopathological features of
breast cancer patients. |
Table I
Clinicopathological features of
breast cancer patients.
| Variable | Clinicopathological
parameter | Samples
(n=129) |
|---|
| Age | 46.81±12.29 | |
| Body surface area,
cm2 | 166.65±11.04 | |
| Breast cancer | | 80 |
| Pre-operative | | 60 |
|
Post-operative | | 20 |
| Benign tumor | | 20 |
| Normal control | | 29 |
| TNM stage |
| I–II | | 41 |
| III–IV | | 19 |
Statistical analysis
Calculations were performed with the SPSS computer
software program (version 19.0 for Windows; SPSS, Inc., Chicago,
IL, USA). Results with normal distributions are presented as the
mean ± standard deviation. The mean values of serum miRNA
expression from the pre-operative and post-operative groups were
compared using paired samples Student’s t-test. Results that did
not show normal distribution are presented as medians and were
compared using the Mann-Whitney U test. Receiver operating
characteristic (ROC) curves were generated and the areas under the
curve (AUCs) were calculated to compare the predictive value of
miRNAs for a breast cancer diagnosis. P<0.05 was considered to
indicate a statistically significant difference.
Total RNA isolation from serum
samples
All serum samples were immediately collected and
stored frozen at −80°C until RNA extraction was conducted. The
frozen sera were thawed and transferred into microcentrifuge tubes
(300 μl of each serum). Each sample was mixed with 1 μl synthetic
cel-miR-67, which served as an internal standard. Isolation of RNA
from all samples of malignant and benign tumors and the
corresponding healthy controls was performed simultaneously using
the miRVana™ PARIS™ kit (Ambion, Austin, TX, USA) for serum,
according to the manufacturer’s instructions. Total RNA, including
small RNAs, was purified following extraction, and the RNA
concentrations and purities of all samples were measured using the
NanoDrop ND8000 Multi-Sample Micro-Volume UV-Vis spectrophotometer
(Thermo Fisher Scientific, Wilmington, DE, USA).
Quantitative reverse transcription
polymerase chain reaction (qRT-PCR)
E. coli polyA polymerase was used to add
polyA tails to the premature and mature forms of the miRNAs. Each
reaction included 0.5 μl E.coli polyA polymerase, 0.5 μl 10X
RT buffer, 2 μl dATPs (Sigma-Aldrich, St. Louis, MO, USA), 0.5 μl
RNasin® ribonuclease inhibitor (Takara) and 10 pg total
RNA. The reaction was incubated at 37°C for 1 h. Next, 1 μl 0.5
μg/μl RT primer was added and the reaction was incubated at 70°C
for 5 min, then placed on ice immediately for at least 2 min to
disrupt the secondary structures formed between the RNA and the
primer. Aliquots of 20 μl from the reverse transcriptase reactions
contained 0.5 μl 5 U/μl M-MLV reverse transcriptase, 4 μl buffer, 1
μl dNTP (10 mM), 10 μl A-plus reaction mix, 0.5 μl RNasin
ribonuclease inhibitor and 4 μl RNase-free water. These reverse
transcription reactions were incubated at 42°C for 1 h. Finally,
for amplification of the cDNA targets, the following 20-μl
reactions were assembled: 1 μl cDNA, 2 μl of 10X universal primer,
2 μl of 10X gene-specific primer (10 μM; Table II), 10 μl of 2X qPCR Mix, 0.4 μl
of 1X ROX reference dye and 4.6 μl ddH2O. The reactions
were heated to 95°C for 5 min, followed by 40 cycles of 95°C for 15
sec and 60°C for 1 min. Cel-miR-356 was used as an internal
standard control for normalization in the qPCR analysis. Gene
expression levels were quantified using the ABI 7900 detection
system (Applied Biosystems, Foster City, CA, USA). All RT reactions
included no-template (no cDNA) and negative (no reverse
transcriptase) controls, and were performed in triplicate. Ct
(threshold cycle for a sample) data were used to evaluate the
precision and reproducibility of the qPCR results. The expression
levels of miRNAs were calculated using 2−ΔΔCt, where ΔCt
= (Ct miRNA − Ct cel-miR-67). ΔΔCt was calculated using the
formula: ΔΔCt = ΔCt of the disease group − ΔCt of the control
group. All reagents/mixes were provided by Quantobio (Beijing,
China).
