1. Introduction
The emergence of multi-drug resistant tuberculosis
(TB) has increased focus on the global prevention and control of
TB. At present, TB control faces a number of challenges, including
low sensitivity, poor specificity and treatment complications, long
detection cycles of traditional diagnostic techniques and a decline
in the immune function of traditional vaccines, including Bacillus
Calmette-Guerin (1). Since 1947,
when mycobacteriophages were first isolated and identified by
Gardner et al (2),
>3,680 types of mycobacteriophage have been isolated from
different sources, of which >544 types of mycobacteriophages
have undergone complete genome sequencing (3). As a member of the bacteriophage
family, which are DNA viruses, mycobacteriophages are able to
infect the host Mycobacteria specifically. L5 (4), D29 (5) and TM4 (6) are the mycobacteriophages, were the
earliest to undergo genome sequencing and are the most widely used
in the investigation of TB. Greater understanding of the structure
and function of the mycobacteriophage genome has increased
awareness of the importance of investigating the diagnosis and
treatment of Mycobacterium tuberculosis.
2. Molecular tools of Mycobacterium
investigation
In 1964, Tokunaga and Sellers (7) were the first to use the D29 phage to
successfully induce outer DNA into M. smegmatis, which
demonstrated the feasibility of mycobacteriophage DNA transfection.
Subsequently, in 1970, I3 was successfully enriched with the use of
M. smegmatis by Raj and Ramakrishnan (8), which again supported the viability of
transduction. Since these early experiments, the rapid development
of genetic engineering has led to the construction of a number of
high-efficiency cloning and expression vectors. Recombinant DNA
technology has also progressed, however, due to a lack of
understanding of the mycobacteriophage genome, progress in
recombinant DNA technology for the application of mycobacteria has
been slow. Mycobacteria have a lipid-rich cell wall, which limits
the ability of the exogenous DNA to pass through the cell wall and,
therefore, foreign DNA are unable to be stably integrated and
expressed in mycobacteria (9).
Jacobs et al (10) succeeded in creating a method of
mycobacterial genome transfer in 1987, overcoming the difficulties
in investigating mycobacterial genes. The plasmid DNA of
Escherichia coli was inserted into the non-essential region
of the TM4 genome, to successfully construct a recombinant shuttle
plasmid vector (Fig. 1). The
vector was a dual function shuttle plasmid vector, which was not
only a plasmid replicated in the E. coli, but also a phage
replicated within the mycobacteria. Therefore, this overcame the
deficiencies of traditional plasmid vectors, carrying a limited
length of exogenous DNA fragments, and the insufficient capacity of
bacterial transformation. The experimental results demonstrated
that the recombinant shuttle vector was transfected into
fast-growing M. smegmatis, however, this experiment was not
successful in slow growing mycobacteria, including Bacillus
Calmette-Guérin (BCG) strains and M. tuberculosis. Despite
this, it demonstrated that recombinant shuttle plasmids may
eventually be suitable for use to induce exogenous DNA into the BCG
vaccine strains to develop a recombinant mycobac-terial vaccine.
Snapper et al (11) also
constructed a shuttle plasmid successfully based on L1 and
demonstrated the stable insertion and replication of exogenous DNA
in M. smegmatis. Lee et al (12) achieved an effective and stable
transformation using the mild site-specific integrated L5
mycobacteriophage. These findings demonstrated the building of an
efficient integration vector by integrating the plasmid sequences
into the mycobacterial genome, with effective integration of the TB
mycobacterium and BCG to obtain stable recombinant DNA.
These previous studies demonstrated that the shuttle
plasmid was of value for specific transduction (13), transposon transfer (14,15)
and the introduction of diagnostic reporter genes (16,17).
The development of this vector system promotes the genetic analysis
Mycobacterium pathogens and the development of a recombinant
vaccine.
3. TB diagnosis and drug sensitivity
assessments based on mycobacteriophages
TB is the most important global public health
problem at present. In 2010, there were 8.8 million incident cases
of TB, 1.1 million deaths from TB among HIV-negative people, and an
additional 0.35 million deaths from HIV-associated TB (18). Therefore, the control of the
condition via rapid and accurate TB diagnosis is important. The
demand for a simple, fast, safe, sensitive and accurate M.
tuberculosis antibiotic susceptibility assessment has become
increasingly urgent, as a result of the emergence and spread of
multidrug-resistant TB and extensively drug-resistant tuberculosis
(XDR-TB). In previous years, molecular techniques for the diagnosis
of TB have been rapidly developed. The nucleic acid amplification
method, involving nucleic acid probes, polymerase chain reaction,
DNA sequencing, Gene Chip and Xpert MTB/RIF enables rapid diagnosis
and assessment of resistance of M. tuberculosis (19). Although the majority of the
techniques are fast with a high sensitivity, the requirement for
specialized instruments and high costs significantly limited its
dissemination and application in the majority of countries with a
high burden of TB. In addition, there was a $1 billion gap in the
funds of the World Health Organization for TB management and
control in 2012, causing financial pressure in the diagnosis and
treatment of TB (18).
