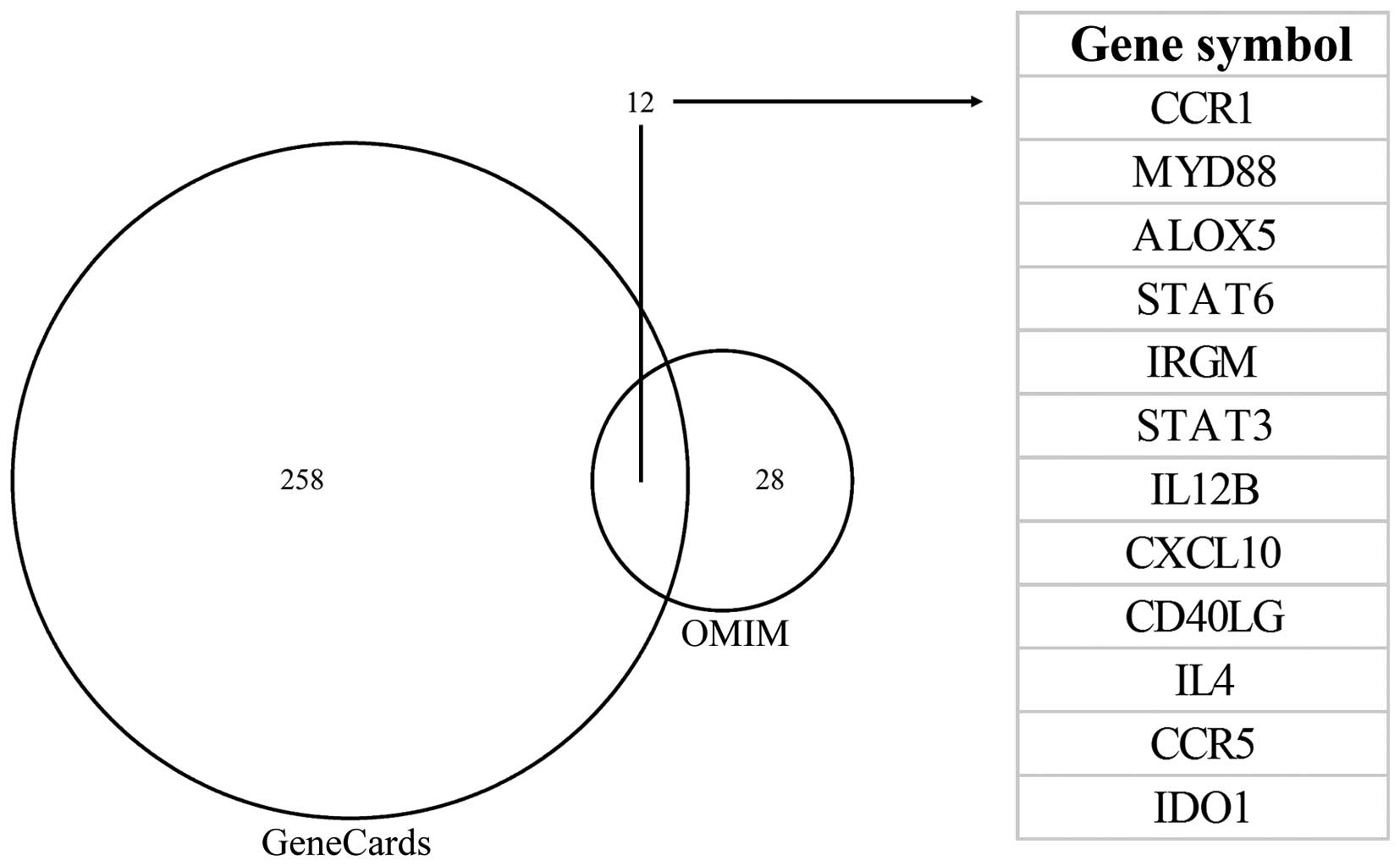
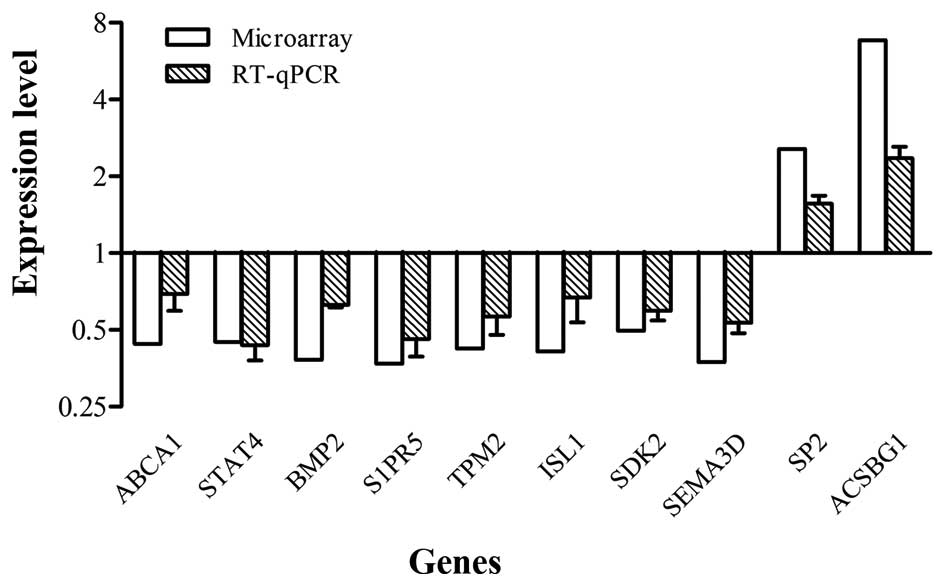
|
1
|
John B, Ricart B, Wojno ED Tait, Harris
TH, Randall LM, Christian DA, Gregg B, De Almeida DM, Weninger W,
Hammer DA and Hunter CA: Analysis of behavior and trafficking of
dendritic cells within the brain during toxoplasmic encephalitis.
PLoS Pathog. 7:e10022462011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Montoya JG and Remington JS: Management of
Toxoplasma gondii infection during pregnancy. Clin Infect Dis.
47:554–566. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tanaka S, Nishimura M, Ihara F, Yamagishi
J, Suzuki Y and Nishikawa Y: Transcriptome analysis of mouse brain
infected with Toxoplasma gondii. Infect Immun. 81:3609–3619. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Boothroyd JC: Have it your way: How
polymorphic, injected kinases and pseudokinases enable Toxoplasma
to subvert host defenses. PLoS Pathog. 9:e10032962013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dubremetz JF: Rhoptries are major players
in Toxoplasma gondii invasion and host cell interaction. Cellular
Microbiol. 9:841–848. 2007. View Article : Google Scholar
|
|
6
|
Mueller C, Klages N, Jacot D, Santos JM,
Cabrera A, Gilberger TW, Dubremetz JF and Soldati-Favre D: The
Toxoplasma protein ARO mediates the apical positioning of rhoptry
organelles, a prerequisite for host cell invasion. Cell Host
Microbe. 13:289–301. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Taylor S, Barragan A, Su C, Fux B,
Fentress SJ, Tang K, Beatty WL, Hajj HE, Jerome M, Behnke MS, et
al: A secreted serine-threonine kinase determines virulence in the
eukaryotic pathogen Toxoplasma gondii. Science. 314:1776–1780.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Boothroyd JC and Dubremetz JF: Kiss and
spit: The dual roles of Toxoplasma rhoptries. Nat Rev Microbiol.
6:79–88. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Saeij JP, Coller S, Boyle JP, Jerome ME,
White MW and Boothroyd JC: Toxoplasma co-opts host gene expression
by injection of a polymorphic kinase homologue. Nature.
