Introduction
It has been demonstrated that heterologous proteins
can be displayed directly on the surface of magnetosomes through
genetic fusion to magnetosome membrane proteins (MMPs) (1). A number of MMP associated with the
synthesis of nanoparticles have been identified, and several of
these are used as anchor proteins, including MpsA, MagA, Mms13 and
Mms6 (MamC, Mam12) (2,3). A previous report indicated that
mms13 (mamC) is a putative membrane anchor gene
(4,5).
Type IV collagenase, also termed gelatinase,
including gelatinase A (MMP-2; 72 kDa) and gelatinase B (MMP-9; 92
kDa), is an important member of the MMP family. Type IV collagenase
is abundantly expressed in proliferating endothelial cells and in
several types of malignant tumor, where it is involved in cancer
invasion, metastasis and angiogenesis (6). Therefore, type IV collagenase is a
potential target in cancer therapy. As reported, the 3G11 type IV
collagenase monoclonal antibody and its single chain Fv fragment
(scFv) exhibit specific binding to target enzymes and can prevent
tumor growth, invasion and metastasis (6,7).
scFv antibodies have potential advantages over whole antibodies,
including their small size, minimal antigenicity, high
penetrability and their ability to be manipulated by genetic
engineering (8). Therefore, these
antibodies present as a relatively ideal tumor-targeting agent.
The present study involved the construction,
expression, purification and characterization of ScFv, Mms13 and
the ScFv-mms13 fusion protein.
Materials and methods
Cell culture
The Magnetospirillum magneticum AMB-1 strain
(American Type Culture Collection, Manassas, VA, USA) was grown
microaerobically at 28°C in modified enriched mangnetic spirillum
growth medium (EMSGM) (9). For
plate cultivation, agar was added (1.5% wt/vol) to the EMSGM. The
Escherichia coli DH5α strain (DH5α-competent cells; Takara
Bio, Inc., Dalian, Liaoning, China) was used for DNA cloning and
the Rosetta (DE3) E. coli strain (Novagen, Heidelberg,
Germany) was used for protein expression. The E. coli DH5α
strain was grown on Luria-Bertani (LB) medium at 37°C, supplemented
with kanamycin (50 μg/ml) or ampicillin (50 μg/ml)
and 1.5% (wt/vol) agar, if appropriate (10).
Human breast carcinoma MCF-7 cells, human hepatoma
SMMC-7721 cells and HepG2 cells were obtained from China Medical
Culture Collection Center (Beijing, China), and grown in RPMI-1640
medium (Gibco-BRL, Carlsbad, CA, USA), supplemented with 10% fetal
bovine serum (Beyotime Biotechnology, Beijing, China), penicillin G
(100 U/ml) and streptomycin (100 mg/ml) at 37°C in an atmosphere of
5% CO2. All cell lines were passaged every 3 days and
were maintained in exponential growth to ~80% confluence for the
subsequent experiments.
Construction of the pET30a(+)-scFv and
pET30a(+)-mms13 expression vectors
The primers and the gene of the anti-type IV
collagenase scFv were synthesized, according to the GenBank
database (accession no. FJ037775 by Takara Bio, Inc. (11) and cloned into the pMD18-T vector
(Thermo Fisher Scientific, San Jose, CA, USA) to create the
pMD-scFv vector. DNA fragments were amplified by polymerase chain
reaction (PCR) for subsequent cloning using a T-Gradient
Thermoblock PCR cycler (Biometra, Gottingen, Germany) and PCR
MasterMix (Hangzhou Biosci Biotech Co., Ltd., Hangzhou, China). The
scFv gene was amplified from the pMD-scFv plasmid by PCR using the
following primers: P1, forward 5′-GGAATTCCATATGCAGGTGAAGCTGCAG-3′,
introducing an NdeI restriction site, and P2, reverse
5′-CCGCTCGAGACGTTTGATTTCCAGCTT-3, introducing an XhoI site
to the 3′ end of the scFv gene. The cycling conditions were as
follows: For mms13, 30 cycles of 94°C for 30 sec, 48°C for
45 sec and 72°C for 1 min; for scFv, 30 cycles of 94°C for 30 sec,
55°C for 45 sec and 72°C for 1 min; and for scFv-mms13, 30
cycles of 94°C for 30 sec, 53°C for 45 sec and 72°C for 1 min. The
763 bp PCR product was purified and digested by
NdeI/XhoI (Thermo Fisher Scientific) and then ligated
into an NdeI/XhoI-cleaved pET-30a(+) (Novagen) to
produce the pET30a(+)-scFv expression vector. Sequences were
analyzed by Sangon Biotech Co., Ltd. (Shanghai, China).
