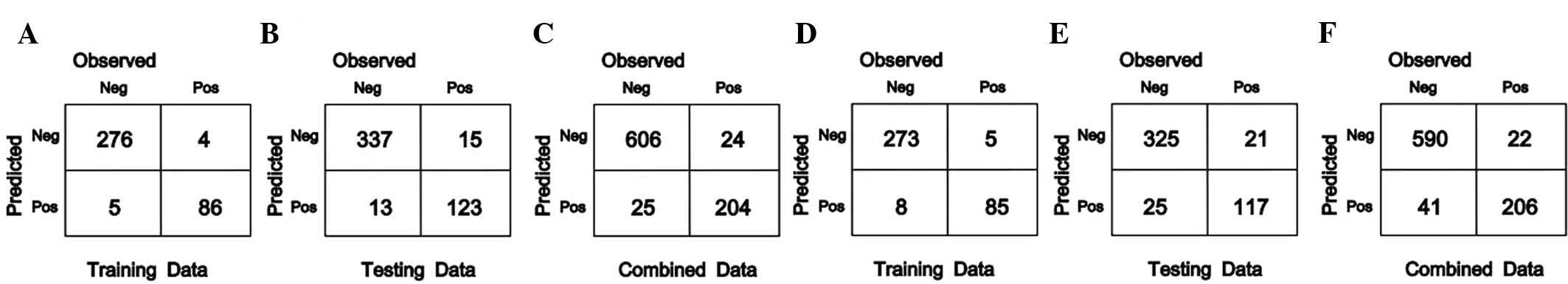
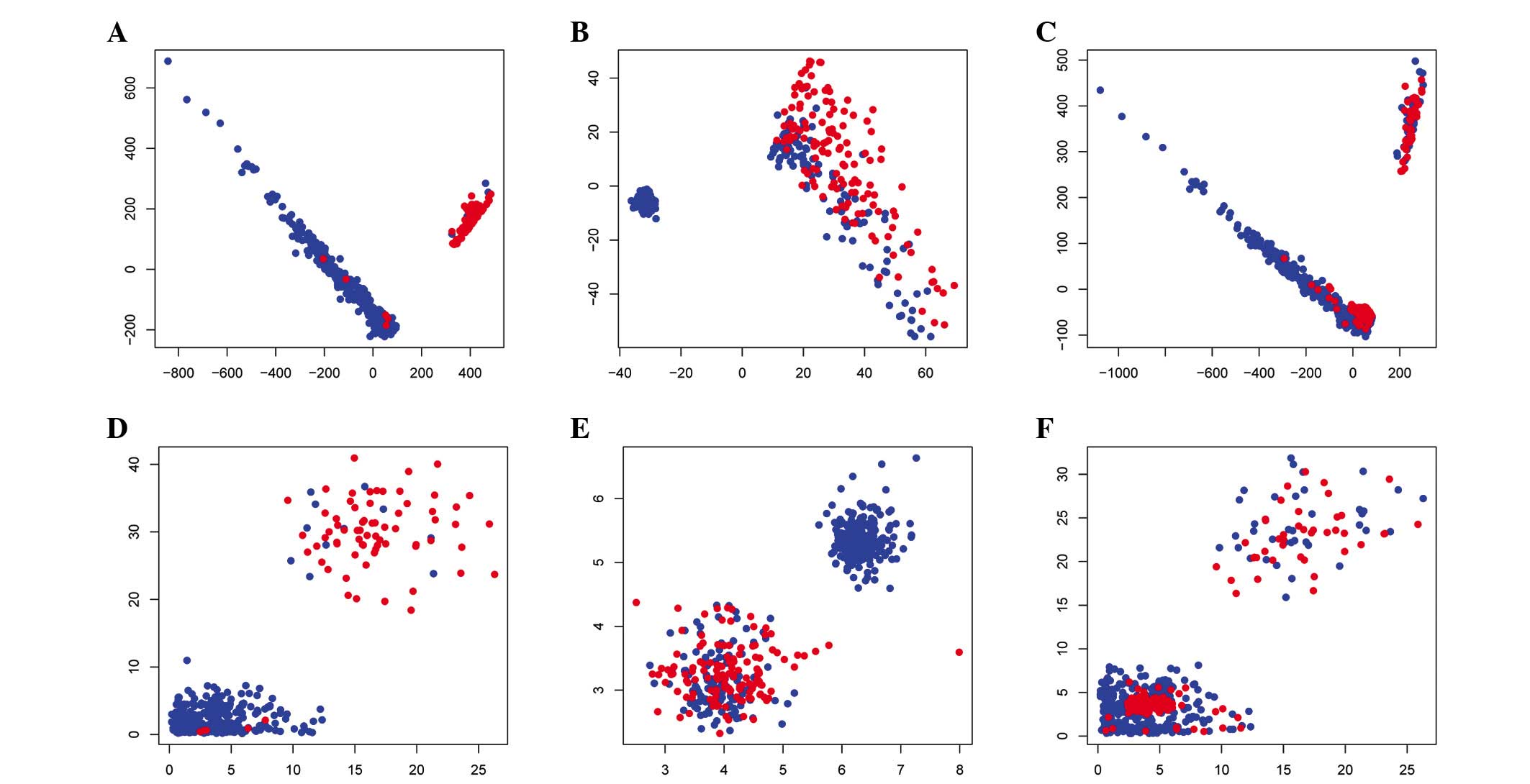
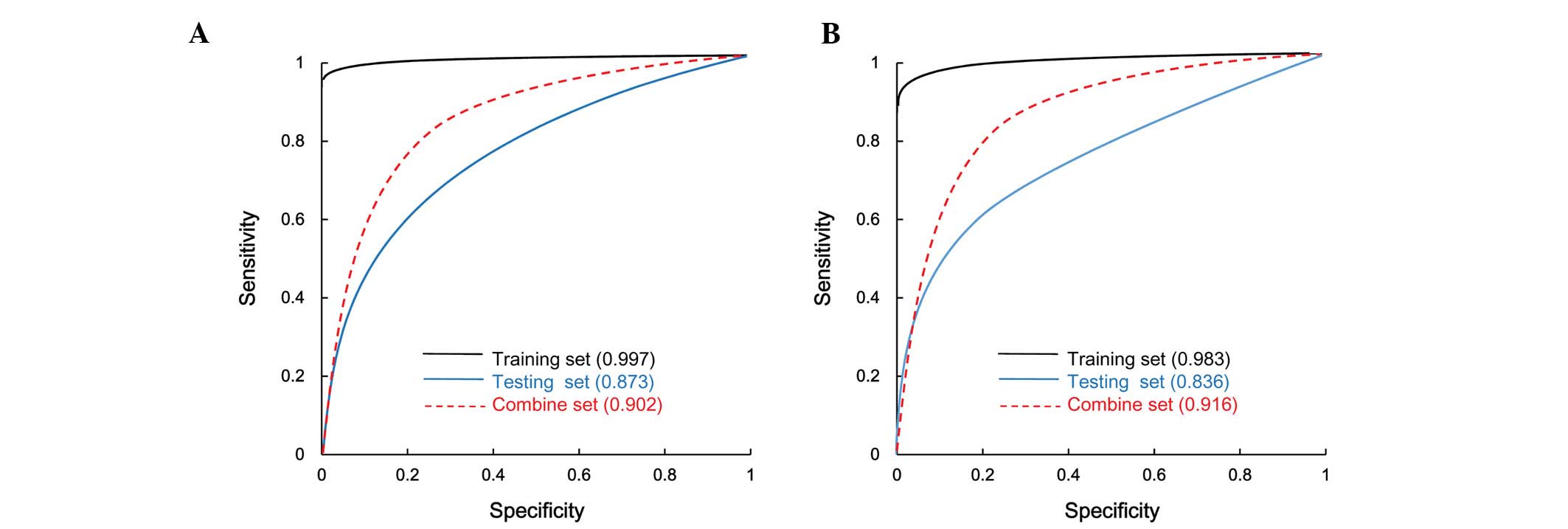
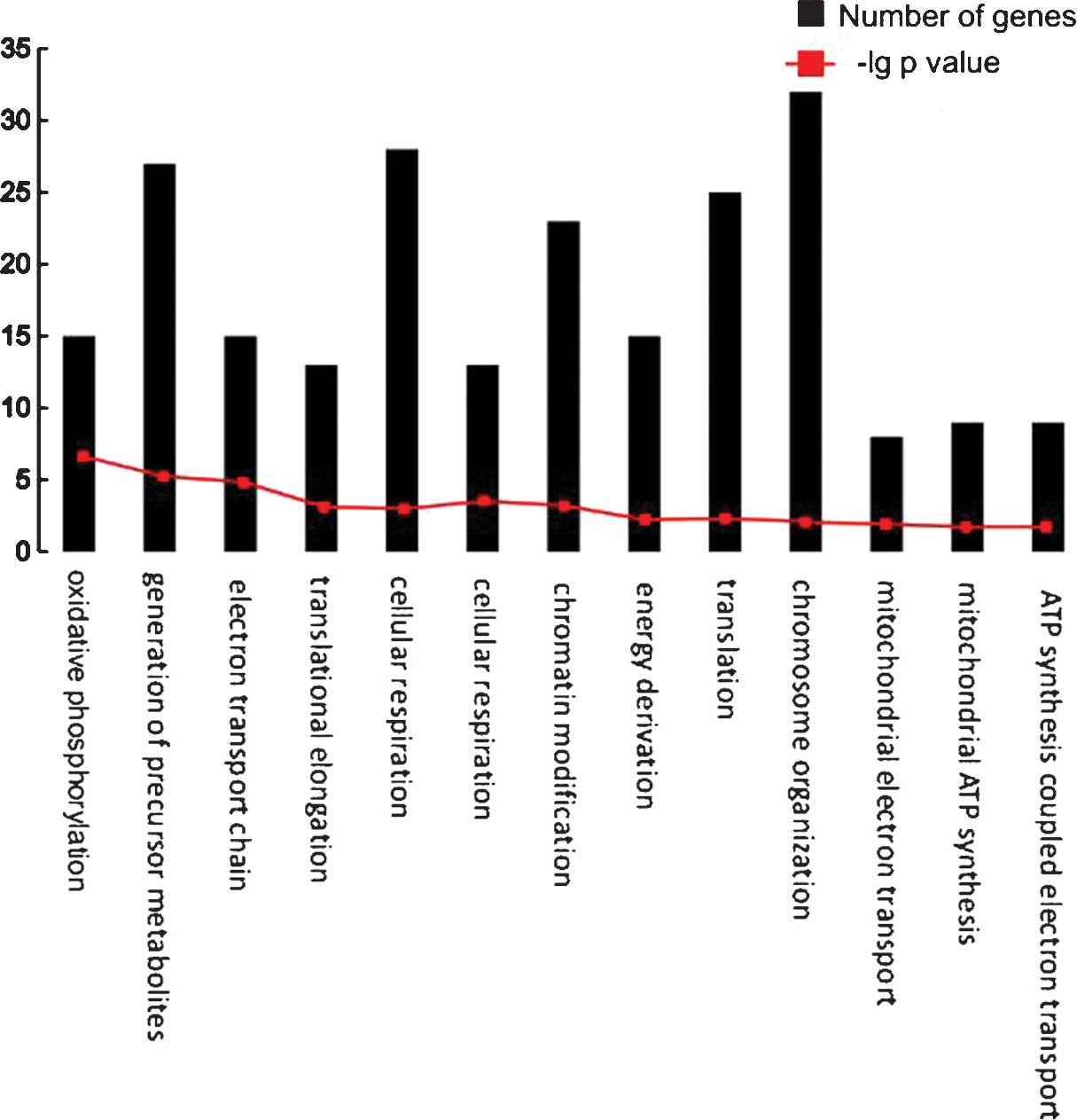
|
1
|
Gilliland DG and Griffin JD: The roles of
FLT3 in hematopoiesis and leukemia. Blood. 100:1532–1542. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li Y, Li H, Wang MN, Lu D, Bassi R, Wu Y,
Zhang H, Balderes P, Ludwig DL, Pytowski B, et al: Suppression of
leukemia expressing wild-type or ITD-mutant FLT3 receptor by a
fully human anti-FLT3 neutralizing antibody. Blood. 104:1137–1144.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen P, Levis M, Brown P, Kim KT, Allebach
J and Small D: FLT3/ITD mutation signaling includes suppression of
SHP-1. J Biol Chem. 280:5361–5369. 2005. View Article : Google Scholar
|
|
4
|
Rausei-Mills V, Chang KL, Gaal KK, Weiss
LM and Huang Q: Aberrant expression of CD7 in myeloblasts is highly
associated with de novo acute myeloid leukemias with FLT3/ITD
