Introduction
Vascular smooth muscle cells (VSMCs), as the primary
cellular components of the normal blood vessel wall, are associated
with the integrity of vascular structure, regulation of vascular
function and stability of vascular lesions (1). Notably, VSMCs are considered to be
vital to the initiation and progression of atherosclerotic lesions
(2). In response to trauma and
altered hemodynamics of vascular reconstruction, the phenotypic
modulation of VSMCs from a contractile to a synthetic phenotype is
observed during atherogenesis, which promotes not only the
migratory and proliferative capacity of VSMCs, but also the
synthesis of extracellular matrix proteins (3,4).
Therefore, it is essential to understand the molecular mechanism
underlying the phenotype transition and cell behavior of VSMCs.
Rab proteins are Ras-related small GTPases, which
may regulate exocytic and endocytic membrane trafficking by vesicle
docking and fusion (5,6). As a key member of the Ras GTPase
superfamily, Rab5a has been shown to be associated with not only
the heterozygous fusion of early endosomes and endocytic vesicles,
but also the homomorphous fusion between early endosomes (7). Ravikumar et al (8) suggested that Rab5a can promote
autophagosome formation, indicating that Rab5a is associated with
autophagy. In addition, Rab5a may influence the morphogenesis and
metastasis of various cancer types, including breast cancer,
cervical cancer, ovarian cancer and hepatocellular carcinoma
(9–12). As the pathogenesis of intimal
hyperplasia is somewhat similar to neoplasia, Rab5a may also be
involved in the intimal hyperplasia and arterial restenosis. A
previous study indicated that Rab5a is involved in VSMC
proliferation and migration (13),
while autophagy induced by platelet-derived growth factor (PDGF)
serves an essential role in the conversion of VSMCs from the
contractile to synthetic phenotype in order to prevent cell death
due to oxidative stress (14).
Therefore, the present study hypothesized that autophagy may be
responsible for the proliferation and migration of VSMCs, and that
Rab5a was essential in this process.
In the present study, a human aorta vasuclar smooth
muscle cell line, referred to as T/G HA-VSMCs, was treated with
small interfering (si)RNA against Rab5a and/or PDGF, and the
phenotype transition and cell behaviors, including proliferation,
cell cycle, migration, apoptosis and autophagy, were assessed. The
present study aimed to reveal the effects of Rab5a on autophagy in
VSMCs, and whether the phenotype transition and cell behaviors of
VSMCs are accompanied by autophagy.
Materials and methods
Cell culture and treatment
T/G HA-VSMCs were obtained from American Type
Culture Collection (Rockefeller, MD, USA). The cells were cultured
in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine
serum (FBS; Hyclone, Logan, UT, USA), penicillin (100 U/ml) and
streptomycin (100 mg/ml) at 37°C with 5% CO2. The cells
were transfected with control siRNA (siC), Rab5a siRNA (siR; a pool
of four siRNAs; Dharmacon Research, Lafayette, CO, USA), siC
combined with PDGF (siC + P; 20 ng/ml; R&D Biosystems,
Minneapolis, MN, USA) and siR combined with PDGF (siR + P; 20
ng/ml) prior to experiments. Transfection was performed using
DharmaFECT transfection reagent in serum-free medium (GE Healthcare
Life Sciences, Chalfont, UK) following manufacturer's protocol.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Following treatment with siRNA and/or PDGF for 24 h,
the total RNA from cells was obtained using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. The RNA (25 nM) was subsequently reverse
transcribed using the RevertAid First-Strand cDNA Synthesis kit
(Fermentas, Vilnius, Lithuania). PCR amplification was performed
with the SYBR Green Premix Ex Taq kit (Takara Bio., Inc., Dalian,
China). Primer sequences are listed in Table I. The PCR program included
denaturation at 95°C for 10 sec, amplification with 40 cycles at
95°C for 5 sec and 60°C for 31 sec, and a final 2 min extension at
72°C. Finally, the 2−ΔΔCq method (15) was used to calculate the expression
levels of genes, normalized against the levels of GAPDH.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Gene | Primer sequence
(5′→3′) |
|---|
| Rab5a |
|
|
Forward |
CAGTTCAAACTAGTACTTCTGG |
|
Reverse |
GCTAGGCTATGGTATCGTTCTTG |
| α-SMA |
|
|
Forward |
GCGTGGCTATTCCTTCGTTA |
|
Reverse |
ATGAAGGATGGCTGGAACAG |
| Calponin |
|
|
Forward |
AGCTAAGAGAAGGGCGGAAC |
|
Reverse |
CATCTGCAGGCTGACATTGA |
| Vimentin |
|
|
Forward |
AAAACACCCTGCAATCTTTCAGA |
|
Reverse |
CACTTTGCGTTCAAGGTCAAGAC |
| Osteopontin |
|
|
Forward |
GGACAGCCGTGGGAAGG |
|
Reverse |
TCAATCACATCGGAATGCTCA |
| GAPDH |
|
|
Forward |
GCACCGTCAAGGCTGAGAAC |
|
Reverse |
TGGTGAAGACGCCAGTGGA |
Western blotting analysis
Following treatment for 48 h with siRNA and/or PDGF,
the cells were lysed in radioimmunoprecipitation buffer (Beyotime
Institute of Biotechnology, Inc., Jiangsu, China) containing 1 mM
phenylmethanesulfonyl fluoride. The supernatants were acquired by
centrifugation at 13,440 × g for 15 min at 4°C. The proteins
(30 µg/lane) were separated and subsequently transferred onto a
polyvinylidene difluoride membrane (EMD Millipore, Billerica, MA,
USA). The membranes were blocked with 5% non-fat milk at room
temperature for 2 h. Following blocking, the membranes were
incubated with specific primary antibodies overnight at 4°C and
then incubated with horserdish peroxidase-conjugated secondary
antibodies (cat. nos. A0181; A0216; A0208; 1:1,000; Beyotime
Institute of Biotechnology, Inc.) at room temperature for 2 h. The
antibodies used were as follows: anti-Rab5a (cat. no. 2143;
1:1,000), anti-survivin (cat. no. 2803), anti-caspase-3 (cat. no.
9662; 1:1,000), anti-Beclin1 (cat. no. 3738; 1:1,000), anti-Light
chain (LC)3-I/II (cat. no. 12741; 1:1,000), anti-phosphorylated
(p-) (cat. no. 4060; 1:1,000) and total (t-)protein kinase B (Akt)
(cat. no. 4691; 1:1,000), anti-p-mammalian target of rapamycin
(mTOR; cat. no 5536; 1:1,000) and anti-t-mTOR (cat. no. 2972;
1:1,000), and p-extracellular signal-regulated kinase (ERK cat. no.
4370; 1:1,000) and t-ERK (cat. no. 4695; 1:1,000; all from Cell
Signaling Technology, Inc., Danvers, MA, USA. Anti-α-smooth muscle
actin (SMA; cat. no. SAB2500963; 0.5 µg/ml) and anti-calponin (cat.
no. C2687; 1:10,000) were obtained from Sigma-Aldrich (St. Louis,
MO, USA). Anti-osteopontin (cat. no. sc-10591; 1:200) was purchased
from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA), and
anti-GAPDH (cat. no. AF0006; 1:1,000) was purchased from Beyotime
Institute of Biotechnology, Inc. Following antibody incubations, an
enhanced chemiluminescence assay (PerkinElmer, Waltham, MA, USA)
was used to detect the proteins. GAPDH was used as a loading
control.