 | Table IIList of all primers used in PCR. |
Table II
List of all primers used in PCR.
| Primer | Sequence
(5′-3′) |
|---|
| miR-374 | S:
ATAATACAACCTGCTA
A: CTCACACGACTCACGA |
| miR-666-5p | S:
GCGGGCACAGCT
A: CTCACACGACTCACGA |
| miR-451 | S:
ACCGTTACCATTACT
A: CTCACACGACTCACGA |
| miR-148a | S:
GGGTCAGTGCACT
A: CTCACACGACTCACGA |
| miR-27a | S:
TCACAGTGGCTAAGT
A: CTCACACGACTCACGA |
| miR-30b | S:
GTAAACATCCTACAC
A: CTCACACGACTCACGA |
| Cel-miR-67 | S:
CAACCTCCTAGA
A: CTCACACGACTCACGA |
Results
Serum miRNA expression levels
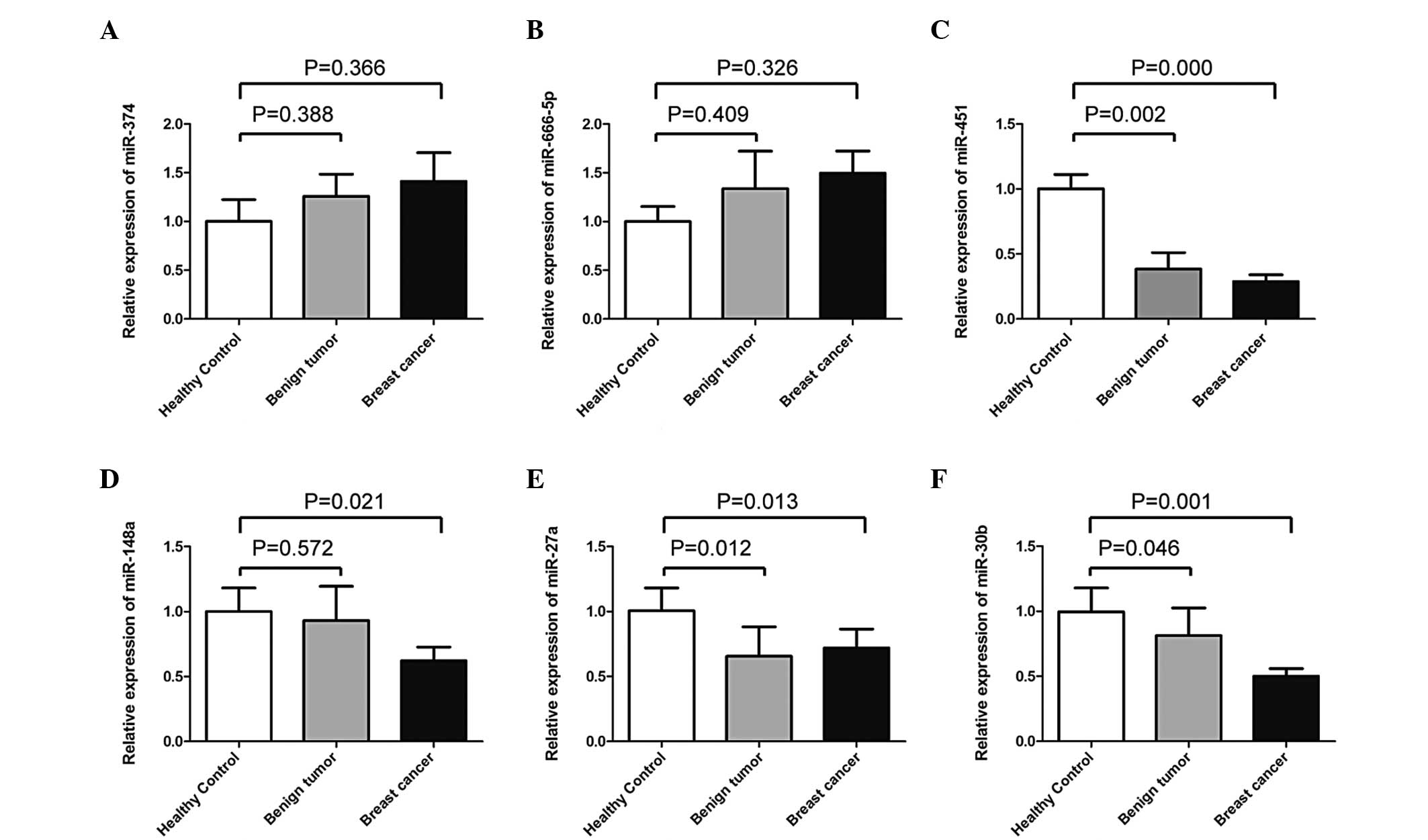
Six target miRNAs (miR-374, miR-666-5p, miR-451,
miR-148a, miR-27a and miR-30b) were selected for further analysis
in serum samples. As shown in Fig.
1, four out of the six candidate miRNAs tended to be
differentially expressed in serum samples between the patients with
breast cancer and the healthy controls: miR-374, P=0.366;
miR-666-5p, P=0.326; miR-451, P=0.000; miR-148a, P=0.021; miR-27a,
P=0.013; and miR-30b, P=0.001. Three out of the six miRNAs
(miR-451, miR-27a and miR-30b) showed significantly different
expression levels between the benign breast tumor groups and the
healthy controls: miR-374, P=0.388; miR-666-5p, P=0.409; miR-451,
P=0.002; miR-148a, P=0.572; miR-27a, P=0.012; and miR-30b
P=0.046.
No significant differences were observed in the