445:324–327. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jensen KD, Hu K, Whitmarsh RJ, Hassan MA,
Julien L, Lu D, Chen L, Hunter CA and Saeij JP: Toxoplasma gondii
rhoptry 16 kinase promotes host resistance to oral infection and
intestinal inflammation only in the context of the dense granule
protein GRA15. Infect Immun. 81:2156–2167. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mi H, Muruganujan A and Thomas PD: PANTHER
in 2013: Modeling the evolution of gene function, and other gene
attributes, in the context of phylogenetic trees. Nucleic Acids
Res. 41:(Database Issue). D377–D386. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mi H, Muruganujan A, Casagrande JT and
Thomas PD: Large-scale gene function analysis with the PANTHER
classification system. Nature Protoc. 8:1551–1566. 2013. View Article : Google Scholar
|
|
13
|
Chen J, Bardes EE, Aronow BJ and Jegga AG:
ToppGene suite for gene list enrichment analysis and candidate gene
prioritization. Nucleic Acids Res (Web Server Issue). 37:W305–W311.
2009. View Article : Google Scholar
|
|
14
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:(Database Issue). D447–D452. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen J, Xu H, Aronow BJ and Jegga AG:
Improved human disease candidate gene prioritization using mouse
phenotype. BMC Bioinformatics. 8:3922007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dugas JC, Mandemakers W, Rogers M, Ibrahim
A, Daneman R and Barres BA: A novel purification method for CNS
projection neurons leads to the identification of brain vascular
cells as a source of trophic support for corticospinal motor
neurons. J Neurosci. 28:8294–8305. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Oh JW, Olman M and Benveniste EN:
CXCL12-mediated induction of plasminogen activator inhibitor-1
expression in human CXCR4 positive astroglioma cells. Biol Pharm
Bull. 32:573–577. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Carruthers VB and Suzuki Y: Effects of
Toxoplasma gondii infection on the brain. Schizophr Bull.
33:745–751. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mori K, Kanemura Y, Fujikawa H, Nakano A,
Ikemoto H, Ozaki I, Matsumoto T, Tamura K, Yokota M and Arita N:
Brain-specific angiogenesis inhibitor 1 (BAI1) is expressed in
human cerebral neuronal cells. Neurosci Res. 43:69–74. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu D, Li C, Swanson AM, Villalba RM, Guo
J, Zhang Z, Matheny S, Murakami T, Stephenson JR, Daniel S, et al:
BAI1 regulates spatial learning and synaptic plasticity in the
hippocampus. J Clin Invest. 125:14975082015. View Article : Google Scholar
|
|
24
|
Nagai T, Aruga J, Minowa O, Sugimoto T,
Ohno Y, Noda T and Mikoshiba K: Zic2 regulates the kinetics of
neurulation. Proc Natl Acad Sci USA. 97:1618–1623. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tsend-Ayush E, O'Sullivan LA, Grützner FS,
Onnebo SM, Lewis RS, Delbridge ML, Graves JA Marshall and Ward AC:
RBMX gene is essential for brain development in zebrafish. Dev Dyn.
234:682–688. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang T, Zhou J, Gan X, Wang H, Ding X,
Chen L, Wang Y, DU J, Shen J and Yu L: Toxoplasma gondii induce
apoptosis of neural stem cells via endoplasmic reticulum stress
pathway. Parasitology. 141:988–995. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nyoman AD and Lüder CG: Apoptosis-like
cell death pathways in the unicellular parasite Toxoplasma gondii
following treatment with apoptosis inducers and chemotherapeutic
agents: A proof-of-concept study. Apoptosis. 18:664–680. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Iwasa H, Kudo T, Maimaiti S, Ikeda M,
Maruyama J, Nakagawa K and Hata Y: The RASSF6 tumor suppressor
protein regulates apoptosis and the cell cycle via MDM2 protein and
p53 protein. J Biol Chem. 288:30320–30329. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Marcar L, MacLaine NJ, Hupp TR and Meek
DW: Mage-A cancer/testis antigens inhibit p53 function by blocking
its interaction with chromatin. Cancer Res. 70:10362–10370. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cantile M, Procino A, D'armiento M,
Cindolo L and Cillo C: HOX gene network is involved in the
transcriptional regulation of in vivo human adipogenesis. J Cell
Physiol. 194:225–236. 2003. View Article : Google Scholar : PubMed/NCBI
|