Assessments, which enable the rapid detection of mycobacteriophages
have numerous advantages, including high speed, simplicity,
specificity, security, no requirement for specialist equipment and
lower costs, and they enable the quantitative detection of viable
cells. Therefore, mycobacteriophages have become an ideal tool for
TB diagnosis and assessment of drug susceptibility.
Phage amplification technology
The investigation of phage amplification technology
can be traced back to 1965. A study by Sellers et al
(20) observed the effects of
anti-TB drugs on mycobacteriophages. The results of the experiments
demonstrated that streptomycin (STR) was able to prevent the phages
copying in M. smegmatis, whilst not affecting the phage
replication of the progeny in resistant strains, or their
subsequent release. Since this observation, other drugs, including
clofazimine, colistin, rifampicin (RIF) and STR have also been
assessed for their effects on the synthesis of D29 (21,22).
In these studies, D29 was able to affect slow-growing pathogenic
mycobacteria and the fast-growing environmental strains, and
visible plaque formed in the fast-growing M. smegmatis
bacteria following overnight incubation. The existence of viable
bacteria can be determined by rapid detection of the release of
progeny phages following infection of the mycobacterium target
using this technique. These experiments laid the foundation for the
subsequent development of phage amplification technology and its
application in assessing anti-mycobacterial drug sensitivity.
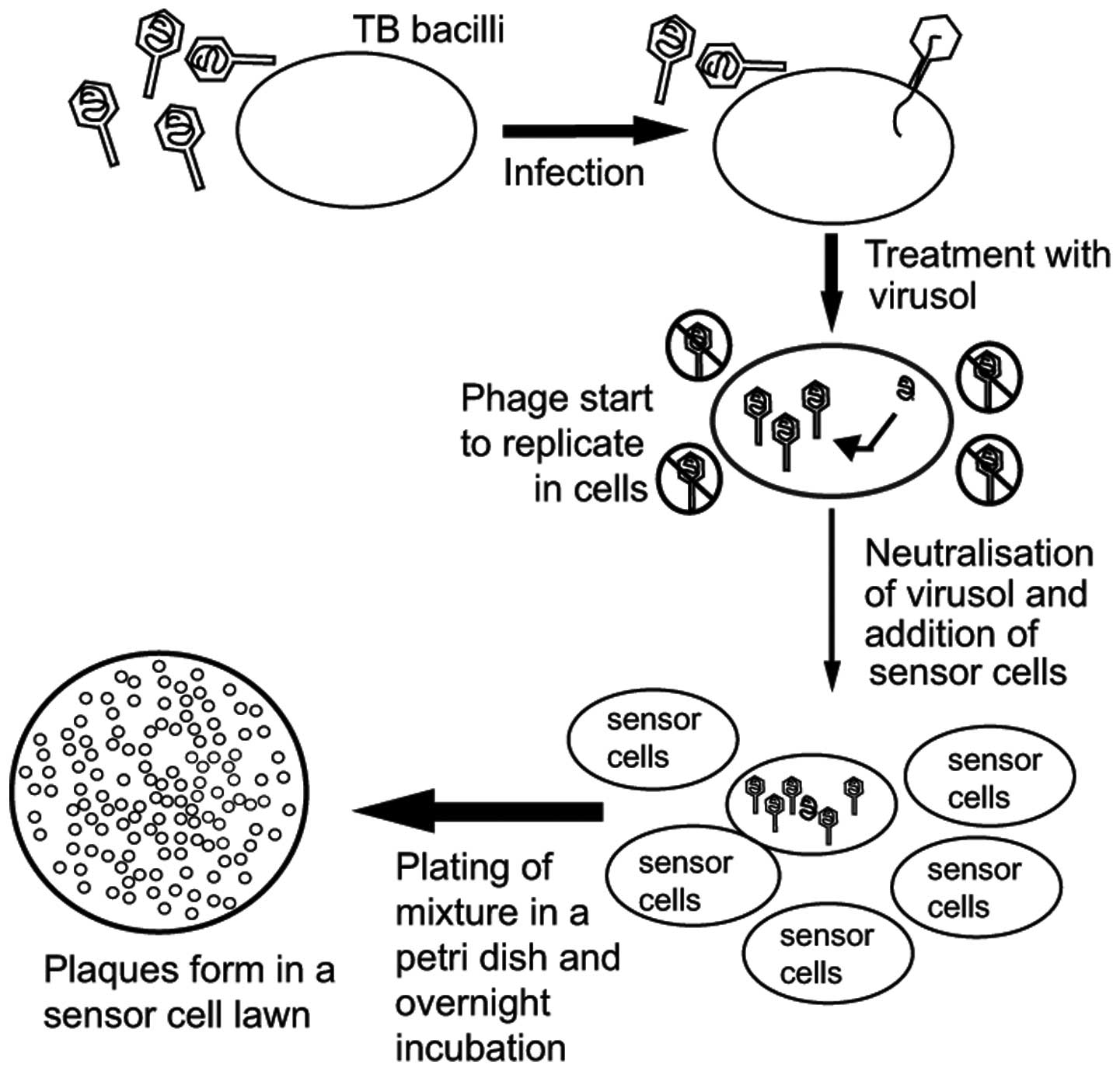
The phage amplification technology, which is in
current clinical use was first described by Wilson et al
(23) in 1997, and further defined
on the basis of further modifications by McNerney et al
(24). Subsequently, Biotec
Laboratories Ltd. (Ipswich, UK) developed corresponding commercial
kits, FASTPlaqueTB™ and FASTPlaqueTB-MDRi™, or
FASTPlaqueTB-RIF™ (25,26),
which were used for the rapid detection of M. tuberculosis
and for the assessment of multi-drug resistance (Fig. 2). Firstly, D29 phages were
introduced into M. tuberculosis, in which they reproduced.
The phages, which did not enter the cell were killed by virucide
agents, however, the phages that entered the viable M.
tuberculosis were not affected. The phages lysed the bacteria
in vivo following replication in the bacteria. Subsequently,
the releasing phages infected and lysed M. smegmatis to form
plaques. As this assessment is reliant on the presence or absence
of plaques to determine the result, this method generally requires