Cell proliferation assay
The proliferation of VSMCs was detected using a
sulforhodamine B (SRB) assay. A total of 3.0×103 T/G
HA-VSMCs were seeded into 96-well plates. The next day, the cells
were assigned into the following four groups: i) siC group, cells
treated with control siRNA; ii) siC + P group, cells treated with
control siRNA and PDGF; iii) siR group, cell treated with Rab5a
siRNA; and iv) siR+P group, cells treated with Rab5a siRNA and
PDGF. Following treatment for 24, 48 and 72 h, the cells were fixed
in 30% (w/v) trichloroacetic acid and were subsequently stained
with 0.4% SRB (Sigma-Aldrich) diluted with 1% acetic acid for 30
min at 25°C. The excess SRB was rinsed off using 1% (v/v) acetic
acid and the proteins were dissolved in 10 mM Tris solution.
Finally, a microplate reader (iMark; Bio-Rad, Hercules, CA, USA)
was used to measure the value of SRB at a wavelength of 570 nm.
Migration analysis
The migration of T/G HA-VSMCs was detected using a
Transwell system with 24-well inserts and 8 µm pores (Corning
Costar, Lowell, MA, USA). Matrigel (30 µl; BD Biosciences, San
Jose, CA, USA) was used to coat the inserts and 600 µl complete
medium was added into the lower chamber. Following treatment for 48
h, trypsin was used to detach the cells and the cells were
resuspended in fresh medium (0.2 ml) without FBS at a concentration
of 1×104 cells. These cell suspensions were seeded into
the upper chamber. Following incubation for 24 h, the cells in the
upper chamber were removed with cotton swabs, while non-migrating
cells were gently removed and rinsed out with phosphate-buffered
saline (PBS) three times. Cells in the lower chamber were fixed
with methanol at room temperature for 30 min and were subsequently
stained using hematoxylin at room temperature for 5 min. Migrating
cells were counted in six randomly selected representative fields
of each insert by light microscopy (BX51; Olympus, Tokyo,
Japan).
Cell cycle and apoptosis analysis
Flow cytometry was performed to detect the cell
cycle and apoptosis in each group. Following treatment for 48 h,
the cells were trypsinized, collected and fixed with pre-cooled 70%
ethanol overnight at 4°C. The cells were subsequently centrifuged
at 1,120 × g for 10 min at 4°C and the ethanol was washed
off with PBS. The cells were trypsinized for 30 min at 37°C.
Propidium iodide (PI; 50 µg/ml; Sigma-Aldrich) was used to stain
the cells for 30 min at 25°C for cell cycle analysis.
Cell apoptosis analysis was performed using a
Vybrant® Apoptosis assay kit (Invitrogen; Thermo Fisher
Scientific, Inc.). Following treatment for 48 h, the cells were
co-stained with annexin V and PI, according to the manufacturer's
protocol. The results were detected using flow cytometry (FACS
Calibur; BD Biosciences, Franklin Lakes, NJ, USA). The data
regarding cell cycle and apoptosis were acquired using CellQuest
7.0 software (Beckman Coulter, Brea, CA, USA) and subsequently
analyzed using ModFit3.0 software (Verity Software House, Maine,
ME, USA).
Acridine orange staining assay
To observe the characteristics of autophagic cell
death, acidic vesicle organelles (AVOs) were stained with acridine
orange (Sigma-Aldrich). Following treatment for 48 h, the cells in
different groups were stained with acridine orange (1 µg/ml) for 15
min at 37°C in the dark. The cells were subsequently rinsed with
PBS, collected with trypsin and then resuspended in PBS,
supplemented with 1% FBS. Finally, the fluorescence emission was
detected using CellQuest 7.0 software.
Detection of autophagy with LC3
localization
To further reveal the autophagy of cells in each
group, the cells were co-transfected with pEGFP-LC3 (National
Institute for Basic Biology, Okazaki, Japan) using
Lipofectamine2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
Following transfection for 48 h, the cells were fixed in 95%
ethanol in the dark and fluorescence was detected by fluorescent
microscopy (BX51; Olympus).
Transmission electron microscopy
(TEM)
TEM was performed to observe the formation of
autophagosomes. Following treatment for 48 h, the cells in each
group were fixed in 3% glutaraldehyde at 25°C for 2 h. The cells
were subsequently fixed in 1% osmium tetroxide, and were sectioned
and embedded in LX112 plastic. Finally, uranyl acetate combined
with lead citrate was used to stain the ultrathin sections, and
electron micrographs were obtained by TEM (Philips Medical Systems,
BG Eindhoven, The Netherlands).
Statistical analysis
Statistical analysis was performed using SPSS
version 15.0 software (SPSS Inc., Chicago, IL, USA). All data were
expressed as the mean ± standard deviation. Comparisons among
different groups were analyzed by one-way analysis of variance,
while comparisons between two groups were analyzed by Student's
t-test. P≤0.05 was considered to indicate a statistically
significantly difference.
Results
Effect of Rab5a siRNA transfection and
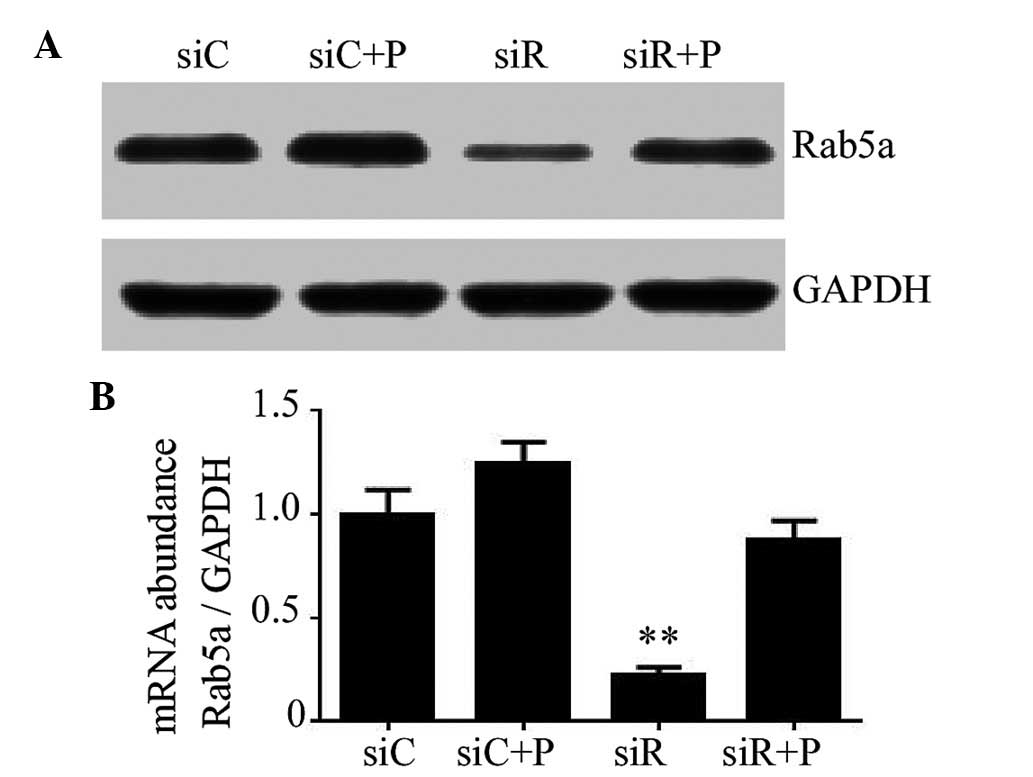
PDGF on the expression of Rab5a in T/G HA-VSMCs
The knockdown efficiency of Rab5a siRNA transfection
and the effects of PDGF on T/G HA-VSMCs were determined using
RT-qPCR and western blotting. As a result, transfection with siRNA
against Rab5a led to a significant decrease in the mRNA and protein
expression levels of Rab5a (P<0.01); however, the expression of
Rab5a in the siR + P group was relatively close to the siC group
(Fig. 1). These data revealed that
Rab5a was effectively downregulated by Rab5a siRNA transfection and
the reduced expression trend of Rab5a can be rescued by the
intervention of PDGF in T/G HA-VSMCs.