mutation. Am J Clin Pathol. 129:624–629. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Okamoto M, Hayakawa F, Miyata Y, Watamoto
K, Emi N, Abe A, Kiyoi H, Towatari M and Naoe T: Lyn is an
important component of the signal transduction pathway specific to
FLT3/ITD and can be a therapeutic target in the treatment of AML
with FLT3/ITD. Leukemia. 21:403–410. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fathi AT, Arowojolu O, Swinnen I, Sato T,
Rajkhowa T, Small D, Marmsater F, Robinson JE, Gross SD, Martinson
M, et al: A potential therapeutic target for FLT3-ITD AML: PIM1
kinase. Leuk Res. 36:224–231. 2012. View Article : Google Scholar :
|
|
7
|
Dalal BI, Mansoor S, Manna M, Pi S, Sauro
GD and Hogge DE: Detection of CD34, TdT, CD56, CD2, CD4 and CD14 by
flow cytometry is associated with NPM1 and FLT3 mutation status in
cytogenetically normal acute myeloid leukemia. Clin Lymphoma
Myeloma Leuk. 12:274–279. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stoesser G, Baker W, van den Broek A,
Camon E, Garcia-Pastor M, Kanz C, Kulikova T, Leinonen R, Lin Q,
Lombard V, et al: The EMBL nucleotide sequence database. Nucleic
Acids Res. 30:21–26. 2002. View Article : Google Scholar :
|
|
9
|
Liu WM, Mei R, Di X, Ryder TB, Hubbell E,
Dee S, Webster TA, Harrington CA, Ho MH, Baid J and Smeekens SP:
Analysis of high density expression microarrays with signed-rank
call algorithms. Bioinformatics. 18:1593–1599. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Smyth GK and Speed T: Normalization of
cDNA microarray data. Methods. 31:265–273. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang S: A comprehensive evaluation of
SAM, the SAM R-package and a simple modification to improve its
performance. Bmc Bioinformatics. 8:2302007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Benjamini Y and Hochberg Y: Controlling
the false discovery Rate: A practical and powerful approach to
multiple testing. J R Stat Soc B. 57:289–300. 1995.
|
|
13
|
Benjamini and Yoav: Discovering the false
discovery rate. Journal of the Royal Statistical Society.
72:405–416. 2010. View Article : Google Scholar
|
|
14
|
Duval B and Hao JK: Advances in
metaheuristics for gene selection and classification of microarray
data. Brief Bioinform. 1:127–141. 2010. View Article : Google Scholar
|
|
15
|
Díaz-Uriarte R and Alvarez de Andrés S:
Gene selection and classification of microarray data using random
forest. Bmc Bioinformatics. 7:32006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Croshaw R, Shapiro-Wright H, Svensson E,
Erb K and Julian T: Accuracy of clinical examination, digital
mammogram, ultrasound and MRI in determining postneoadjuvant
pathologic tumor response in operable breast cancer patients. Ann
Surg Oncol. 18:3160–3163. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fawcett T: An introduction to ROC
analysis. Pattern Recogn Lett. 27:861–874. 2006. View Article : Google Scholar
|
|
18
|
Huang DW, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
19
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar
|
|
20
|
Basak NP and Banerjee S: Mitochondrial
dependency in progression of acute myeloid leukemia. Mitochondrion.