1–2 days to produce results. As there is a proportional association
between the quantity of plaques and the quantity of M.
tuberculosis in the specimens, the content of M.
tuberculosis in the sample can be calculated according to the
number of plaques. As M. tuberculosis is cleaved during the
experiment, the experiment has fewer safety concerns for the
individuals involved.
The clinical effects of this assay have been
evaluated in several countries, including Egypt (27), Pakistan (28), South Africa (29) and Spain (30). Kalantri et al (31) performed a meta-analysis of the
detection of M. tuberculosis in clinical samples, based on
phage amplification technology in 2005 by examining the literature
from databases, including Medline, EMBASE (http://www.elsevier.com/online-tools/embase), Web
of Science (http://wok.mimas.ac.uk/) and BIOSIS
Previews (http://biosispreviews.isihost.com/). A total of 13
studies were included, which complied with designated standards.
The specificity and sensitivity of these assays were between 0.83
and 1.00, and between 0.21 and 0.94, respectively, with sputum
culture as a reference standard. The results revealed that the
assay had a high specificity and a moderate/variable sensitivity,
which required improvement. The predominant reasons for the lower
sensitivity included anti-TB treatment prior to the experiment,
sample transportation, environmental conditions and the selection
of detergents. Therefore, further investigations are required to
improve the sensitivity of the phage-based assessment.
The Foundation for Innovative New Diagnostics (FIND)
extensively evaluated the role of the FASTPlaque assessment
technique in rifampin resistance in 2007 (32). The FASTPlaque assay failed
to achieve the desired objectives in two trial sites in South
Africa. Therefore, FIND terminated the FASTPlaque assessment
pilot program until a satisfactory improvement had been made
(32). Therefore, although
FASTPlaque assessment can be widely used for the rapid
diagnosis of TB, however, further improvement of the optimization
techniques is required.
Luciferase reporter phage
The fluorescent reporter phage is a rapid detection
system for M. tuberculosis susceptibility and drug
susceptibility based on recombinant DNA technology. The first
generation of luciferase reporter phages (LRPs) were developed
successfully by Jacobs et al in 1993 (16). These were constructed from the
phAE39 plasmid shuttle, on the basis of TM4, and the firefly
luciferase (FFlux) gene was inserted using a potent promoter
of heat shock protein 60 (hsp60; Fig.