Effect of Rab5a siRNA transfection and
PDGF on the phenotype transition of T/G HA-VSMCs
The expression of molecular markers of the T/G
HA-VSMC phenotype, including α-SMA, calponin, osteopontin and
vimentin, were detected to evaluate the phenotypic transition of
T/G HA-VSMCs in different groups. As a result, the mRNA expression
levels of α-SMA and calponin were significantly increased, while
osteopontin and vimentin were significantly reduced in the siR
group compared with the siC group (P<0.01). However, the
expression of these proteins was restored to normal levels by the
intervention of PDGF, which was relatively close to siC (Fig. 2A). Similarly, the changes in the
protein expression levels of α-SMA, calponin and osteopontin were
consistent with that of the mRNA level (Fig. 2B). These results indicated that
Rab5a may promote the phenotypic transition of T/G HA-VSMCs.
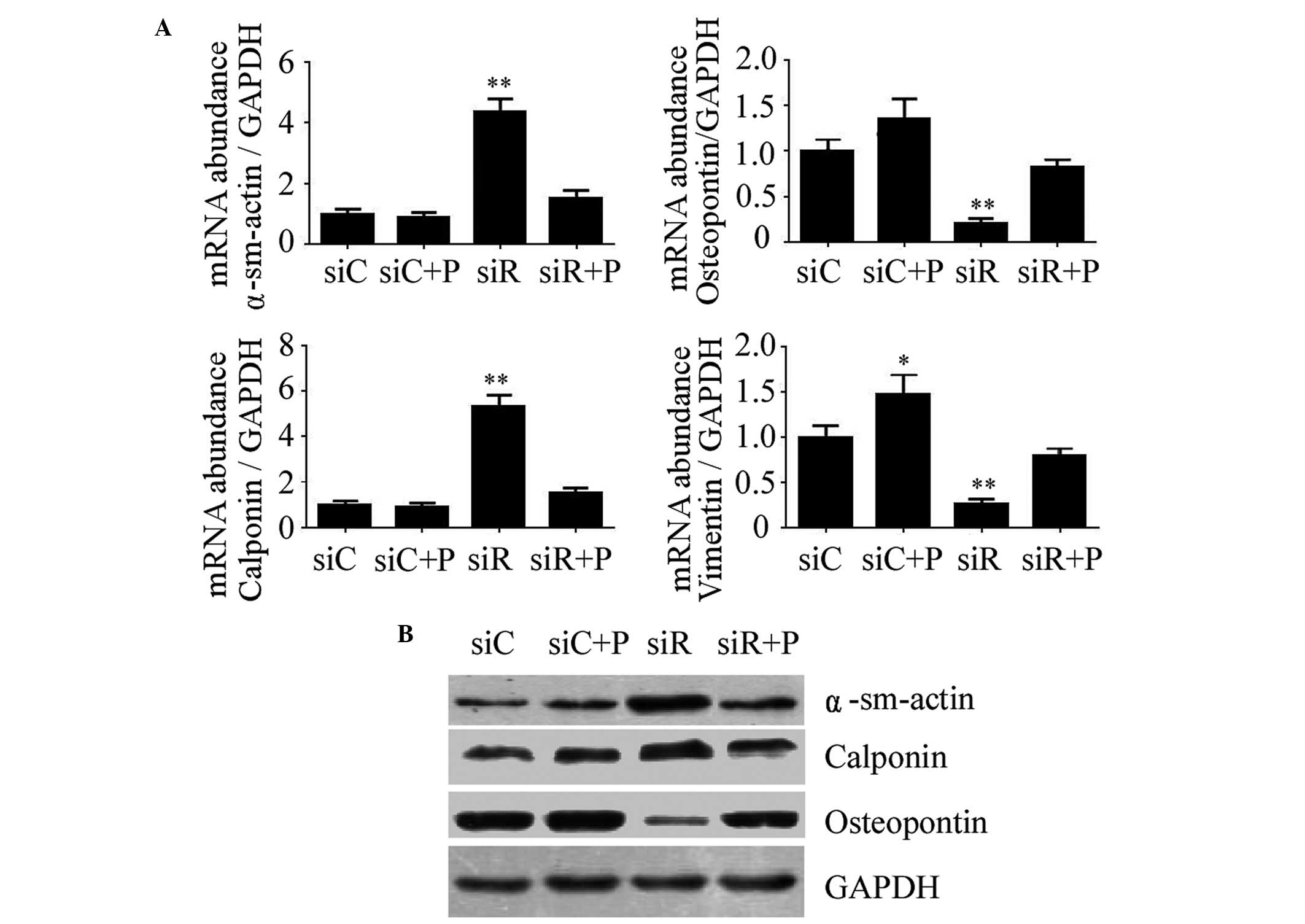 | Figure 2.Expression of molecular markers of the
VSMC phenotype following various treatments. (A) The mRNA
expression levels of α-sm-actin, Calponin, Osteopontin and Vimentin
were determined by reverse transcription-quantitative polymerase
chain reaction in T/G HA-VSMCs treated with siC, siR, siC + P, and
siR + P. The mRNA expression levels were normalized against that of
GAPDH. The data are presented as the mean ± standard deviation
(*P<0.05 and **P<0.01 compared with the siC group.) (B) The
protein expression levels of α-sm-actin, Calponin and Osteopontin
were determined by western blotting analysis. The protein and mRNA
expression changes were confirmed to be the same. GAPDH was used as
a loading control. si, siRNA; C, control; R, Rab5a; P, PDGF; PDGF,
platelet-derived growth factor; T/G HA-VSMC, human aorta-vascular
smooth muscle cell line; α-sm-actin, α-smooth muscle actin. |
Effect of Rab5a siRNA transfection and
PDGF on the proliferation, cell cycle and migration of T/G
HA-VSMCs
In addition to the phenotypic transition of T/G
HA-VSMCs, the proliferation, cell cycle and migration of T/G
HA-VSMCs were investigated in the different groups. Compared with
the siC group, the growth of the cells was significantly reduced in
the siR group at 24 (P<0.05), 48 (P<0.01) and 72 h
(P<0.01). This growth trend was rescued by the intervention of
PDGF in the siR + P group (Fig.