21:41–48. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schimmer AD and Skrtić M: Therapeutic
potential of mitochondrial translation inhibition for treatment of
acute myeloid leukemia. Expert Rev Hematol. 5:117–119. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yamaguchi S, Iwanaga E, Tokunaga K, Nanri
T, Shimomura T, Suzushima H, Mitsuya H and Asou N: IDH1 and IDH2
mutations confer an adverse effect in patients with acute myeloid
leukemia lacking the NPM1 mutation. Eur J Haematol. 92:471–477.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shi J, Wang E, Zuber J, Rappaport A,
Taylor M, Johns C, Lowe SW and Vakoc CR: The Polycomb complex PRC2
supports aberrant self-renewal in a mouse model of MLL-AF9;Nras
(G12D) acute myeloid leukemia. Oncogene. 32:930–938. 2013.
View Article : Google Scholar
|
|
24
|
Tiacci E, Grossmann V, Martelli MP,
Kohlmann A, Haferlach T and Falini B: The corepressors BCOR and
BCORL1: Two novel players in acute myeloid leukemia. Haematologica.
97:3–5. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zagaria A, Anelli L, Casieri P, Coccaro N,
Tota G, Minervini CF, Minervini A, Impera L, Brunetti C, Orsini P,
et al: BCOR gene dysregulation due to chromosomal translocation in
acute myeloid leukemia: A new mechanism based on long non-coding
RNA dislocation? Leuk Lymphoma. 55:2199–2201. 2014. View Article : Google Scholar
|
|
26
|
Osaki H, Walf-Vorderwülbecke V, Mangolini
M, et al: The AAA+ ATPase RUVBL2 is a critical mediator of MLL-AF9
oncogenesis. Leukemia. 27:1461–1468. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sroczynska P, Cruickshank VA, Bukowski JP,
Miyagi S, Bagger FO, Walfridsson J, Schuster MB, Porse B and Helin
K: shRNA screening identifies JMJD1C as being required for leukemia
maintenance. Blood. 123:1870–1882. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Martinsubero JI, Harder L, Gesk S, Schoch
R, Novo FJ, Grote W, Calasanz MJ, Schlegelberger B and Siebert R:
Amplification of ERBB2, RARA, and TOP2A genes in a myelodysplastic
syndrome transforming to acute myeloid leukemia. Cancer Genet
Cytogenet. 127:174–176. 2001. View Article : Google Scholar
|
|
29
|
Aggerholm A and Hokland P: DAP-kinase CpG
island methylation in acute myeloid leukemia: Methodology versus
biology? Blood. 95:2997–2998. 2000.PubMed/NCBI
|
|
30
|
Wang L, Luo J, Nian Q, Xiao Q, Yang Z and
Liu L: Ribosomal protein S14 silencing inhibits growth of acute
myeloid leukemia transformed from myelodysplastic syndromes via
activating p53. Hematology. 19:225–231. 2014. View Article : Google Scholar
|
|
31
|
Hariri F, Arguello M, Volpon L,
Culjkovic-Kraljacic B, Nielsen TH, Hiscott J, Mann KK and Borden
KL: The eukaryotic translation initiation factor eIF4E is a direct
transcriptional target of NF-κB and is aberrantly regulated in
acute myeloid leukemia. Leukemia. 10:2047–2055. 2013. View Article : Google Scholar
|
|
32
|
Assouline S, Culjkovic B, Cocolakis E,
Rousseau C, Beslu N, Amri A, Caplan S, Leber B, Roy DC, Miller WH
Jr and Borden KL: Molecular targeting of the oncogene eIF4E in
acute myeloid leukemia (AML): A proof-of-principle clinical trial
with ribavirin. Blood. 2:257–260. 2009. View Article : Google Scholar
|