3). LRPs are able to transfer recombinant DNA into
mycobacteria, including the M. smegmatis and M.
tuberculosis BCG vaccine strains, In the presence of adenosine
triphosphate and luciferin, FFlux is able to continuously
express and generate an optical signal following mycobacterial
infection. If there are at least 104/milliliter of M.
tuberculosis in the sample, the relative light units can be
detected within a few minutes following LRP infection of the live
mycobacteria. This method reduced the reporting duration
considerably compared with the traditional detection methods. LRPs
based on L5 (33) and D29
(34) have been subsequently
constructed, however, various defects remain. The mild L5
mycobacteriophage is unable to infect the M. tuberculosis
complex, which limits its application in the drug resistance
detection of clinical samples. The lytic characteristics of D29 and
TM4 result in the loss of light output and reduced sensitivity.
Since the characteristics of lytic phages may reduce light output,
Kumar et al (35)
constructed new LRPs using the mild Che12 bacteriophage to increase
light output and improve the sensitivity of the assessment.
Carriere et al (36)
addressed the problem using a number of strategies, including
changing the position of FFlux in the phage genome,
isolating host-range mutant phages and inducing
temperature-sensitive mutants of phages to screen more sensitive
mutants compared with the first generation LRPs. Although the
sensitivity of LRPs has improved, these LRPs can infect
mycobacteria with the exception of M. tuberculosis, leading
to misdiagnosis in clinical prac-tice, therefore, it is necessary
to improve the experimental program to confirm the presence of the
M. tuberculosis complex. Considering these problems, Riska
et al (37) added
ρ-nitro-α-acetylamino-β-hydroxy propiophenone to the substrate to
selectively inhibit the M. tuberculosis complex bacteria,
and combined the corrected program with the ordinary LRPs to
accurately distinguish strains of the M. tuberculosis
complex and non-TB mycobacteria, which improved the accuracy of the
anti-TB drug susceptibility assessment.
As the phages only replicate in living cells, the
limitations of the above methods include the ability to detect only
viable cells in the sample. However, M. tuberculosis is
dormant in the bodies of numerous patients with clinically latent
infections (38), presenting a
challenge in detecting dormant M. tuberculosis. Dusthackeer
(39) used the hsp60,
isocitrate lyase and α crystal protein (α-crystallin) gene
promoters to promote the gene expression of FFlux, and
successfully detected the dormant M. tuberculosis bacteria.
Dusthackeer et al (40)
improved the experimental method further by detecting the sputum
samples without the primary culture. It was suggested that this
provided a better simulation of the natural state of dormant
bacteria. The results of this study supported this hypothesis,
which demonstrated the possibility of potential TB detection.
Banaiee et al (41) compared the assessment of the drug
susceptibility of LRPs with the BACTEC 460 assay as a reference in
clinical applications. The BACTEC 460 assay is a semi-automated
phage-based antibiotic susceptibility assay. The results revealed
that the diagnostic accuracy of LRPs reached 98.4%, and the drug
detection accuracy rate was 100%. The sensitivity and specificity
for the detection of RIF drug resistance were 100%, and for
isoniazid (INH) were 100 and 97.7%, respectively. The duration
required to perform an LRP trial was considerably reduced, just 2
days, compared with the BACTEC 460 assessment, which required 9
days. In addition, its economic cost is low, at $0.40 for each
strain. This semi-automated LRP assessment technique is ideal for
laboratories with limited funds, enabling assessments in
economically underdeveloped countries experiencing a high burden of
TB. Minion and Pai (42) performed
a meta-analysis of the phage-based assessments of RIF resistance
prior to 2009, a total of 31 studies were included in a sample of
3,085 studies, and the phage amplified biological assessment and
LRP assessment were compared. The results revealed that the
sensitivity and specificity of the LRPs were 99.3 and 98.6%,
marginally higher than the phage amplified biologically assessment
at 98.5 and 97.5%. However, a similar investigation with a larger
LRP sample size is required.