3A). According to the cell cycle analysis, the percentage of
cells in the siR group was significantly decreased in the G0/G1
phase and increased at the G2/M and S phases; however, the
percentage of cells at the G0/G1, G2/M and S phases were not
significantly changed in the siR + P group compared with the siC
group (Fig. 3B). These data
indicated that Rab5a modulated T/G HA-VSMC proliferation at
different phases of the cell cycle. In addition, compared with the
siC group, the average number of migrated cells in the siR group
was revealed to be significantly decreased (P<0.01). However,
the percentage of migrated cells was not markedly changed in the
siR + P group. Additionally, more migrated cells were observed in
the siC + P compared with the siC group (P<0.05; Fig. 3C). These results indicated that the
knockdown of Rab5a can inhibit T/G HA-VSMCs migration and that PDGF
treatment can reverse the decreased migration level in Rab5a siRNA
transfected cells.
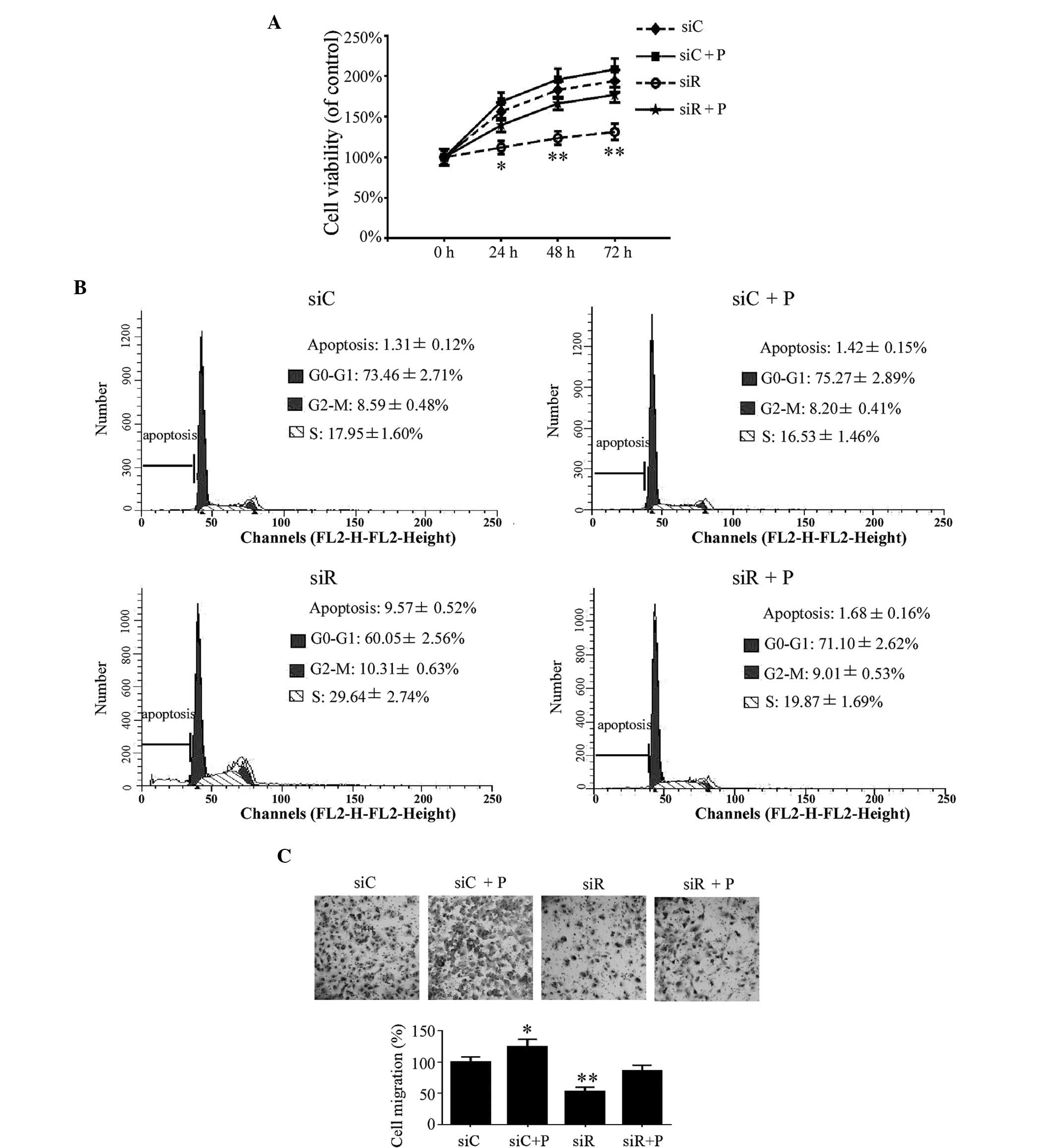 | Figure 3.Proliferation, cell cycle and
migration in T/G HA-VSMCs following various treatments. (A) A
sulforhodamine B assay was used to demonstrate the growth of the
cells following treatment that compared with siC, siR, siC + P and
siR + P at 24, 48, and 72 h,. (B) Cell cycle analysis was performed
by flow cytometry following treatment with siC, siR, siC + P and
siR + P. The percentage of apoptotic, G0-G1, G2-M or S phase cells
were calculated. (C) Microscopy observation (magnification, ×20) of
the cells was performed following migration assay. The percentage
of cells migrating was calculated. Quantified data are presented as
the mean ± standard deviation (**P<0.01 and *P<0.05 compared
with the siC group.) si, siRNA; C, control; R, Rab5a; P, PDGF;
PDGF, platelet-derived growth factor; T/G HA-VSMC, human
aorta-vascular smooth muscle cell line. |
Effect of Rab5a siRNA transfection and
PDGF on apoptosis in T/G HA-VSMCs
The early apoptotic rate was quantitatively analyzed
by flow cytometry and the results demonstrated that cells
transfected with siRNA against Rab5a exhibit a significantly higher
apoptotic rate compared with the siC group (P<0.01), while the
percentage of apoptotic cells was significantly reduced by the
intervention of PDGF (Fig. 4A). In
addition, the expression of the apoptotic proteins survivin and
caspase-3 was detected by western blotting. These results revealed
reduced expression of survivin and increased expression of cleaved
caspase-3 in the siR group compared with the siC group, while a
combination treatment with PDGF and siRNA (siCon or siRab)
transfection did not alter the expression levels of survivin and
caspase-3 (Fig. 4B).