The genomic DNA of the M. magneticum AMB-1
was extracted using a MiniBEST Bacterial Genomic DNA Extraction kit
(Takara Bio, Inc.). The mms13 gene was amplified from the
genome of M. magneticum using the following primers: P3,
forward 5′-GGAATTCCATATGCCCTTTCACCTTG-3′, introducing an
NdeI restriction site, and P4, reverse
5′-CCGCTCGAGGGCCAGTTCGTCCCG-3′, introducing an XhoI site to
the 3′ end of the mms13 gene. The 388 bp PCR product was
cloned into pET-30a(+) to produce pET30a(+)-mms13, similar
to the previously constructed pET30a(+)-scFv.
Construction of the pET30a(+)-scFv-mms13
expression vector
The scFv-mms13 fusion gene was constructed
using the splicing by overlap-extension PCR (SOE-PCR) technique.
The scFv gene was amplified by PCR using the following primers: P5,
forward 5′-ATGCAGGTGAAGCTGCAG-3′, and P6, reverse
5′-GGAGCCGCCGCCGCCAGAACCACCACCACCACGTTTGATTTCCAGCTT-3′, and the
mms13 gene was amplified using the following primers: P7,
forward 5′-GGCGGCGGCG-GCTCCGGTGGTGGTGGTTCTATGCCCTTTCACCTTG-3′ and
P8 reverse 5′-GGCCAGTTCGTCCCG-3′. The PCR products of scFv and
mms13 were purified using the TIANquick Midi Purification
kit (Tiangen Biotech (Beijing) Co., Ltd., Beijing, China) and
mixed. The assembly reaction included one cycle of denaturation,
annealing and extension, in the absence of primers, to facilitate
the assembly of chains, followed by 30 cycles of amplification
reactions, in the presence of the scFv forward primer and
mms13 reverse primers. The scFv reverse primer and
mms13 forward primer included an artificial region of
overlap to enable the formation of a flexible segment,
corresponding to the G4S3 linker. The product of the SOE-PCR was
purified and cloned into the pMD18-T vector to create the
pMD-scfv-mms13 expression vector. The scFv-mms13
fusion gene was then amplified from the pMD-scfv-mms13 using
the following primers: P1, forward
5′-GGAATTCCATATGCAGGTGAAGCTGCAG-3′, introducing an NdeI
restriction site, and P4, reverse 5′-CCGCTCGAGGGCCAGTTCGTCCCG-3′,
introducing an XhoI restriction site. The PCR product was
cloned into pET-30a(+) to produce pET30a(+)-scFv-mms13.
Expression and cellular location of the
ScFv, Mms13 and ScFv-mms13 proteins by western blot analysis
The procedures for the growth of the DE3 E.
coli strain, transformed with pET30a(+)-scFv,
pET30a(+)-mms13 or pET30a(+)-scFv-mms13 were
performed, according to standard protocol. The strain was cultured
in LB medium at 37°C, supplemented with kanamycin (50
μg/ml). Following induction of the target proteins at 37°C
for 4 h with 0.2 mM isopropylthio-β-D-thiogalactopyranoside (IPTG;
Takara Bio, Inc.), the four fractions, including the medium,
periplasmic, soluble cytoplasmic and insoluble samples, were
obtained, according to the pET system manual (10th ed; http://www.merckmillipore.com/). The samples were
electrophoresed on 15% SDS-polyacrylamide gels, and the proteins
were then transblotted onto a nitrocellulose membrane (Millipore,
Bedford, MA, USA), blocked with 5% bovine serum albumin
(BSA)/Tris-buffered saline with Tween 20 (TBST; 25 mM Tris-HCl at
pH 8.0, 125 mM NaCl and 0.05% Tween-20; Beyotime Institute of
Biotechnology) for 1 h and incubated with a 1:1,000 dilution of
anti-His-Tag monoclonal mouse primary antibody (cat. no. AH367;
Beyotime Institute of Biotechnology) overnight at 4°C, followed by
incubation with 1:1,000 horseradish peroxidase-labeled goat
anti-mouse polyclonal immunoglobulin (Ig)G secondary antibody (cat.
no. AO216; Beyotime Institute of Biotechnology) at 37°C for 1 h.