expression levels of miR-374, miR-666-5p, miR-451, miR-148a,
miR-27a or miR-30b between the malignant and benign breast tumors
(miR-374, P=0.870; miR-666-5p, P=0.100; miR-451, P=0.839; miR-148a,
P=0.121; miR-27a, P=0.875; and miR-30b, P=0.511).
Paired sample t-tests were performed to compare
miRNA expression in the serum samples prior to treatment and three
days after the surgery from 20 breast cancer patients who underwent
surgery. miRNA expression was not significantly different between
the pre- and post-operative groups (miR-374, P=0.514; miR-666-5p,
P=0.873; miR-451, P=0.154; miR-148a, P=0.740; miR-27a, P=0.588; and
miR-30b, P=0.091).
Comparisons of the predictive values of
miRNAs
A ROC curve analysis was used to compare the
predictive diagnostic values of the miRNAs studied for the breast
cancer, benign breast tumor and healthy control groups. These
results, presented in Table III,
showed that miR-451 had the highest AUC of 0.915 [asymptotic 95%
confidence interval (CI), 0.850–0.979; P<0.000]. miR-27a is
inferior to miR-451 and showed an AUC of 0.909 (asymptotic 95% CI,
0.841–0.976; P<0.000). miR-451 had a sensitivity of 93% and a
specificity of 79.3%, while miR-27a had a sensitivity of 88.1% and
a specificity of 86.2%.
 | Table IIIParameters for predictive value of
panel 1. |
Table III
Parameters for predictive value of
panel 1.