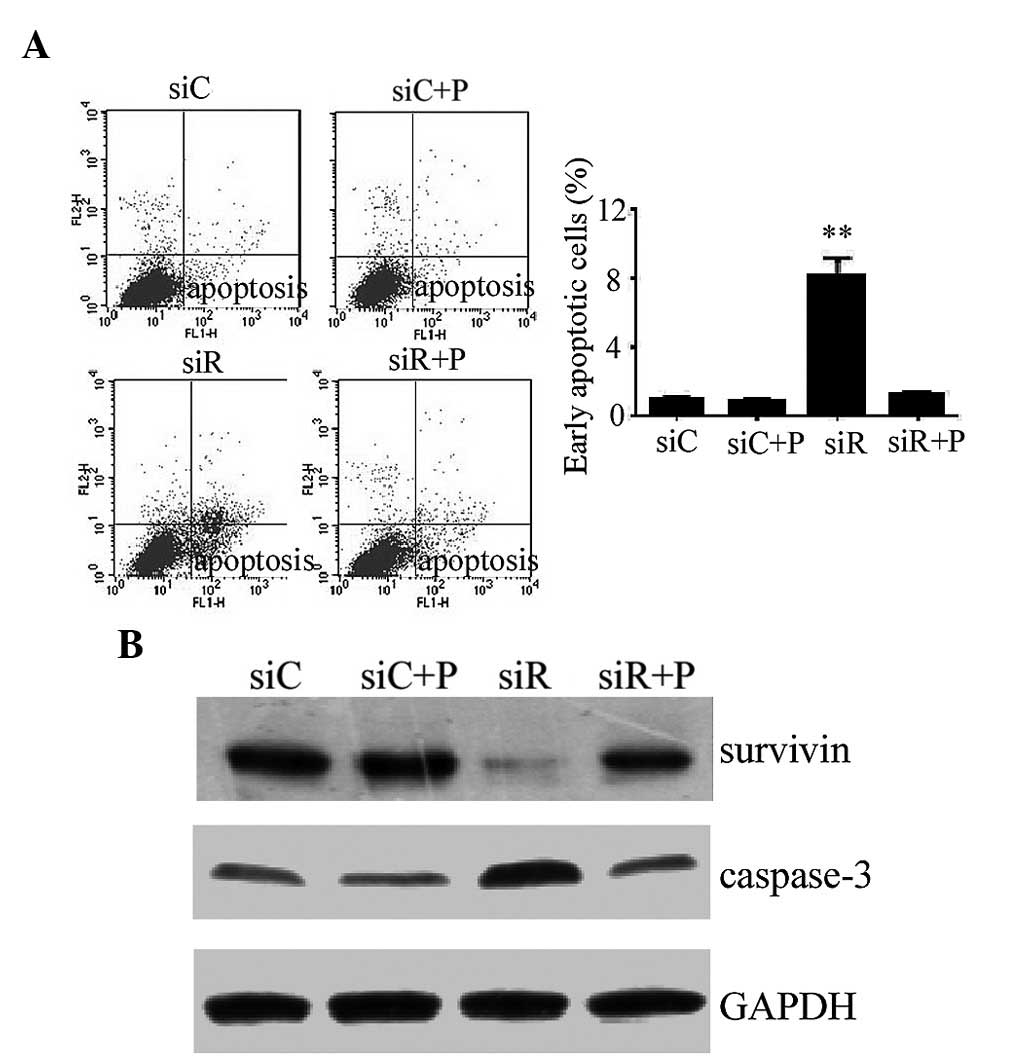 | Figure 4.Apoptotic response in T/G HA-VSMCs
following various treatments. (A) Flow cytometry was performed to
detemin the percentage of cells undergoing apoptosis following
treatment with siC, siR, siC + P and siR + P. The data are
presented as the mean ± standard deviation (**P<0.01 compared
with the siC group.) (B) The protein expression levels of
apoptosis-associated proteins, survivin and caspase-3, were
determined by western blotting analysis. GAPDH was used as a
loading control. si, siRNA; C, control; R, Rab5a; P, PDGF; PDGF,
platelet-derived growth factor; T/G HA-VSMC, human aorta-vascular
smooth muscle cell line. |
Effect of Rab5a siRNA transfection and
PDGF on autophagy in T/G HA-VSMCs
The autophagy in T/G HA-VSMCs from each group was
first detected by flow cytometry. As a result, the percentage of
cells with AVOs were significantly decreased (P<0.01), while
autophagy returned to a normal degree following treatment with PDGF
(Fig. 5A). In addition, the
autophagy in these cells was also analyzed by fluorescence
localization of LC3. As a result, the percentage of cells with
punctate fluorescence was significantly reduced in the siR group
(P<0.05), while no difference was observed in the other three
groups (Fig. 5B). Additionally,
VSMCs were visualized by TEM to examine the formation of
autophagosomes. It was revealed that Rab5a siRNA transfection
caused a significantly reduced number of autophagosomes with
typical scattered double-membrane vacuolar structures (P<0.05),
while the number of phagosomes in cells treated with PDGF remained
unchanged compared with the siC group (Fig. 5C). Furthermore, the expression
levels of the autophagic proteins Beclin1, LC3-I and LC3-II were
also analyzed by western blotting. As an autophagy-related protein,
Beclin1 is required for the initiation of autophagy. Additionally,
LC3-II formation is also a confirmed marker of autophagy. As the
formation of LC3-II is transient and the protein can be rapidly
degraded in the lysosome, a conversion of LC3-I to LC3-II may also
indicate autophagy in cells. Compared with the siC group, the
protein expression of Beclin1 and the conversion of LC3-I to LC3-II
was significantly reduced in the siR group, whereas it was elevated
to a normal level following treatment with PDGF (Fig. 5D).
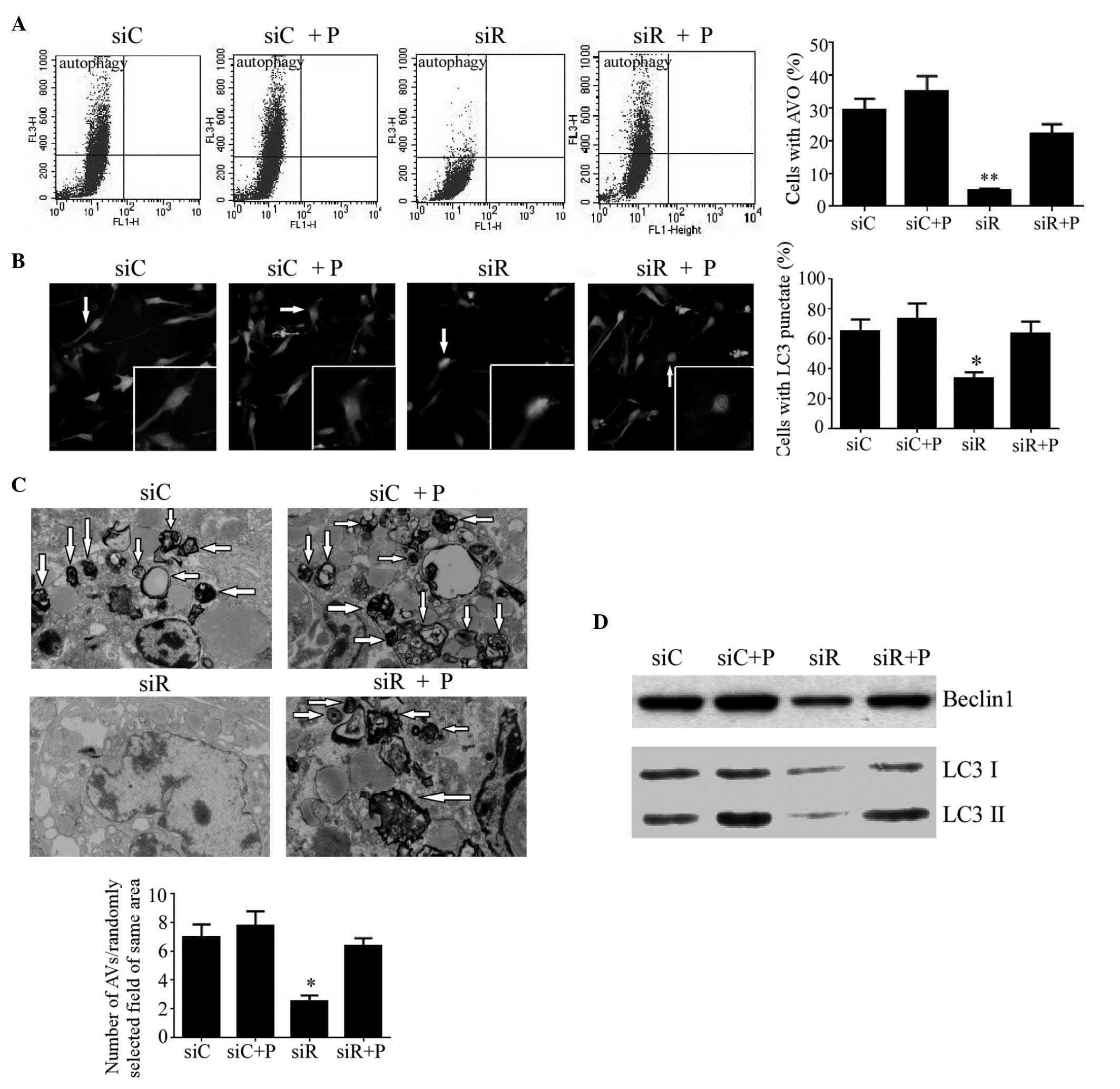 | Figure 5.Autophagy in T/G HA-VSMCs following
various treatments. (A) Flow cytometry was performed to determine
the percentage of cells with AVOs as a sign of autophagy following
treatment with siC, siR, siC + P and siR + P. (B) Representative
microscopy images (magnification, ×40) of fluorescence localization
of LC3 were captured and the percentage of cells with punctate
fluorescence was calculated. (C) Transmission electron microscopy
(magnification, 1,000x) was performed to determine the number of
autophagosomes with typical scattered double-membrane vacuolar
structures (arrows). (D) The protein expression levels of Beclin1,
LC3 I and LC3 II were determined by western blotting analysis.