The membrane was washed with TBST 5 times for 5 min each time and
the antibody reactions were visualized using a Super signal West
Pico Trial kit (Thermo Fisher Scientific).
Purification and refolding of the ScFv,
Mms13 and ScFv-mms13 proteins
The induced bacterial cells (~108
cells/ml) were centrifuged at 10,000 g for 10 min (HC-2518R; Anhui
USTC Zonkia Scientific Instruments Co., Ltd., Hefei, China). The
cells pellet was resuspended in binding buffer, containing 20
mmol/l imidazole, 0.5 mol/l NaCl and 20 mmol/l
NaH2PO4 (pH 7.5), and sonicated (JY92-2D;
Ningbo Scientz Bio, Inc., Ningbo, China), followed by
centrifugation of the cell lysate at 12,000 g for 10 min at 4°C.
The pellet was then resuspended and incubated in binding buffer,
containing 8 mol/l urea, on ice for 1 h. The insoluble material was
then removed by centrifugation at 12,000 g for 20 min. The
supernatant was filtered through a 0.45 nm membrane and purified
using HisTrap affinity columns (GE Healthcare, Amersham, UK), under
denaturing conditions, according to the manufacturer’s
instructions. The column was washed with distilled water with a
flow rate of 1 ml/min and was equilibrated with binding buffer (20
mM NaH2PO4, 0.5 M NaCl, 20 mM imidazole, 8 M
urea, pH 7.4). The column was then washed with binding buffer and
eluted with elution buffer (20 mM NaH2PO4,
0.5 M NaCl, 500 mM imidazole, 8 M urea, pH 7.4). The purified ScFv
and ScFv-mms13 proteins were refolded using step-wise
dialysis, as reported previously (12). In brief, β-mercaptoethanol (Tianjin
Guangfu Fine Chemical Research Institute, Tianjin, China) was added
to the protein solution at 1 M. The protein solution was dialysed
with 50 fold refolding buffer (50 mM Tris-HCl, 1 mM EDAT, 200 mM
NCl, pH 8.0) with 8 M urea (Tianjin Kemiou Chemical Reagent Co.,
Ltd., Tianjin, China) at 4°C for 12 h. The refolding buffer was
replaced with step-wise concentrations of urea (4, 2, 1, 0.5 and 0
M). Oxidized glutathione (Beijing Solarbio Science & Technology
Co., Ltd., Beijing, China) was added at 50 μM and L-arginine
(Sangon Biotech Co., Ltd.) at 400 mM was added with the 1 M urea
step.
The protein concentrations of the fractions was
determined using a Bicinchoninic Acid Protein Assay kit (Beyotime
Institute of Biotechnology) using various concentrations of BSA (2,
1.5, 1, 0.75, 0.5, 0.25, 0.125, 0.025 and 0 mg/m1; 25
μl/well; Beyotime Institute of Biotechnology) as a standard.
The proteins were analyzed throughout using SDS-PAGE and the gels
were stained with Coomassie brilliant blue (2.5 g with 500 ml
methanol and 100 ml glacial acetic acid made up with 1 L
ddH2O; Sinopharm Chemical Reagent Co., Ltd., Shanghai,
China).