| miRNA | AUC | CI | Sensitivity | Specificity | P-value |
|---|
| miR-451a | 0.915 | 0.850–0.979 | 0.930 | 0.793 | 0.000 |
| miR-148aa | 0.889 | 0.821–0.957 | 0.763 | 0.966 | 0.000 |
| miR-27aa | 0.909 | 0.841–0.976 | 0.881 | 0.862 | 0.000 |
| miR-30ba | 0.900 | 0.829–0.971 | 0.881 | 0.828 | 0.000 |
| Panel 1 | 0.953 | 0.915–0.992 | 0.947 | 0.828 | 0.000 |
The results shown in Table IV demonstrated that miR-451 also
had the highest AUC for differentiating between the benign breast
tumor cases and healthy controls, with a sensitivity of 100% and a
specificity of 69%. Thus, miR-451 is a highly accurate predictor
for the initial screening of breast diseases.
 | Table IVParameters for predictive value of
panel 2. |
Table IV
Parameters for predictive value of
panel 2.
| miRNA | AUC | CI | Sensitivity | Specificity | P-value |
|---|
| miR-451a | 0.904 | 0.823–0.985 | 1 | 0.690 | 0.000 |
| miR-27aa | 0.791 | 0.635–0.948 | 0.400 | 0.966 | 0.001 |
| miR-30ba | 0.706 | 0.552–0.860 | 0.800 | 0.862 | 0.017 |
| Panel 2 | 0.880 | 0.891–0.983 | 0.850 | 0.862 | 0.000 |
These results also show that miR-148a levels
differentiated between malignant and benign breast masses, with an
AUC of 69.8% (asymptotic 95% CI, 0.561–0.835), a sensitivity of
56.1% and a specificity of 78.9%.
Multiple marker assays
Multiple marker assays evaluated by multivariate
regression analysis may significantly improve the sensitivity and
specificity of detecting tumors compared with single marker
assays.
The combined panel of miRNA markers (panel 1;
miR-451, miR-148a, miR-27a and miR-30b) achieved 94.7% sensitivity,
82.8% specificity and a 95.3% AUC for distinguishing breast cancer
cases from healthy controls (Table
III; Fig. 2A).
A panel of three combined markers (panel 2; miR-451,
miR-27a and miR-30b) achieved 85% sensitivity, 86.2% specificity
and attained an 88% AUC for distinguishing benign breast tumor
cases from healthy controls (Table
IV; Fig. 2B).
The single marker miR-148a showed a potential
predictive value in differentiating between malignant and benign
breast masses (P=0.01), with a sensitivity of 56.1%, a specificity
of 78.9% and an AUC of 69.8% (Fig.
2C).
Discussion
The present study was designed to investigate the
expression and correlation of a selected panel of miRNAs associated
with breast tumors in serum samples. Indeed, miRNAs are known to be
involved in several critical oncogenic cellular processes,
including proliferation, differentiation and apoptosis.
Serum-derived miRNAs have the following characteristics: i) miRNAs
are tremendously stable in serum due to their small size (23); ii) circulating miRNAs may be easily
and reproducibly measured (24);
and iii) miRNAs have rich information content and tumor-specific
profiles (25), and individual
miRNAs show unique expression levels in the varying organs and
stages of cellular processes. In addition, clinical specimens of
serum are more abundant and more conveniently collected. Therefore,
serum-derived miRNAs are expected to be clinically useful as novel
and minimally invasive tools to aid in the early detection and
monitoring of breast cancer.
Six miRNAs from the expression profile (miR-374,
miR-666-5p, miR-451, miR-148a, miR-27a and miR-30b) were
empirically selected for the qPCR analysis of serum samples
collected from breast cancer patients, benign tumor patients and
healthy controls. The aberrant expression of candidate miRNAs in
breast cancer has been reported by several previous studies. For
example, it has been demonstrated that miR-451 levels are
significantly reduced in tamoxifen-resistant breast cancer cells
(26). In MCF-7/DOX cells, the
enforced increase of miR-451 levels downregulates the expression
levels of mdr1 and increases the sensitivity of the MCF-7-resistant
cancer cells to DOX (27).