Fluoromycobacteriophages
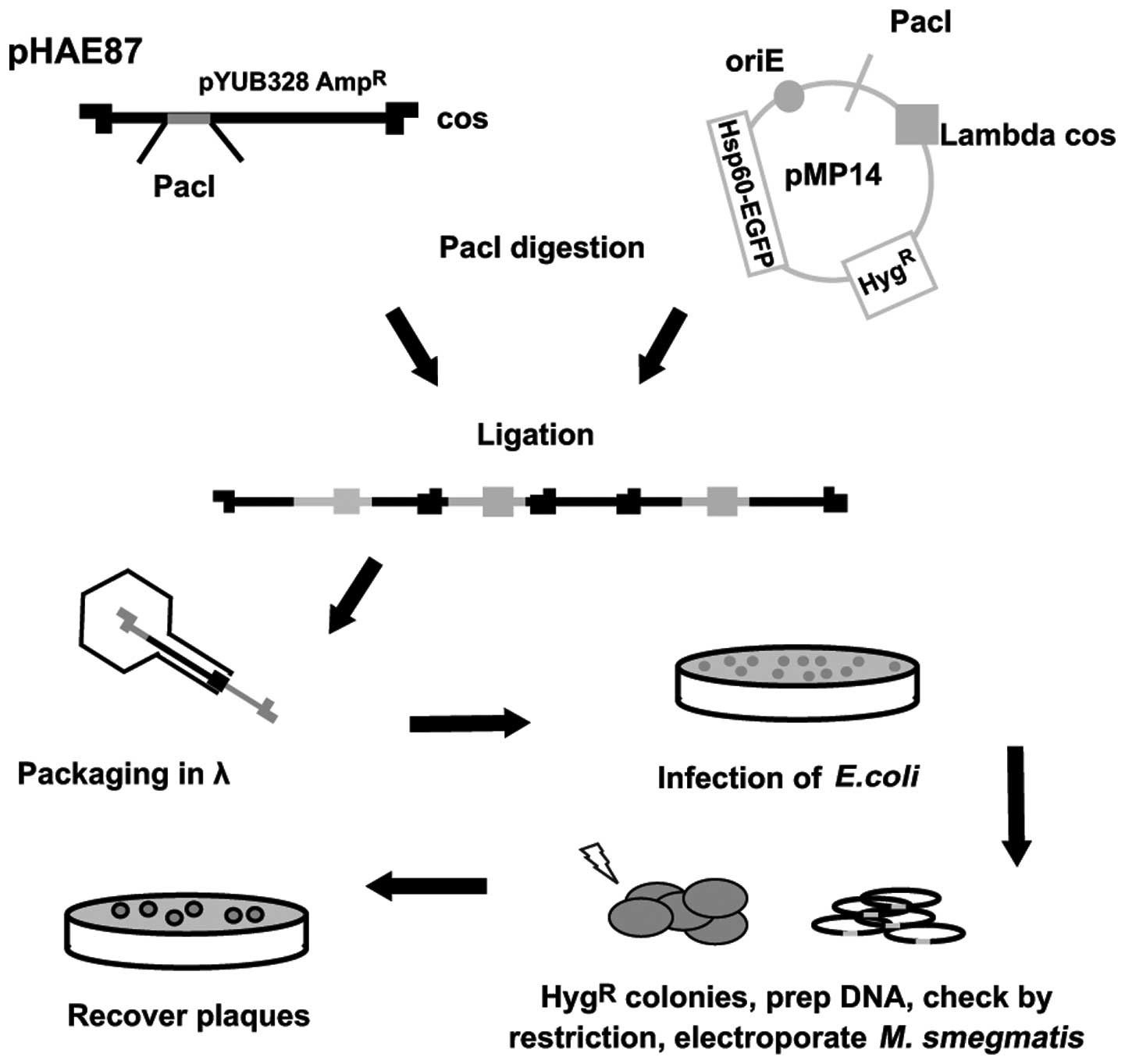
Fluoromycobacteriophages, a novel phage, were
identified in 2009, and differ from the previously reported LRPs.
Piuri et al (17)
constructedthe fluoromycobacteriophages phAE87::hsp60-EGFP
and phAE87::hsp60-ZsYellow (Fig. 4). Green fluorescent protein
(GFP) or the ZsYellow fluorescent markers were
introduced into M. tuberculosis, to detect the drug
susceptibility using fluorescence microscopy or flow cytometry
within 24 h. The technique has several advantages compared with the
LRPs, as no substrate is required, <100/ml M.
tuberculosis can be identified, drug-resistant strains may be
detected in the mixed population and the biosecurity of the samples
is enhanced by polyformalin-fixed processing. Rondón et al
(43) also designed an enhanced
GRP (EGFP) phage, phAE87:: hsp60-EGFP, containing
EGFP on the basis of TM4. This technique was used to detect
drug resistance of M. tuberculosis strains to INH, RIF and
STR, and the results revealed that the sensitivity of this
technique to all antibiotics was 94%, and the specificities of INH,
RIF and STR were 90, 93 and 95%, respectively, compared with the
resazurin microplate technique. The results of the resazurin
microplate assay also exhbibited 94% sensitivity for INH and RIF,
whereas sensitivity for STR was higher at 98%. The reporting
time-period of this technique was 2–3 days and the costs were ~$2.
