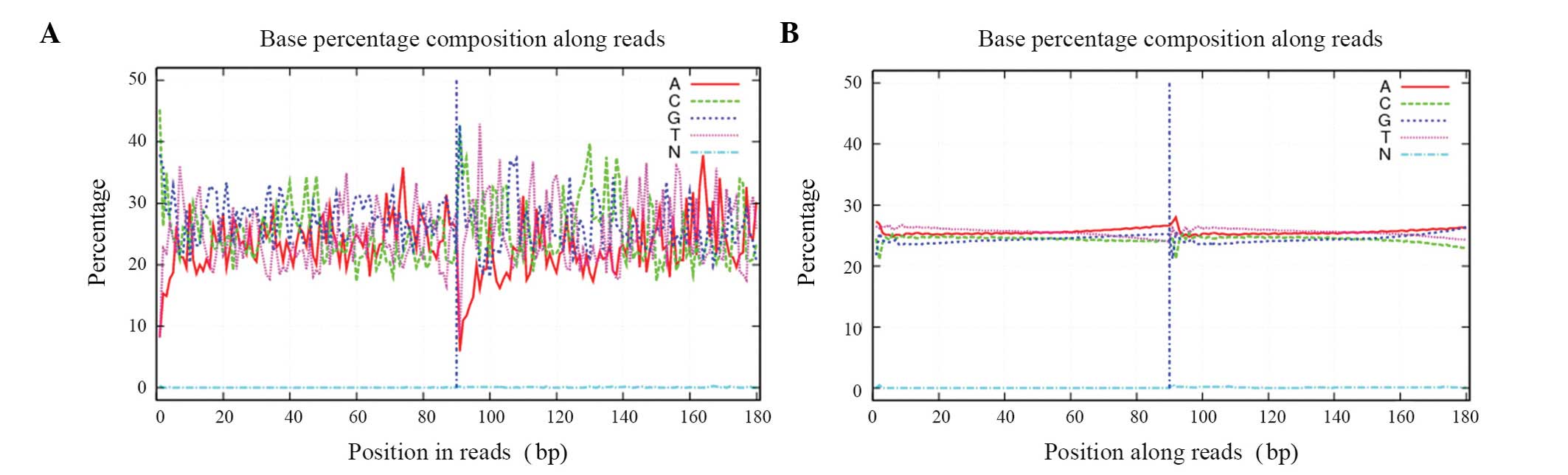
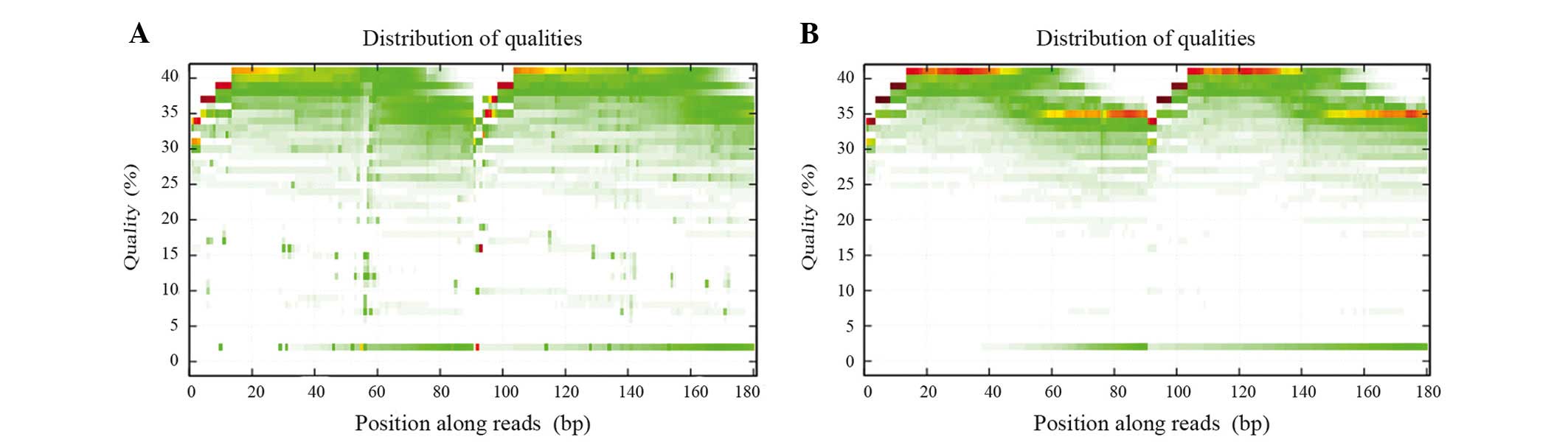
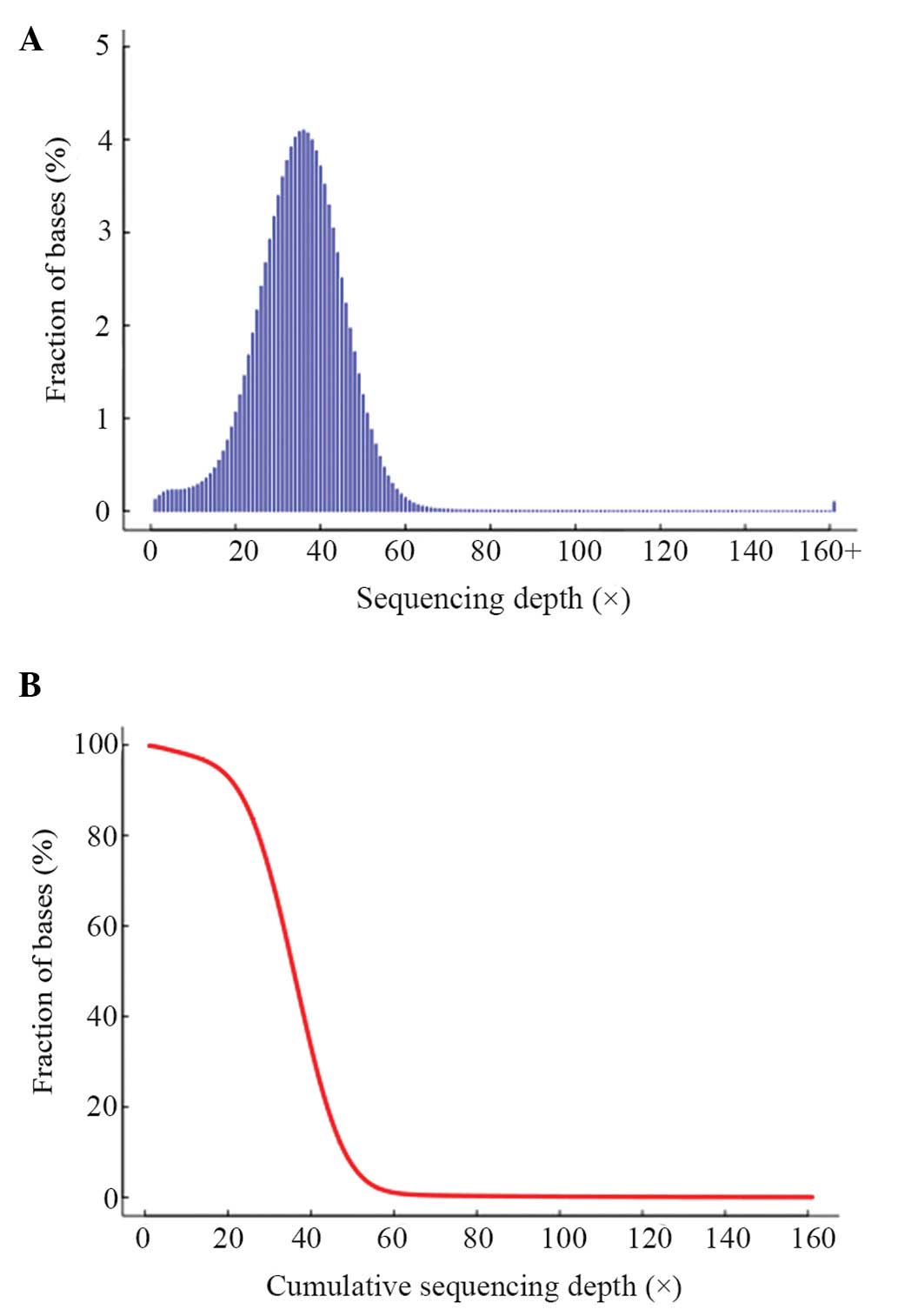
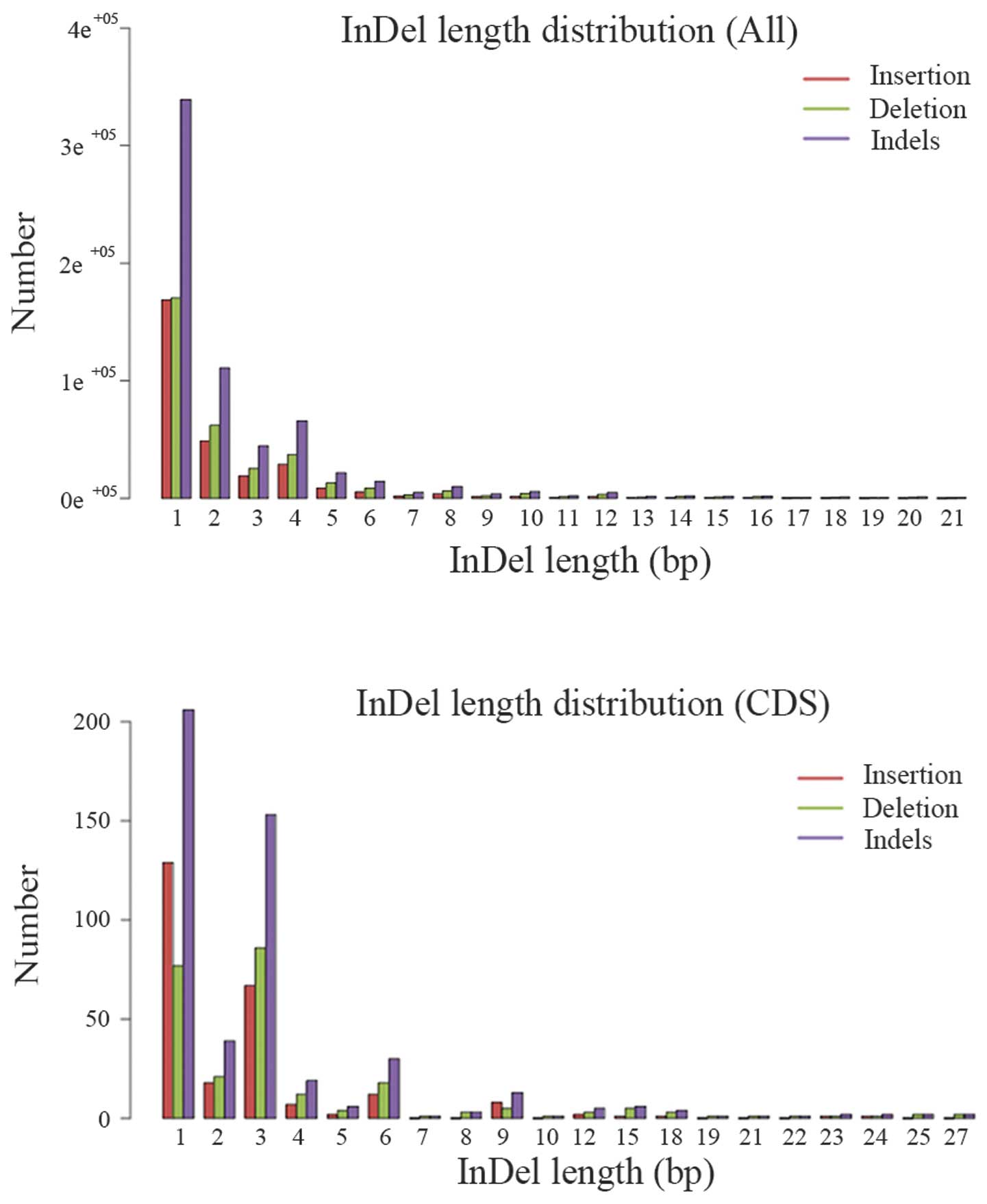
|
1
|
Chen L, Magliano DJ and Zimmet PZ: The
worldwide epidemiology of type 2 diabetes mellitus-present and
future perspectives. Nat Rev Endocrinol. 8:228–236. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tsai FJ, Yang CF, Chen CC, Chuang LM, Lu
CH, Chang CT, Wang TY, Chen RH, Shiu CF, Liu YM, et al: A
genome-wide association study identifies susceptibility variants
for type 2 diabetes in Han Chinese. PLoS Genet. 6:e10008472010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Scott LJ, Mohlke KL, Bonnycastle LL,
Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS,
Jackson AU, et al: A genome-wide association study of type 2
diabetes in Finns detects multiple susceptibility variants.