However, miR-451 shows higher expression levels in breast invasive
micropapillary carcinoma than in unspecified invasive ductal
carcinomas (28). The expression
levels of miR-148a are decreased in breast cancer cells and tissues
(29). miR-27a appears to be
significant in breast cancer by suppressing the expression of a
transcription factor, ZBTB10 (30). Furthermore, the single nucleotide
polymorphism in miR-27a has been associated with the risk of
familial breast cancer and has also been correlated with lymph node
metastasis (31). miR-30b, a
member of the miR-30 family, is a tumor suppressor miRNA that binds
to the 3′-UTR of the catalase mRNA and inhibits its expression
(32). In addition, miR-30b has
been identified as a trastuzumab-inducible miRNA in cancer cells
(33), and expression levels of
miR-30b have been shown to correctly predict the nature of
inflammatory breast cancer phenotypes (34). To the best of our knowledge, there
are few studies on the expression of miR-666-5p and miR-374 in
breast cancer. miR-374, together with four other miRNAs (miR-21,
miR-31, miR-125a and miR-181c), has been described as a signature
marker of the human natural T-reg cell (35), and it is abundantly expressed in
transdifferentiated neuronal progenitors (36). Furthermore, the majority of the
studies previously undertaken have focused on comparing miRNA
expression patterns in breast tumor tissues or in cancer cell
lines. Thus, to a certain extent, the present research involving
the testing of serum miRNA expression profiling and its use as a
diagnostic tool is a pioneering approach.
The present results show that miR-451, miR-27a and
miR-30b are differentially expressed in the serum samples in benign
and malignant breast tumors compared with the samples of healthy
patients. Therefore, these miRNAs may be used to discriminate
breast cancer from healthy controls. In addition, miR-148a may be
used to distinguish benign breast tumors from malignant tumors due
to its differential serum levels in these two groups. The results
of the present study confirm that miR-451 expression provides the
greatest sensitivity in detecting human breast cancers, which is
consistent with data that has been previously reported (37).
Certain studies have indicated that the removal of
the primary tumor leads to the loss of elevated levels of
circulating miRNAs (38,39). In the present study, no significant
differences were observed in the expression levels of miR-374,
miR-666-5p, miR-451, miR-148a, miR-27a and miR-30b between the
pre-operative and post-operative serum samples of the breast cancer
patients. However, as the study had a relatively small sample size
of only 20 paired patients, a greater number of breast cancer
patients should be investigated to confirm these results. In
addition, the post-operative serum samples used in this study were
collected only three days after surgery. Thus, the levels of
circulating miRNAs at various periods subsequent to surgery should
be investigated.
Furthermore, the present study also attempted to
evaluate the expression levels of the panel of miRNAs at differing
tumor stages. Certain previous studies have shown that differential
miRNA expression in serum is more frequently observed in the
advanced, rather than early, stages of breast cancer (40). Therefore, the expression levels of
miRNAs have been linked to the tumor stage. In the present study,
miRNA expression profiling was not demonstrated to show a higher
sensitivity for patients with a higher tumor-node-metastasis stage
and lymph node metastasis.
The present study of miRNAs with downregulated
expression in disease patient samples demonstrated that the serum
levels of cancer-associated miRNAs can distinguish patients with
breast cancer from healthy controls. However, miR-27a is regarded
as a risk factor for colorectal cancer (41) and has also shown to be correlated
with lymph node metastasis in gastric cancer (42). Furthermore, miR-451 levels are
decreased in serum samples from renal cell carcinoma patients
(43), and miR-148a expression is
suppressed >4-fold in gastric cancer. The role of miR-148a is to
function as a tumor metastasis suppressor in gastric cancer, hence,
downregulation of miR-148a contributes to gastric cancer lymph node
metastasis and progression (44).
Finally, the expression of miR-30b is suppressed in invasive
bladder cancer (45) and lung
squamous cell carcinoma (46).