Quantified data are presented as the mean ± standard deviation
(**P<0.01 and *P<0.05 compared with the siC group.) AVOs,
acidic vesicular organelles; LC3, light chain 3; si, siRNA; C,
control; R, Rab5a; P, PDGF; PDGF, platelet-derived growth factor;
T/G HA-VSMC, human aorta-vascular smooth muscle cell line. |
Effect of Rab5a siRNA transfection and
PDGF on the PI3K/AKT/mTOR and ERK1/2 signaling pathways in T/G
HA-VSMCs
To assess the mechanism of Rab5a siRNA
transfection-related autophagy, the PI3K/AKT/mTOR and ERK1/2
signaling pathways in T/G HA-VSMCs were assessed by western
blotting. The result revealed that compared with the siC group, the
expression levels of p-AKT, p-mTOR and p-ERK were markedly reduced
in the siR group, while PDGF recovered the normal expression of
these proteins. In addition, no differences were observed in the
expression levels of t-AKT, t-mTOR and t-ERK among these four
groups (Fig. 6). The results
suggested that the MAPK/ERK pathway may be involved in
Rab5a-related autophagy, which did not occur via the inhibition of
the classical PI3K/AKT/mTOR pathway.
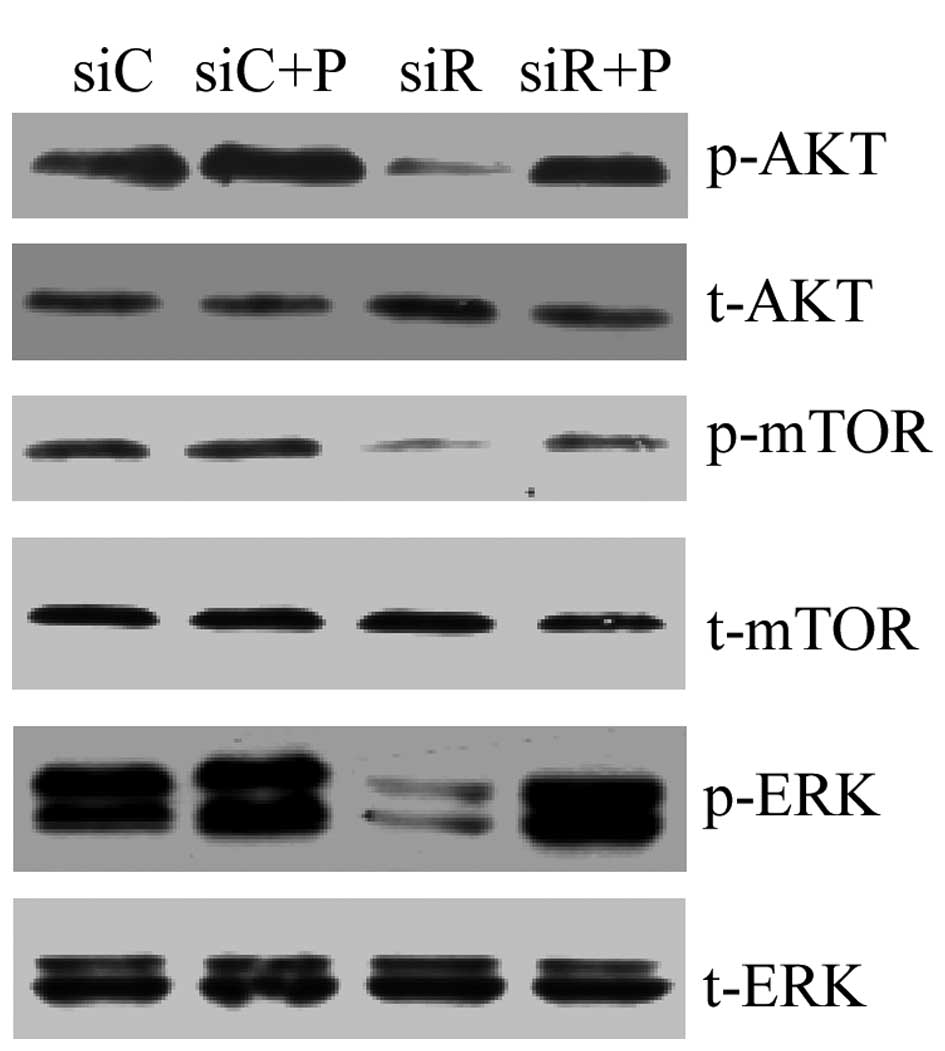 | Figure 6.PI3K/AKT/mTOR and ERK1/2 signaling
pathways in T/G HA-VSMCs following various treatments. The mRNA
expression levels of p-AKT, t-AKT, p-mTOR, t-mTOR, p-ERK and t-ERK
were determined by western blotting analysis following treatment
with siC, siR, siC + P and siR + P. p-, phosphorylated; t-, total;
Akt, protein kinase B; mTOR, mammalian target of rapamycin; ERK,
extracellular signal-regulated kinase; si, siRNA; C, control; R,
Rab5a; P, PDGF; PDGF, platelet-derived growth factor; T/G HA-VSMC,
human aorta-vascular smooth muscle cell line. |
Discussion
It is generally accepted that accelerated
proliferation and migration of VSMCs are essential for the
development of neointimal hyperplasia, which is also a key
pathological feature of atherosclerosis and vascular reconstruction
with restenosis (16). The present
study revealed that downregulated expression of Rab5a inhibited the
phenotypic transition, proliferation, migration and autophagy of
T/G HA-VSMCs, while it increased apoptosis in T/G HA-VSMCs.