Science. 316:1341–1345. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Diabetes Genetics Initiative of Broad
Institute of Harvard and MIT; Lund University; Novartis Institutes
of BioMedical Research; Saxena R, Voight BF, Lyssenko V, Burtt NP,
de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, et al:
Genome-wide association analysis identifies loci for type 2
diabetes and triglyceride levels. Science. 316:1331–1336. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sladek R, Rocheleau G, Rung J, Dina C,
Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, et al:
A genome-wide association study identifies novel risk loci for type
2 diabetes. Nature. 445:881–885. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zeggini E, Weedon MN, Lindgren CM,
Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW,
Freathy RM, et al: Replication of genome-wide association signals
in UK samples reveals risk loci for type 2 diabetes. Science.
316:1336–1341. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wellcome Trust Case Control Consortium:
Genome-wide association study of 14,000 cases of seven common
diseases and 3,000 shared controls. Nature. 447:661–678. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zeggini E, Scott LJ, Saxena R, Voight BF,
Marchini JL, Hu T, de Bakker PI, Abecasis GR, Almgren P, Andersen
G, et al: Meta-analysis of genome-wide association data and
large-scale replication identifies additional susceptibility loci
for type 2 diabetes. Nat Genet. 40:638–645. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gudmundsson J, Sulem P, Steinthorsdottir
V, Bergthorsson JT, Thorleifsson G, Manolescu A, Rafnar T,
Gudbjartsson D, Agnarsson BA, Baker A, et al: Two variants on
chromosome 17 confer prostate cancer risk and the one in TCF2
protects against type 2 diabetes. Nat Genet. 39:977–983. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Saxena R, Elbers CC, Guo Y, Peter I, Gaunt
TR, Mega JL, Lanktree MB, Tare A, Castillo BA, Li YR, et al:
Large-scale gene-centric meta-analysis across 39 studies identifies
type 2 diabetes loci. Am J Hum Genet. 90:410–425. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Baum AE, Akula N, Cabanero M, Cardona I,
Corona W, Klemens B, Schulze TG, Cichon S, Rietschel M, Nöthen MM,
et al: A genome-wide association study implicates diacylglycerol
kinase eta (DGKH) and several other genes in the etiology of
bipolar disorder. Mol Psychiatry. 13:197–207. 2008. View Article : Google Scholar :
|
|
12
|
Galvan A, Falvella FS, Frullanti E,
Spinola M, Incarbone M, Nosotti M, Santambrogio L, Conti B,
Pastorino U, Gonzalez-Neira A and Dragani TA: Genome-wide
association study in discordant sibships identifies multiple
inherited susceptibility alleles linked to lung cancer.