Therefore, the specificity of a single miRNA as a
tumor marker is somewhat less than that of multiple miRNAs
together. Multiple marker assays may significantly improve the
sensitivity and specificity of tumor detection compared with single
marker assays. However, this improvement in detection is dependent
upon the selection of markers. In the present study, miR-451,
miR-148a, miR-27a and miR-30b were combined into a panel of markers
for breast cancer diagnosis. The sensitivities of miR-451,
miR-148a, miR-27a and miR-30b individually were 93, 76.3, 88.1 and
88.1%, respectively. In addition, their specificities were 79.3,
96.6, 86.2 and 82.8%, respectively. By comparison, the sensitivity
and specificity of the four combined markers were significantly
higher. The ROC curve analysis using the four combined markers
yielded an AUC of 0.953, a sensitivity of 94.7% and a specificity
of 82.8%.
In conclusion, qPCR analysis of miRNAs in the serum
samples of breast cancer patients may have a clinical significance
for the detection of breast cancer. Moreover, the present results
indicate that a panel of miRNAs may be useful as a sensitive and
specific tool for the detection of breast cancer. However, studies
with long-term follow-up and a larger number of patients are
required in order to confirm the clinical application of these
molecular markers.
References
|
1
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar
|
|
2
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complimentary to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kim VN, Han J and Siomi MC: Biogenesis of
small RNAs in animals. Nat Rev Mol Cell Biol. 10:126–139. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang ZX, Lu BB, Wang H, Cheng ZX and Yin
YM: MicroRNA-21 modulates chemosensitivity of breast cancer cells
to doxorubicin by targeting PTEN. Arch Med Res. 42:281–290. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Di Leva G and Croce CM: Roles of small
RNAs in tumor formation. Trends Mol Med. 16:257–267.
2010.PubMed/NCBI
|
|
6
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Selcuklu SD, Donoghue MT and Spillane C:
miR-21 as a key regulator of oncogenic processes. Biochem Soc
Trans. 37:918–925. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Faraoni I, Antonetti FR, Cardone J and
Bonmassar E: miR-155 gene: a typical multifunctional microRNA.
Biochim Biophys Acta. 1792:497–505. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang Y and Lee CG: MicroRNA and cancer -
focus on apoptosis. J Cell Mol Med. 13:12–23. 2009. View Article : Google Scholar
|
|
10
|
Yan LX, Huang XF, Shao Q, Huang MY, Deng
L, Wu QL, Zeng YX and Shao JY: MicroRNA miR-21 overexpression in
human breast cancer is associated with advanced clinical stage,
lymph node metastasis and patient poor prognosis. RNA.
14:2348–2360. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lodygin D, Tarasov V, Epanchintsev A,
Berking C, Knyazeva T, Körner H, Knyazev P, Diebold J and Hermeking
H: Inactivation of miR-34a by aberrant CpG methylation in multiple
types of cancer. Cell Cycle. 7:2591–2600. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sempere LF, Christensen M, Silahtaroglu A,
Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S and Cole CN:
Altered MicroRNA expression confined to specific epithelial cell
subpopulations in breast cancer. Cancer Res. 67:11612–11620. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kondo N, Toyama T, Sugiura H, Fujii Y and
Yamashita H: miR-206 expression is down-regulated in estrogen
receptor alpha -positive human breast cancer. Cancer Res.
68:5004–5008. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang Q, Gumireddy K, Schrier M, le Sage
C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, et al:
The microRNAs miR-373 and miR-520c promote tumour invasion and
metastasis. Nat Cell Biol. 10:202–210. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Valastyan S, Reinhardt F, Benaich N,
Calogrias D, Szász AM, Wang ZC, Brock JE, Richardson AL and
Weinberg RA: A pleiotropically acting microRNA, miR-31, inhibits
breast cancer metastasis. Cell. 137:1032–1046. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu XX, Li XJ, Zhang B, Liang YJ, Zhou CX,
Cao DX, He M, Chen GQ, He JR and Zhao Q: MicroRNA-26b is
underexpressed in human breast cancer and induces cell apoptosis by
targeting SLC7A11. FEBS Lett. 585:1363–1367. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang F, Zheng Z, Guo J and Ding X:
Correlation and quantitation of microRNA aberrant expression in
tissues and serum from patients with breast tumor. Gynecol Oncol.