However, PDGF treatment reversed these changes. Furthermore,
Rab5a-related autophagy may be induced via the ERK pathway,
however, not the PI3K/AKT/mTOR pathway.
Previous studies have demonstrated that phenotype
transition in VSMCs is regulated by numerous factors, including
contractile agonists and extracellular matrix components (17–19).
Notably, the present study found that downregulation of Rab5a
resulted in increased expression levels of α-SMA and calponin, as
well as decreased expression levels of osteopontin and vimentin.
These results indicated that Rab5a may be associated with the
phenotype transition from contractile to synthetic in VSMCs;
however, the underlying mechanism remains unclear. Su et al
(20) suggested that Rab5a was
associated with autophagy induced by hepatitis C virus
non-structural protein 4B, while autophagy was an essential
regulator for the phenotype transition of VSMCs (14). Therefore, it was speculated that
autophagy may serve a vital role in the effect of Rab5a on the
phenotype transition of VSMCs. In addition, it was revealed that
PDGF treatment can significantly reverse the phenotype switching of
VSMCs caused by Rab5a siRNA transfection. Notably, PDGF is usually
overexpressed in atherosclerotic lesions and can promote VSMC
proliferation (21,22), which may induce autophagy and
phenotype switching in VSMCs (14). These results further indicated that
the phenotypic transition of VSMCs induced by Rab5a may be
regulated by autophagy.
Furthermore, the present study demonstrated that
Rab5a promoted the proliferation and migration of T/G HA-VSMCs.
Similarly, our previous study also suggested a critical role for
Rab5a in rat VSMC proliferation and migration (13). However, the exact mechanism remains
to be elucidated. Autophagy was considered to be the basic
catabolic mechanism that is essential for cell survival,
differentiation, development and the cellular response to stress
via the regulation of the actions of lysosomes (23). Du et al (24) suggested that autophagy can promote
angiogenesis by AKT activation in aortic endothelial cells, while
increased angiogenesis may promote tube formation and endothelial
cell migration (25). In addition,
in lung cancer cells, autophagy also facilitated cell
proliferation, migration and invasion by increasing the expression
levels of various cytokines, including interleukin 6, vascular
endothelial growth factor A and matrix metallopeptidase 2 (26). Therefore, the present study
speculated that the proliferation and migration of T/G HA-VSMCs
induced by Rab5a may also be regulated by autophagy.
The crosstalk between apoptosis and autophagy is
very complex. Autophagy is a physiological process accompanied by
the degradation of disordered cellular components, while apoptosis
is programmed cell death. Under stress circumstances, cells exhibit
a stress adaptation by inducing low levels of autophagy, and the
intrinsic and extrinsic pathways of apoptosis are inhibited in
order to avoid cell death (27).
However, in certain circumstances, autophagy acts as a key
regulator to activate the programmed cell death pathway, causing
massive cell death and eventually leading to functional impairment
in vivo (28). In the
present study, Rab5a was revealed to inhibit the apoptosis of
VSMCs, which exhibited reduced expression of survivin, high
expression levels of caspase-3 and a higher apoptosis rate.
However, Rab5a was demonstrated to induce autophagy in VSMCs that
exhibited significantly reduced expression of Beclin1, to
downregulated the conversion of LC3-I to LC3-II, reduce AVOs, LC3
punctate fluorescence and autophagosomes with typical scattered
double-membrane vacuolar structures in Rab5a siRNA transfected
cells. This may be explained by the fact that autophagy confers
cytoprotection to VSMCs. In addition, Shen et al (29) observed that high autophagy levels
induced cell death in tumor cells receiving anticancer treatments,
while low levels of autophagy may promote cell survival by removing
disordered intracellular proteins and organelles (30). These two previous studies indicated
that the cell survival or death was associated with the level of
autophagy, which may also explain the present results. Therefore,
Rab5a-induced autophagy may protect against the onset of apoptosis,
and the inhibition of autophagy by Rab5a siRNA transfection can
trigger the apoptosis of T/G HA-VSMCs, and subsequently regulate
the proliferation and migration of T/G HA-VSMCs.
The present study further examined various signaling
pathways involved in autophagy. Previously, the PI3K/AKT/mTOR
signaling pathway has been considered to be crucial in the
regulation of cellular survival, differentiation, proliferation,
metabolism, migration and angiogenesis (31). Additionally, it was also a
well-known pathway in the regulation of autophagy in mammalian
cells, which can activate autophagy by the inhibition of AKT and
mTOR (32,33). However, the present results
demonstrated that the expression of phosphorylated AKT and mTOR was
reduced by the knockdown of Rab5a, demonstrating that Rab5a-related
autophagy was independent of the PI3K/AKT/mTOR pathway. Notably, a
reduced expression trend of Rab5a was also observed, to be
consistent with the phosphorylation of ERK1/2. The MAPK/ERK pathway
served a primary role in the promotion of cellular proliferation in
response to growth factors (34)
and the induction of autophagy by the activation of the ERK1/2
pathway (35,36). These results indicated that
Rab5a-related autophagy correlated with the ERK1/2 pathway;
however, not the PI3K/AKT/mTOR pathway.
Rab5a is pivotal for autophagy in VSMCs, and the
phenotype transition, proliferation, cell cycle, migration and
apoptosis induced by Rab5a may be regulated by autophagy via the
activation of the ERK1/2 pathway. The present results supported a
critical role for Rab5a in atherogenesis. However, further studies
are required to detail the underlying molecular mechanisms of
Rab5a-induced autophagy.
Acknowledgements
The authors would like to thank the Electron
Microscopy Laboratory of Fudan University for technical assistance
in the TEM analysis. This work was supported by the Outstanding
Leaders Training Program of Pudong Health Bureau of Shanghai
(PWRI2011-02), the Project of Science and Technology Commission of
Shanghai Municipality (12jc1408101) and the Shanghai Pudong
District Science Project (PKJ2012-Y68).
References
|
1
|
Owens GK, Kumar MS and Wamhoff BR:
Molecular regulation of vascular smooth muscle cell differentiation
in development and disease. Physiol Rev. 84:767–801. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pyle AL and Young PP: Atheromas feel the
pressure: Biomechanical stress and atherosclerosis. Am J Pathol.
177:4–9. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rudijanto A: The role of vascular smooth
muscle cells on the pathogenesis of atherosclerosis. Acta Med
Indones. 39:86–93. 2007.PubMed/NCBI
|
|
4
|
Mack CP: Signaling mechanisms that
regulate smooth muscle cell differentiation. Arterioscler Thromb
Vasc Biol. 31:1495–1505. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jordens I, Marsman M, Kuijl C and Neefjes
J: Rab proteins, connecting transport and vesicle fusion. Traffic.
6:1070–1077. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Grosshans BL, Ortiz D and Novick P: Rabs
and their effectors: Achieving specificity in membrane traffic.