Carcinogenesis. 31:462–465. 2010. View Article : Google Scholar
|
|
13
|
Forstbauer LM, Brockschmidt FF, Moskvina
V, Herold C, Redler S, Herzog A, Hillmer AM, Meesters C, Heilmann
S, Albert F, et al: Genome-wide pooling approach identifies SPATA5
as a new susceptibility locus for alopecia areata. Eur J Hum Genet.
20:326–332. 2012. View Article : Google Scholar :
|
|
14
|
Wong LP, Ong RT, Poh WT, Liu X, Chen P, Li
R, Lam KK, Pillai NE, Sim KS, Xu H, et al: Deep whole-genome
sequencing of 100 southeast asian malays. Am J Hum Genet. 92:52–66.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kong A, Steinthorsdottir V, Masson G,
Thorleifsson G, Sulem P, Besenbacher S, Jonasdottir A, Sigurdsson
A, Kristinsson KT, Jonasdottir A, et al: Parental origin of
sequence variants associated with complex diseases. Nature.
462:868–874. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Voight BF, Scott LJ, Steinthorsdottir V,
Morris AP, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS,
Thorleifsson G, et al: Twelve type 2 diabetes susceptibility loci
identified through large-scale association analysis. Nat Genet.
42:579–589. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dupuis J, Langenberg C, Prokopenko I,
Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji
N, Gloyn AL, et al: New genetic loci implicated in fasting glucose
homeostasis and their impact on type 2 diabetes risk. Nat Genet.
42:105–116. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qi L, Cornelis MC, Kraft P, Stanya KJ,
Linda Kao WH, Pankow JS, Dupuis J, Florez JC, Fox CS, Paré G, et
al: Genetic variants at 2q24 are associated with susceptibility to
type 2 diabetes. Hum Mol Genet. 19:2706–2715. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yamauchi T, Hara K, Maeda S, Yasuda K,
Takahashi A, Horikoshi M, Nakamura M, Fujita H, Grarup N, Cauchi S,
et al: A genome-wide association study in the Japanese population
identifies susceptibility loci for type 2 diabetes at UBE2E2 and
C2CD4A-C2CD4B. Nat Genet. 42:864–868. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shu XO, Long J, Cai Q, Qi L, Xiang YB, Cho
YS, Tai ES, Li X, Lin X, Chow WH, et al: Identification of new
genetic risk variants for type 2 diabetes. PLoS Genet.
6:e10011272010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kooner JS, Saleheen D, Sim X, Sehmi J,
Zhang W, Frossard P, Been LF, Chia KS, Dimas AS, Hassanali N, et
al: Genome-wide association study in individuals of South Asian
ancestry identifies six new type 2 diabetes susceptibility loci.
Nat Genet. 43:984–989. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cho YS, Chen CH, Hu C, Long J, Ong RT, Sim
X, Takeuchi F, Wu Y, Go MJ, Yamauchi T, et al: Meta-analysis of
genome-wide association studies identifies eight new loci for type
2 diabetes in east Asians. Nat Genet. 44:67–72. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Alberti KG and Zimmet PZ: Definition,
diagnosis and classification of diabetes mellitus and its
complications. Part 1: diagnosis and classification of diabetes
mellitus provisional report of a WHO consultation. Diabet Med.