119:586–593. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Turchinovich A, Weiz L and Burwinkel B:
Extracellular miRNAs: the mystery of their origin and function.
Trends Biochem Sci. 37:460–465. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang JX, Song W, Chen ZH, Wei JH, Liao
YJ, Lei J, Hu M, Chen GZ, Liao B, Lu J, Zhao HW, Chen W, He YL,
Wang HY, Xie D and Luo JH: Prognostic and predictive value of a
microRNA signature in stage II colon cancer: a microRNA expression
analysis. Lancet Oncol. 14:1295–1306. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Frank GA, Danilova NV, Andreeva IuIu and
Nefedova NA: WHO classification of tumors of the breast, 2012. Arkh
Patol. 75:53–63. 2013.(In Russian).
|
|
23
|
Wang H, Peng W, Ouyang X, Li W and Dai Y:
Circulating microRNAs as candidate biomarkers in patients with
systemic lupus erythematosus. Transl Res. 160:198–206. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tsui NB, Ng EK and Lo YM: Stability of
endogenous and added RNA in blood specimens, serum, and plasma.
Clin Chem. 48:1647–1653. 2002.PubMed/NCBI
|
|
25
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant
KC, Allen A, et al: Circulating microRNAs as stable blood based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang H, Peng W, Ouyang X, Li W and Dai Y:
miR-451 were upregulated in the patients with SLE and were also
significantly increased in the patients with RA. J Transl Med.
10:552012.PubMed/NCBI
|
|
27
|
Kovalchuk O, Filkowski J, Meservy J,
Ilnytskyy Y, Tryndyak VP, Chekhun VF and Pogribny IP: Involvement
of microRNA-451 in resistance of the MCF-7 breast cancer cells to
chemotherapeutic drug doxorubicin. Mol Cancer Ther. 7:2152–2159.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li S, Yang C, Zhai L, Zhang W, Yu J, Gu F,
Lang R, Fan Y, Gong M, Zhang X and Fu L: Deep sequencing reveals
small RNA characterization of invasive micropapillary carcinomas of
the breast. Breast Cancer Res Treat. 136:77–87. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu Q, Jiang Y, Yin Y, Li Q, He J, Jing Y,
Qi YT, Xu Q, Li W, Lu B, Peiper SS, Jiang BH and Liu LZ: A
regulatory circuit of miR-148a/152 and DNMT1 in modulating cell
transformation and tumor angiogenesis through IGF-IR and IRS1. J
Mol Cell Biol. 5:3–13. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tang W, Zhu J, Su S, Wu W, Liu Q, Su F and
Yu F: MiR-27 as a prognostic marker for breast cancer progression
and patient survival. PLoS One. 7:e517022012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Catucci I, Verderio P, Pizzamiglio S,
Bernard L, Dall’olio V, Sardella D, Ravagnani F, Galastri L, Barile
M, Peissel B, et al: The SNP rs895819 in miR-27a is not associated
with familial breast cancer risk in Italians. Breast Cancer Res
Treat. 133:805–807. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Haque R, Chun E, Howell JC, Sengupta T,
Chen D and Kim H: MicroRNA-30b-mediated regulation of catalase
expression in human ARPE-19 cells. PLoS One. 7:e425422012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ichikawa T, Sato F, Terasawa K, Tsuchiya
S, Toi M, Tsujimoto G and Shimizu K: Trastuzumab produces
therapeutic actions by upregulating miR-26a and miR-30b in breast
cancer cells. PLoS One. 7:e314222012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Van der Auwera I, Limame R, van Dam P,
Vermeulen PB, Dirix LY and Van Laere SJ: Integrated miRNA and mRNA
expression profiling of the inflammatory breast cancer subtype. Br
J Cancer. 103:532–541. 2010.PubMed/NCBI
|
|
35
|
Rouas R, Fayyad-Kazan H, El Zein N,
Lewalle P, Rothé F, Simion A, Akl H, Mourtada M, El Rifai M, Burny
A, et al: Human natural Treg microRNA signature: role of
microRNA-31 and microRNA-21 in FOXP3 expression. Eur J Immunol.