Enzyme-linked immunosorbent assays
(ELISA)
The cells (2×104 cells/well) were grown
in 96-well plates to confluence, washed with phosphate-buffered
saline (PBS; Beyotime Institute of Biotechnology) and fixed for 30
min with 50 μl/well methanol at 4°C. For gelatinase, 96-well
microtiter plates were coated with 100 μl/well of 10
μg/ml type IV collagenase (Thermo Fisher Scientific) at 4°C
overnight. The plates containing type IV collagenase or the fixed
cells were washed three times with PBS and blocked with 1% BSA/PBS
at 4°C overnight. The wells were emptied and 50 μl ScFv,
Mms13 or ScFv-mms13 were added in two-fold serial dilutions
at concentrations ranging between 0.1 and 50 μmol/l for 2 h
at 37°C. Following washing with PBS, the wells were incubated with
50 μl/well 1:1,500 anti-His tag mAb (Beyotime Institute of
Biotechnology), as a primary antibody, at 37°C 1 h. The wells were
then overlaid with 50 μl/well 1:2,000 horseradish
peroxidase-labeled goat anti-mouse IgG (Beyotime Institute of
Biotechnology), as a secondary antibody, at 37°C 1 h following
washing with PBS. Color development was achieved using 100
μl o-Phenylenediamine solution, which was terminated after
10 min with 100 μl 2 mol/l H2SO4. The
absorbance was measured at 490 nm using a Multiskan MK3 microplate
reader (Thermo Fisher Scientific). All assays were performed in
triplicate.
Immunofluorescent cytochemical staining
of the SMMC-7721 and MCF-7 cells
Immunofluorescent staining was performed on the
antigen-positive SMMC-7721 and MCF-7 cells. The cells were grown on
slides and fixed in ice-cold methanol for 30 min. Nonspecific
binding was inhibited using 200 μl/well 1% BSA/PBS at 4°C
overnight. Following washing with PBS, the cells were incubated
with 50 μl/well ScFv, Mms13 or ScFv-mms13. The cells
were then overlaid with 1:1,500 mouse anti-His tag monoclonal
antibodies (Beyotime Institute of Biotechnology) following washing
with PBS. A final washing step with PBS was performed, and the
slides were mounted with 1:2,000 fluorescein
isothiocyanate-conjugated goat anti-mouse antibody (Beyotime
Institute of Biotechnology) and fluorescence images were captured
using an Olympus BX60 microscope (magnification, ×100), equipped
with an Olympus DP71 camera and Olympus DP-Controller software,
version 2.1 (Olympus Corporation, Tokyo, Japan).
Results
Construction of the pET30a(+)-mms13,
pET30a(+)-scFv and pET30a(+)-scFv-mms13 expression vectors
The DNA sequences encoding the mms13 gene and
the scFv of the 3G11 mAb were cloned into the
NdeI/XhoI restriction sites of pET-30a(+), producing
the pET30a(+)-mms13 (Fig.
1A) and pET30a(+)-scFv (Fig.
1B) expression vectors. The mms13 and scFv genes were
amplified by PCR, and were linked using SOE-PCR to yield the
scFv-mms13 fusion gene. A 15 amino acid spacer
(G4S)3 was present between the C-terminus of
the scFv and the N-terminus of the mms13 genes. The
scFv-mms13 fusion gene was cloned into the
NdeI/XhoI restriction sites of pET-30a(+) to create
the pET30a(+)-scfv-mms13 expression vector (Fig. 1C). Sequence analyses confirmed all
the expected DNA sequences. All the three genes were under the
control of the T7 promoter, and a (His)6-tag was
introduced at the C-terminus of the constructs to facilitate
purification using immobilized metal affinity chromatography. The
Mms13, ScFv and ScFv-mms13 fusion protein were composed of
132, 253 and 394 amino acids (Fig.
1D), with theoretical molecular weights of 13.4, 27.4 and 40.9
kDa respectively.
Expression, purification and refolding of
the ScFv, Mms13 and ScFv-mms13 proteins
The three expression vectors were transformed into
the DE3 E. coli strain, and the target proteins were induced
by the addition of IPTG. The results of the Coomassie Blue-stained
gel indicated that Mms13 was present in the cytoplasmic soluble
fraction, ScFv was present in the insoluble fractions (data not
shown) and the ScFv-mms13 fusion protein was present in the
cytoplasmic soluble fraction and insoluble fractions (Fig. 2A). The three proteins were further
confirmed using western blot analysis with an anti-His-Tag antibody
(Fig. 2B). The three proteins were
purified using immobilized metal-affinity chromatography resin
under denaturing conditions, and the purified ScFv and
ScFv-mms13 proteins were refolded using step-wise dialysis,
as reported previously (12).