15:539–553. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mardis ER: Next-generation DNA sequencing
methods. Annu Rev Genomics Hum Genet. 9:387–402. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang J, Wang W, Li R, Li Y, Tian G,
Goodman L, Fan W, Zhang J, Li J, Zhang J, et al: The diploid genome
sequence of an Asian individual. Nature. 456:60–65. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wheeler DA, Srinivasan M, Egholm M, Shen
Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, et al:
The complete genome of an individual by massively parallel DNA
sequencing. Nature. 452:872–876. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li R, Li Y, Fang X, Yang H and Wang J,
Kristiansen K and Wang J: SNP detection for massively parallel
whole-genome resequencing. Genome Res. 19:1124–1132. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li H, Handsaker B, Wysoker A, Fennell T,
Ruan J, Homer N, Marth G, Abecasis G and Durbin R: 1000 Genome
project data processing subgroup: The sequence alignment/Map format
and SAMtools. Bioinformatics. 25:2078–2079. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
McKenna A, Hanna M, Banks E, Sivachenko A,
Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly
M and DePristo MA: The Genome Analysis Toolkit: A MapReduce
framework for analyzing next-generation DNA sequencing data. Genome
Res. 20:1297–1303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
DePristo MA, Banks E, Poplin R, Garimella
KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA and
Hanna M: A framework for variation discovery and genotyping using
next-generation DNA sequencing data. Nat Genet. 43:491–498. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Abyzov A, Urban AE, Snyder M and Gerstein
M: CNVnator: An approach to discover, genotype, and characterize
typical and atypical CNVs from family and population genome
sequencing. Genome Res. 21:974–984. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mills RE, Walter K, Stewart C, Handsaker
RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, et al:
Mapping copy number variation by population-scale genome
sequencing. Nature. 470:59–65. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Koboldt DC, Chen K, Wylie T, Larson DE,
McLellan MD, Mardis ER, Weinstock GM, Wilson RK and Ding L:
VarScan: Variant detection in massively parallel sequencing of
individual and pooled samples. Bioinformatics. 25:2283–2285. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen K, Wallis JW, McLellan MD, Larson DE,
Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, et
al: BreakDancer: An algorithm for high-resolution mapping of
genomic structural variation. Nat Methods. 6:677–681. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chiang DY, Getz G, Jaffe DB, O'Kelly MJ,
Zhao X, Carter SL, Russ C, Nusbaum C, Meyerson M and Lander ES:
High-resolution mapping of copy-number alterations with massively
parallel sequencing. Nat Methods. 6:99–103. 2009. View Article : Google Scholar :
|
|
37
|
Wang K, Li M and Hakonarson H: ANNOVAR:
Functional annotation of genetic variants from high-throughput
sequencing data. Nucleic Acids Res. 38:e1642010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kumar P, Henikoff S and Ng PC: Predicting
the effects of coding non-synonymous variants on protein function
using the SIFT algorithm. Nat Protoc. 4:1073–1081. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Adzhubei IA, Schmidt S, Peshkin L,
Ramensky VE, Gerasimova A, Bork P, Kondrashov AS and Sunyaev SR: A
method and server for predicting damaging missense mutations. Nat
Methods. 7:248–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yasuda K, Miyake K, Horikawa Y, Hara K,
Osawa H, Furuta H, Hirota Y, Mori H, Jonsson A, Sato Y, et al:
Variants in KCNQ1 are associated with susceptibility to type 2
diabetes mellitus. Nat Genet. 40:1092–1097. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Vélez JI, Chandrasekharappa SC, Henao E,
Martinez AF, Harper U, Jones M, Solomon BD, Lopez L, Garcia G,
Aguirre-Acevedo DC, et al: Pooling/bootstrap-based GWAS (pbGWAS)
identifies new loci modifying the age of onset in PSEN1 p.Glu280Ala
Alzheimer's disease. Mol Psychiatry. 18:568–575. 2013. View Article : Google Scholar :
|
|
42
|
Clayton DG, Walker NM, Smyth DJ, Pask R,
Cooper JD, Maier LM, Smink LJ, Lam AC, Ovington NR, Stevens HE, et
al: Population structure, differential bias and genomic control in
a large-scale, case-control association study. Nat Genet.
37:1243–1246. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zondervan KT and Cardon LR: The complex
interplay among factors that influence allelic association. Nat Rev
Genet. 5:89–100. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Neel JV: The Genetics of Diabetes
Mellitus. Creutzfeldt W, Köbberling J and Neel JV: Springer;
Berlin: 1976, pp. 1–11. View Article : Google Scholar
|