39:1608–1618. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chang SJ, Weng SL, Hsieh JY, Wang TY,
Chang MD and Wang HW: MicroRNA-34a modulates genes involved in
cellular motility and oxidative phosphorylation in neural
precursors derived from human umbilical cord mesenchymal stem
cells. BMC Med Genomics. 4:652011. View Article : Google Scholar
|
|
37
|
Ng EK, Li R, Shin VY, Jin HC, Leung CP, Ma
ES, Pang R, Chua D, Chu KM, Law WL, Law SY, Poon RT and Kwong A:
Circulating microRNAs as specific biomarkers for breast cancer
detection. PLoS One. 8:e531412013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Iguchi H, Kosaka N and Ochiya T: Secretory
microRNAs as a versatile communication tool. Commun Integr Biol.
3:478–481. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kosaka N, Iguchi H and Ochiya T:
Circulating microRNA in body fluid: a new potential biomarker for
cancer diagnosis and prognosis. Cancer Sci. 101:2087–2092. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Catucci I, Verderio P, Pizzamiglio S,
Bernard L, Dall’olio V, Sardella D, Ravagnani F, Galastri L, Barile
M, Peissel B, Zaffaroni D, Manoukian S, Radice P and Peterlongo P:
The SNP rs895819 in miR-27a is not associated with familial breast
cancer risk in Italians. Breast Cancer Res Treat. 133:805–807.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hezova R, Kovarikova A, Bienertova-Vasku
J, Sachlova M, Redova M, Vasku A, Svoboda M, Radova L, Kiss I,
Vyzula R and Slaby O: Evaluation of SNPs in miR-196-a2, miR-27a and
miR-146a as risk factors of colorectal cancer. World J
Gastroenterol. 18:2827–2831. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Katada T, Ishiguro H, Kuwabara Y, Kimura
M, Mitui A, Mori Y, Ogawa R, Harata K and Fujii Y: microRNA
expression profile in undifferentiated gastric cancer. Int J Oncol.
34:537–542. 2009.PubMed/NCBI
|
|
43
|
Redova M, Poprach A, Nekvindova J, Iliev
R, Radova L, Lakomy R, Svoboda M, Vyzula R and Slaby O: Circulating
miR-378 and miR-451 in serum are potential biomarkers for renal
cell carcinoma. J Transl Med. 10:552012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zheng B, Liang L, Wang C, Huang S, Cao X,
Zha R, Liu L, Jia D, Tian Q, Wu J, et al: MicroRNA-148a suppresses
tumor cell invasion and metastasis by downregulating ROCK1 in
gastric cancer. Clin Cancer Res. 17:7574–7583. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wszolek MF, Rieger-Christ KM, Kenney PA,
Gould JJ, Silva Neto B, Lavoie AK, Logvinenko T, Libertino JA and
Summerhayes IC: A MicroRNA expression profile defining the invasive
bladder tumor phenotype. Urol Oncol. 29:794–801. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gao W, Shen H, Liu L, Xu J, Xu J and Shu
Y: MiR-21 overexpression in human primary squamous cell lung
carcinoma is associated with poor patient prognosis. J Cancer Res
Clin Oncol. 137:557–566. 2011. View Article : Google Scholar : PubMed/NCBI
|