Although EGFP phage technology for rapid screening of
combined drug resistance is of potential economic value, it
requires further simplification to suit clinical requirements as a
rapid and economic way to detect multidrug-resistant or extensively
drug-resistant strains of TB in resource-poor settings with minimal
infrastructure, and improve sensitivity.
One problem of fluoromycobacteriophages is that, as
a potent mycobacteriophage, TM4 initially infects bacteria, and
then cleaves it, terminating the expression of EGFP. Therefore, the
sensitivity of fluoromycobacteriophages is reduced as the duration
of EGFP expression is shortened (43). To address this problem using
bacteriophage recombineering of electroporated DNA, da Silva et
al (44) inserted a
Phsp60-egfp cassette into the D29 mycobacterio-phage genome
to construct a novel reporter phage. Based on the this novel
reporter phage, an attempt was made to construct a lysis-defective
mutant by deleting the lysA gene, however, it was not possible to
purify the mutant. Despite this, the attempt provided a novel
strategy for the development of a more sensitive reporter
phage.
Another problem of fluoromycobacteriophages is that
the adsorption of TM4 is relatively inefficient. However, mutants
can be isolated with enhanced adsorption, which may provide a
strategy for improving the efficiency of recovery. Piuri et
al (45) constructed a plasmid
expressing the major capsid protein gene (gp9) of TM4, and
containing Strep-tag II (STAG II). Particles with capsids composed
of wild-type and STAG-tagged subunit mixtures were able to grow to
high titers, exhibited good infectivity and were suitable for used
to isolate phage-bacterium complexes. Reporter phage technology
based on the fluorescent protein emitting principle requires
further evaluation of its clinical effects.
4. Conclusion
Since mycobacteriophages were identified 50 years
ago, >2,439 types of mycobacteriophages have been isolated and
the genome sequences of >363 types of mycobacteriophages have
been completed. Mycobacteriophage genomes have several features,
including diversity and mosaicism, a simple structure and
amenability to genetic manipulation. Based on these
characteristics, a shuttle plasmid was constructed for TB
investigation using recombinant DNA technology. With improvements
in genomics, shuttle plasmids have also been used to build
different luciferase reporter phages and fluoromycobacteriophages,
which have contributed to the investigation of mycobacteria and TB.
Following several years of limited studies, phage therapy is again
an active area of investigation, particularly in bacteriophage
lyase. As investigation into mycobacterial phages progresses,
improvements in the current understanding of its role in TB, and
particularly its diagnosis and treatment, is expected.
References
|
1
|
Weir RE, Gorak-Stolinska P, Floyd S, et
al: Persistence of the immune response induced by BCG vaccination.
BMC Infect Dis. 8:1–9. 2008. View Article : Google Scholar
|
|
2
|
Gardner GM and Weiser RS: A bacteriophage
for Mycobacterium smegmatis. Proc Soc Exp Biol Med. 66:2051947.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mycobacteriophage database. http://www.phagesdb.org/.
Accessed 16 May, 2014.
|
|
4
|
Hatfull GF and Sarkis GJ: DNA sequence:
structure and gene expression of mycobacteriophage L5: a phage
system for mycobacterial genetics. Mol Microbiol. 7:395–405. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ford ME, Sarkis GJ, Belanger AE, et al:
Genome structure of mycobacteriophage D29: implications for phage
evolution. J Mol Biol. 279:143–164. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ford ME, Stenstrom C, Hendrix RW, et al:
Mycobacteriophage TM4: genome structure and gene expression. Tuber
Lung Dis. 79:63–73. 1998. View Article : Google Scholar
|
|
7
|
Tokunaga T and Sellers M: Infection of
Mycobacterium smegmatis with D29 phage DNA. J Exp Med. 119:139–149.
1964. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Raj CV and Rama krishnan T: Transduction
in Mycobacterium smegmatis. Nature. 228:280–281. 1970. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jacobs WR Jr, Snapper SB, Tuckman M and
Bloom BR: Mycobacteriophage vector systems. Rev Infect Dis.