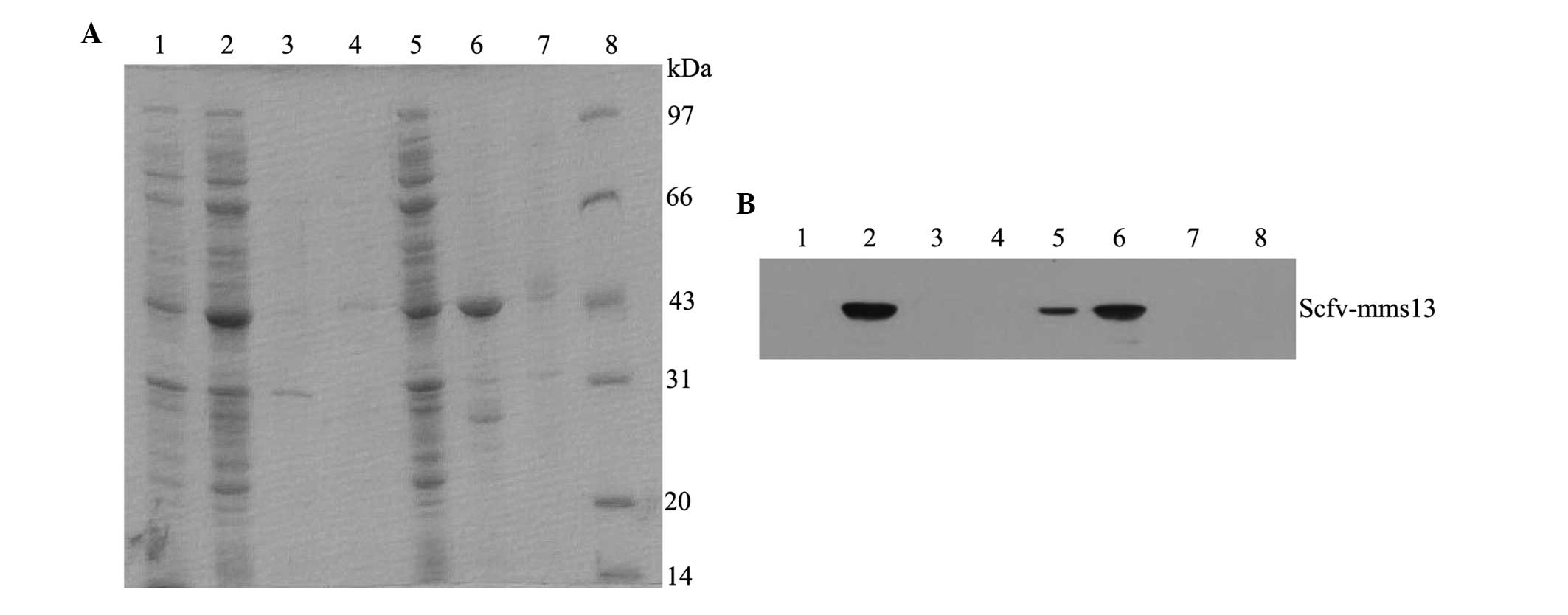 | Figure 2SDS-PAGE assay and western blot
analysis of the ScFv-mms13 fusion protein. The (A) SDS-PAGE
and (B) western blot of each fraction of the E. coli Rosetta
(DE3) cells expressing the ScFv-mms13 fusion protein. Lane
1, total proteins of E. coli containing the pET-30a(+)
plasmid following IPTG induction; Lane 2, total proteins of E.
coli containing the pET-30a(+)-scFv-mms13 plasmid
following IPTG induction; Lane 3, medium sample; Lane 4,
periplasmic fraction; Lane 5, cytoplasmic soluble fraction; Lane 6,
cytoplasmic insoluble fraction; Lane 7, cytoplasmic insoluble
fraction of E. coli containing the
pET30a(+)-scFv-mms13 plasmid without IPTG induction; Lane 8,
molecular weight marker. ScFv, single chain Fv; IPTG,
isopropylthio-β-D-thiogalacto pyranoside. |
ELISA and immunofluorescent cytochemical
staining
To confirm the correct folding and functional
binding of the fusion protein, the abilities of ScFv, Mms13 and
ScFv-mms13 to bind to the target antigen type IV collagenase
or antigen-relevant tumor cells were examined using ELISA. The data
indicated that ScFv and ScFv-mms13 bound to the type IV
collagenase or the antigen-associated cancer cells, including
SMMC-7721, MCF-7 and HepG2 cells, in a dose-dependent and saturable
manner (Fig. 3A). Although the
immunoreactivities of ScFv-mms13 to the type IV collagenase
and associated tumor cells were marginally lower than the
corresponding scFv (3G11), there remained considerable binding
ability to the antigen by ScFv-mms13 (Fig. 3A).