Proc Natl Acad Sci USA. 103:11821–11827. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hutagalung AH and Novick PJ: Role of Rab
GTPases in membrane traffic and cell physiology. Physiol Rev.
91:119–149. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ravikumar B, Imarisio S, Sarkar S, O'Kane
CJ and Rubinsztein DC: Rab5 modulates aggregation and toxicity of
mutant huntingtin through macroautophagy in cell and fly models of
Huntington disease. J Cell Sci. 121:1649–1660. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang PS, Yin PH, Tseng LM, Yang CH, Hsu
CY, Lee MY, Horng CF and Chi CW: Rab5A is associated with axillary
lymph node metastasis in breast cancer patients. Cancer Sci.
102:2172–2178. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu SS, Chen XM, Zheng HX, Shi SL and Li
Y: Knockdown of Rab5a expression decreases cancer cell motility and
invasion through integrin-mediated signaling pathway. J Biomed Sci.
18:582011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao Z, Liu XF, Wu HC, Zou SB, Wang JY, Ni
PH, Chen XH and Fan QS: Rab5a overexpression promoting ovarian
cancer cell proliferation may be associated with APPL1-related
epidermal growth factor signaling pathway. Cancer Sci.
101:1454–1462. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fukui K, Tamura S, Wada A, Kamada Y, Igura
T, Kiso S and Hayashi N: Expression of Rab5a in hepatocellular
carcinoma: Possible involvement in epidermal growth factor
signaling. Hepatol Res. 37:957–965. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ma Z, Wang H, Wu L, Zhui L, Shi W, Ma D,
Chen Z and Yu B: RNAi-mediated Rab5a suppression inhibits
proliferation and migration of vascular smooth muscle cells. Acta
Cardiol. 65:507–514. 2010.PubMed/NCBI
|
|
14
|
Salabei JK, Cummins TD, Singh M, Jones SP,
Bhatnagar A and Hill BG: PDGF-mediated autophagy regulates vascular
smooth muscle cell phenotype and resistance to oxidative stress.
Biochem J. 451:375–388. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Butoi E, Gan AM and Manduteanu I:
Molecular and functional interactions among monocytes/macrophages
and smooth muscle cells and their relevance for atherosclerosis.
Crit Rev Eukaryot Gene Expr. 24:341–355. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Garat C, Van Putten V, Refaat ZA, Dessev
C, Han SY and Nemenoff RA: Induction of smooth muscle alpha-actin
in vascular smooth muscle cells by arginine vasopressin is mediated
by c-Jun amino-terminal kinases and p38 mitogen-activated protein
kinase. J Biol Chem. 275:22537–22543. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hautmann MB, Thompson MM, Swartz EA, Olson
EN and Owens GK: Angiotensin II-induced stimulation of smooth
muscle alpha-actin expression by serum response factor and the
homeodomain transcription factor MHox. Circ Res. 81:600–610. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang L, Zheng J, Du Y, Huang Y, Li J, Liu
B, Liu CJ, Zhu Y, Gao Y, Xu Q, et al: Cartilage oligomeric matrix
protein maintains the contractile phenotype of vascular smooth
muscle cells by interacting with alpha(7)beta(1) integrin. Circ
Res. 106:514–525. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Su WC, Chao TC, Huang YL, Weng SC, Jeng KS
and Lai MM: Rab5 and class III phosphoinositide 3-kinase Vps34 are
involved in hepatitis C virus NS4B-induced autophagy. J Virol.
85:10561–10571. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Myllärniemi M, Calderon L, Lemström K,
Buchdunger E and Häyry P: Inhibition of platelet-derived growth
factor receptor tyrosine kinase inhibits vascular smooth muscle
cell migration and proliferation. FASEB J. 11:1119–1126.
1997.PubMed/NCBI
|
|
22
|
Yu JC, Lokker NA, Hollenbach S, Apatira M,
Li J, Betz A, Sedlock D, Oda S, Nomoto Y, Matsuno K, et al:
Efficacy of the novel selective platelet-derived growth factor
receptor antagonist CT52923 on cellular proliferation, migration,
and suppression of neointima following vascular injury. J Pharmacol
Exp Ther. 298:1172–1178. 2001.PubMed/NCBI
|
|
23
|
Levine B and Klionsky DJ: Development by
self-digestion: Molecular mechanisms and biological functions of
autophagy. Dev Cell. 6:463–477. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Du J, Teng RJ, Guan T, Eis A, Kaul S,
Konduri GG and Shi Y: Role of autophagy in angiogenesis in aortic
endothelial cells. Am J Physiol Cell Physiol. 302:C383–C391. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Slevin M, Krupinski J, Rovira N, Turu M,
Luque A, Baldellou M, Sanfeliu C, de Vera N and Badimon L:
Identification of pro-angiogenic markers in blood vessels from
stroked-affected brain tissue using laser-capture microdissection.
BMC Genomics. 10:1132009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhan Z, Xie X, Cao H, Zhou X, Zhang XD,
Fan H and Liu Z: Autophagy facilitates TLR4-and TLR3-triggered
migration and invasion of lung cancer cells through the promotion
of TRAF6 ubiquitination. Autophagy. 10:257–268. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lum JJ, DeBerardinis RJ and Thompson CB:
Autophagy in metazoans: Cell survival in the land of plenty. Nat
Rev Mol Cell Biol. 6:439–448. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Platini F, Pérez-Tomás R, Ambrosio S and
Tessitore L: Understanding autophagy in cell death control. Curr
Pharm Des. 16:101–113. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shen S, Kepp O, Michaud M, Martins I,
Minoux H, Métivier D, Maiuri MC, Kroemer RT and Kroemer G:
Association and dissociation of autophagy, apoptosis and necrosis
by systematic chemical study. Oncogene. 30:4544–4556. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Amaravadi RK: Autophagy-induced tumor
dormancy in ovarian cancer. J Clin Invest. 118:3837–3840.
2008.PubMed/NCBI
|
|
31
|
Hsieh AC, Liu Y, Edlind MP, Ingolia NT,
Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et
al: The translational landscape of mTOR signalling steers cancer
initiation and metastasis. Nature. 485:55–61. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Saiki S, Sasazawa Y, Imamichi Y, Kawajiri
S, Fujimaki T, Tanida I, Kobayashi H, Sato F, Sato S, Ishikawa K,
et al: Caffeine induces apoptosis by enhancement of autophagy via
PI3K/Akt/mTOR/p70S6K inhibition. Autophagy. 7:176–187. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang RC, Wei Y, An Z, Zou Z, Xiao G,
Bhagat G, White M, Reichelt J and Levine B: Akt-mediated regulation
of autophagy and tumorigenesis through Beclin 1 phosphorylation.
Science. 338:956–959. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shaul YD and Seger R: The MEK/ERK cascade:
From signaling specificity to diverse functions. Biochim Biophys
Acta. 1773:1213–1226. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang J, Whiteman MW, Lian H, Wang G, Singh
A, Huang D and Denmark T: A non-canonical MEK/ERK signaling pathway
regulates autophagy via regulating Beclin 1. J Biol Chem.
284:21412–21424. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cagnol S and Chambard JC: ERK and cell
death: Mechanisms of ERK-induced cell death-apoptosis, autophagy
and senescence. FEBS J. 277:2–21. 2010. View Article : Google Scholar : PubMed/NCBI
|