11:S404–S410. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jacobs WR Jr, Tuckman M and Bloom BR:
Introduction of foreign DNA into mycobacteria using a shuttle
phasmid. Nature. 327:532–535. 1987. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Snapper SB, Lugosi L, Jekkelt A, et al:
Lysogeny and transformation in mycobacteria: stable expression of
foreign genes. Proc Natl Acad Sci USA. 85:6987–6991. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee MH, Pascopella L, Jacobs WR Jr and
Hatfull GF: Site-specific integration of mycobacteriophage L5:
integration-proficient vectors for Mycobacterium smegmatis,
Mycobacterium tuberculosis and bacille Calmette-Guérin. Proc Natl
Acad Sci USA. 88:3111–3115. 1991. View Article : Google Scholar
|
|
13
|
Bardarov S, Bardarov Jr S Jr, Pavelka Jr
MS Jr, et al: Specialized transduction: an efficient method for
generating marked and unmarked targeted gene disruptions in M.
tuberculosis, M. bovis BCG and M. smegmatis. Microbiology.
148:3007–3017. 2002.PubMed/NCBI
|
|
14
|
Bardarov S, Kriakov J, Carriere C, et al:
Conditionally replicating mycobacteriophages: a system for
transposon delivery to M. tuberculosis. Proc Natl Acad Sci USA.
94:10961–10966. 1997. View Article : Google Scholar
|
|
15
|
Sassetti CM, Boyd DH and Rubin EJ: Genes
required for mycobacterial growth defined by high density
mutagenesis. Mol Microbiol. 48:77–84. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jacobs WR Jr, Barletta RG, Udani R, et al:
Rapid assessment of drug susceptibilities of M. tuberculosis by
means of luciferase reporter phages. Science. 260:819–822. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Piuri M, Jacobs WR Jr and Hatfull GF:
Fluoromycobacteriophages for rapid, specific and sensitive
antibiotic susceptibility testing of M. tuberculosis. PLoS One.
4:e48702009. View Article : Google Scholar
|
|
18
|
World Health Organization: Global
tuberculosis control: WHO report 2011. WHO, Geneva: 2011
|
|
19
|
Pholwat S, Ehdaie B, Foongladda S, Kelly K
and Houpt E: Real-time PCR using mycobacteriophage DNA for rapid
phenotypic drug susceptibility results for Mycobacterium
tuberculosis. J Clin Microbiol. 50:754–761. 2012. View Article : Google Scholar :
|
|
20
|
Tokunaga T and Sellers MI: Streptomycin
induction of premature lysis of bacteriophage-infected
mycobacteria. J Bacteriol. 89:537–538. 1965.PubMed/NCBI
|
|
21
|
Phillips LM and Sellers MI: Effects of
ethambutol, actinomycin D and mitomycin C on the biosynthesis of
D29-infected mycobacterium smegmatis. Host-virus relationships in
mycobacterium, nocardia and actinomyces. Juhasz SE and Plummer G:
Charles C. Thomas; Springfield: pp. 80–102. 1970
|
|
22
|
David HL, Clavel S, Clement F and
Moniz-Pereira J: Effects of antituberculosis and antileprosy drugs
on mycobacteriophage D29 growth. Antimicrob Agents Chemother.
18:357–359. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wilson SM, al-Suwaidi Z, McNerney R, et
al: Evaluation of a new rapid bacteriophage-based method for the
drug susceptibility testing of Mycobacterium tuberculosis. Nat Med.
3:465–468. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
McNerney R, Wilson SM, Sidhu AM, et al:
Inactivation of mycobacteriophage D29 using ferrous ammonium
sulphate as a tool for the detection of viable Mycobacterium
smegmatis and M. tuberculosis. Res Microbiol. 149:487–495. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mole RJ and Maskell TW: Phage as a
diagnostic - the use of phage in TB diagnosis. J Chem Technol
Biotechnol. 76:683–688. 2001. View
Article : Google Scholar
|
|
26
|
Seaman T, Trollip A, Mole R, et al: The
use of a novel phage-based technology as a practical tool for the
diagnosis of tuberculosis in Africa. Afr J Biotechnol. 2:40–45.
2003. View Article : Google Scholar
|
|
27
|
Marei AM, El-Behedy EM, Mohtady HA and
Afify AF: Evaluation of a rapid bacteriophage-based method for the
detection of Mycobacterium tuberculosis in clinical samples. J Med
Microbiol. 52:331–335. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Muzaffar R, Batool S, Aziz F, et al:
Evaluation of the FASTPlaqueTB assay for direct detection of
Mycobacterium tuberculosis in sputum specimens. Int J Tuberc Lung
Dis. 6:635–640. 2002.PubMed/NCBI
|
|
29
|
Albert H, Trollip AP, Mole RJ, et al:
Rapid indication of multidrug-resistant tuberculosis from liquid
cultures using FASTPlaqueTB-RIF™, a manual phage-based test. Int J
Tuberc Lung Dis. 6:523–528. 2002.PubMed/NCBI
|
|
30
|
Alcaide F, Galí N, Domínguez J, et al:
Usefulness of a new mycobacteriophage-based technique for rapid
diagnosis of pulmonary tuberculosis. J Clin Microbiol.