Immunofluorescence staining was performed on the
ScFv-, mms13- and ScFv-mms13-treated SMMC-7721 and
MCF-7 cells. As shown in Fig. 3B,
the ScFv and ScFv-mms13 fusion protein exhibited green
fluorescence, whereas no fluorescence was observed in the PBS
control or Mms13, which further confirmed the immunoreactivity of
ScFv-mms13.
Discussion
ScFv antibodies retaining the binding
characteristics of the parent immunoglobulin have been preferred in
clinical and diagnostic applications due to their prominent
advantages, including lower molecular weight, superior penetration
of tumor tissue, improved pharmacokinetics and a reduction in
immunogenicity (13). The
single-chain antibody of type IV collagenase, which is associated
with the invasion, metastasis and angiogenesis of malignant types
of tumor, may not only serve as a tumor targeting vehicle, but also
exert its own anti-tumor activity by inhibiting target enzymes
(6,7). A number of heterologous proteins have
been fused to certain MMPs and displayed on the surface of
magnetosomes (4,5,14,15).
As the most abundant magnetosome protein, Mms13 has been
demonstrated as an efficient anchor protein (4,5).
It has been previously reported that ScFv antibodies
have low solubility, which imposes a significant limitation in
their diagnostic and clinical implication (16). The co-expression of ScFv with
affinity tags or molecular chaperones, including glutathione
S-transferase (17), green
fluorescent protein (18) or
maltose binding protein (19) can
enhance the solubility of a number of the fusion proteins.
Therefore, the preset study hypothesized that the difference in the
solubility of ScFv-mms13 from the general isolated ScFv may
be associated with the occurrence of Mms13. Mms13 itself was highly
soluble in aqueous environments (data not shown), and the present
study demonstrated that the ScFv-mms13 fusion protein was
present in the cytoplasmic soluble fraction (~30%) and insoluble
fractions (Fig. 2a). It is
possible that the existence of Mms13 may affect the solubility
behavior and partly prevent the aggregation of the
ScFv-mms13 fusion protein. A putative explanation is that
the linking of ScFv to Mms13 partly covers the exposed hydrophobic
surface of ScFv and, therefore, prevents its aggregation.
Rosenblum et al (20) previously described immunotoxins, in
which the single-chain antibody of ZME-018 was fused to the
ribosome-inactivating plant toxin, gelonin, and found that the
recombinant immunotoxin preserved the cytotoxicity and
antigen-binding activity. The results of the present study were
consistent with these findings. The ELISA results revealed that the
ScFv-mms13 fusion protein retained the antigen binding
activity of ScFv and interacted with type IV collagenase and
several antigen-relevant tumor cells. The results of the
immunofluorescence analysis also demonstrated the immunoreactivity
of the ScFv-mms13 fusion protein with various
antigen-associated cancer cells. These data suggested that the
ScFv-mms13 fusion protein contained sufficient structural
information for specific antigen recognition.
In conclusion, the engineered ScFv-mms13
fusion protein demonstrated antigen-binding activity, and presents
as a promising candidate for the display of ScFv on magneto-some
surfaces.
Acknowledgments
This study was supported by the Science and
Technology Brainstorm Project of Shandong Province (grant no.
2009GG10002079) and the Higher Educational Science and Technology
Program of Shandong Province (grant no. 7J12LE51).
Abbreviations:
|
scFv
|
single chain Fv
|
|
SOE-PCR
|
splicing by overlap extension
polymerase chain reaction
|
|
MMPs
|
magnetosome membrane proteins
|
|
EMSGM
|
enriched mangnetic spirillum growth
medium
|
References
|
1
|
Yan L, Zhang S, Chen P, et al:
Magnetotactic bacteria, magnetosomes and their application.