41:2867–2871. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kalantri S, Pai M, Pascopella L, et al:
Bacteriophage-based tests for the detection of Mycobacterium
tuberculosis in clinical specimens: a systematic review and
meta-analysis. BMC Infect Dis. 5:592005. View Article : Google Scholar
|
|
32
|
Foundation for Innovative New Diagnostics
(FIND): FIND interrupts demonstration projects using phage-based
assays for detection of rifampin resistance. Geneva, Switzerland:
FIND; 3–October. 2007, http://www.finddiagnostics.org/media/news/index.jsp?year=2007&domain.
Accessed 16 May, 2014.
|
|
33
|
Sarkis GJ, Jacobs WR Jr and Hatfull GF: L5
luciferase reporter mycobacteriophages: a sensitive tool for the
detection and assay of live mycobacteria. Mol Microbiol.
15:1055–1067. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pearson RE, Jurgensen S, Sarkis GJ, et al:
Construction of D29 shuttle phasmids and luciferase reporter phages
for detection of mycobacteria. Gene. 183:129–136. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kumar V, Loganathan P, Sivaramakrishnan G,
et al: Characterization of temperate phage Che12 and construction
of a new tool for diagnosis of tuberculosis. Tuberculosis (Edinb).
88:616–623. 2008. View Article : Google Scholar
|
|
36
|
Carrière C, Riska PF, Zimhony O, et al:
Conditionally replicating luciferase reporter phages: improved
sensitivity for rapid detection and assessment of drug
susceptibility of Mycobacterium tuberculosis. J Clin Microbiol.
35:3232–3239. 1997.PubMed/NCBI
|
|
37
|
Riska PF, Jacobs WR Jr, Bloom BR, et al:
Specific identification of M. tuberculosis with the luciferase
reporter mycobacte-riophage: use of
p-nitro-alpha-acetylamino-beta-hydroxy propiophenone. J Clin
Microbiol. 35:3225–3231. 1997.PubMed/NCBI
|
|
38
|
Dye C, Scheele S, Dolin P, Pathania V and
Raviglione MC: Consensus statement. Global burden of tuberculosis:
Estimated incidence, prevalence, and mortality by country WHO
Global Surveillance and Monitoring Project. JAMA. 282:677–686.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dusthackeer A, Kumar V, Subbian S, et al:
Construction and evaluation of luciferase reporter phages for the
detection of active and non-replicating tubercle bacilli. J
Microbiol Methods. 73:18–25. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dusthackeer VN, Balaji S, Gomathi NS, et
al: Diagnostic luciferase reporter phage assay for active and
non-replicating persistors to detect tubercle bacilli from sputum
samples. Clin Microbiol Infect. 18:492–496. 2012. View Article : Google Scholar
|
|
41
|
Banaiee N, January V, Barthus C, et al:
Evaluation of a semi-automated reporter phage assay for
susceptibility testing of Mycobacterium tuberculosis isolates in
South Africa. Tuberculosis (Edinb). 88:64–68. 2008. View Article : Google Scholar
|
|
42
|
Minion J and Pai M: Bacteriophage assays
for rifampicin resistance detection in Mycobacterium tuberculosis:
updated meta-analysis. Int J Tuberc Lung Dis. 14:941–951.
2010.PubMed/NCBI
|
|
43
|
Rondón L, Piuri M, Jacobs WR Jr, et al:
Evaluation of fluoromycobacteriophages for detecting drug
resistance in Mycobacterium tuberculosis. J Clin Microbiol.
49:1838–1842. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
da Silva JL, Piuri M, Broussard G, et al:
Application of BRED technology to construct recombinant D29
reporter phage expressing EGFP. FEMS Microbiol Lett. 344:166–172.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Piuri M, Rondón L, Urdániz E and Hatfull
GF: Generation of affinity-tagged fluoromycobacteriophages by mixed
assembly of phage capsids. Appl Environ Microbiol. 79:5608–5615.
2013. View Article : Google Scholar : PubMed/NCBI
|