Microbiol Res. 167:507–519. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Matsunaga T, Sato R, Kamiya S, et al:
Chemiluminescence enzyme immunoassay using protein A-bacterial
magnetite complex. J Magn Magn Mater. 194:126–134. 1999. View Article : Google Scholar
|
|
3
|
Yoshino T, Takahashi M, Takeyama H, et al:
Assembly of G protein-coupled receptors onto nanosized bacterial
magnetic particles using Mms16 as an anchor molecule. Appl Environ
Microbiol. 70:2880–2885. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yoshino T and Matsunaga T: Efficient and
stable display of functional proteins on bacterial magnetic
particles using mms13 as a novel anchor molecule. Appl Environ
Microbiol. 72:465–471. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lang C and Schüler D: Expression of green
fluorescent protein fused to magnetosome proteins in
microaerophilic magnetotactic bacteria. Appl Environ Microbiol.
74:4944–4953. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhong G, Zhang S, Li Y, et al: A tandem
scFv-based fusion protein and its enediyne-energized analogue show
intensified therapeutic efficacy against lung carcinoma xenograft
in athymic mice. Cancer Lett. 295:124–133. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li L, Huang YH, Li Y, et al: Antitumor
activity of anti-type IV collagenase monoclonal antibody and its
lidamycin conjugate against colon carcinoma. World J Gastroenterol.
11:4478–4483. 2005.PubMed/NCBI
|
|
8
|
Cao L, Shen G, Zhu Y, et al:
Characterization of a single-chain variable fragment (scFv)
antibody directed against the human asialoglycoprotein receptor.
Biotechnol Appl Biochem. 44:65–72. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang C, Takeyama H, Tanaka T, et al:
Effects of growth medium composition, iron sources and atmospheric
oxygen concentrations on production of luciferase-bacterial
magnetic particle complex by a recombinant Magnetospirillum
magneticum AMB-1. Enzyme Microb Technol. 29:13–19. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sambrook J and Russell DW: Molecular
cloning: a laboratory manual. 3rd ed. Cold Spring Harbor Laboratory
Press; Cold Spring Harbor, New York: pp. 1595–1599. 2001
|
|
11
|
Zhong GS, Zhang SH, Li Y, et al: A tandem
scFv-based fusion protein and its enediyne-energized analogue show
intensified therapeutic efficacy against lung carcinoma xenograft
in athymic mice. Cancer Lett. 295:124–133. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Miao QF, Shang BY, Li L, et al: Expression
of single-chain Fv fragment directed against type IV collagenase
produced in Escherichia Coliand its antitumor activity. J Med Res.
36:25–29. 2007.In Chinese.
|
|
13
|
Raju TS and Strohl WR: Potential
therapeutic roles for antibody mixtures. Expert Opin Biol Ther.
13:1347–1352. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pollithy A, Romer T, Lang C, et al:
Magnetosome expression of functional camelid antibody fragments
(nanobodies) in Magnetospirillum gryphiswaldense. Appl Environ
Microbiol. 77:6165–6171. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ohuchi S and Schüler D: In vivo display of
a multisubunit enzyme complex on biogenic magnetic nanoparticles.
Appl Environ Microbiol. 75:7734–7738. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang R, Xiang S, Feng Y, et al:
Engineering production of functional scFv antibody in E. coli by
co-expressing the molecule chaperone Skp. Front Cell Infect
Microbiol. 3:722013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
DelProposto J, Majmudar CY, Smith JL and
Brown WC: Mocr: a novel fusion tag for enhancing solubility that is
compatible with structural biology applications. Protein Expr
Purif. 63:40–49. 2009. View Article : Google Scholar
|
|
18
|
Petrausch U, Dernedde J, Coelho V, et al:
A33scFv-green fluorescent protein, a recombinant single-chain
fusion protein for tumor targeting. Protein Eng Des Sel.
20:583–590. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nallamsetty S and Waugh DS:
Solubility-enhancing proteins MBP and NusA play a passive role in
the folding of their fusion partners. Protein Expr Purif.
45:175–182. 2006. View Article : Google Scholar
|
|
20
|
Rosenblum MG, Cheung LH, Liu Y and Marks
JW III: Design, expression, purification and characterization, in
vitro and in vivo, of an antimelanoma single-chain Fv antibody
fused to the toxin Gelonin. Cancer Res. 63:3995–4002.
2003.PubMed/NCBI
|

















