Introduction
Blood pressure regulation is a complex,
multifactorial process, which is associated with physiological,
biochemical and molecular mechanisms. An increase in Na+
and water retention is required for the development of most forms
of hypertension (1). The
epithelial sodium channel (ENaC) mediates the initial step of
active sodium reabsorption, which is essential for the maintenance
of body salt and water homeostasis (2). The ENaC is composed of three
different subunits (3), one of
which is the amiloride-sensitive sodium channel beta subunit
(SCNN1B). Mutations in SCNN1B may cause Liddle's syndrome
(4–6), which is an autosomal dominant
disorder that is characterized by early, and frequently severe,
hypertension (7). Essential
hypertension (EH), the form of hypertension that by definition has
no identifiable cause, tends to be familial and may be due to an
interaction between environmental and genetic factors.
DNA methylation refers to the addition of a methyl
group to the cytosine or adenine DNA nucleotides in mammalian
cells, and usually occurs at CpG islands, which contain clusters of
CpG dinucleotides. Promoter hypermethylation often silences gene
transcription and is an important event during disease progression
(8). Our previous study
demonstrated that reduced α-adducin gene promoter methylation
increased the risk of essential hypertension (EH) in Chinese men
and women (9). Furthermore,
hypo/hypermethylation of the sulfatase 1 gene may serve an
important role in the pathogenesis of hypertension in young African
American men (10). Altered gene
methylation has also been observed in other cardiovascular diseases
(11,12), as well as in type 2 diabetes
(13–16).
DNA methylation has been reported to mediate some of
the effects of environmental exposure and lifestyle factors on
disease risk (17). DNA
methylation is heritable, but can also be altered by medical
therapy. It has previously been reported that drugs can alter the
gene methylation status in patients with cancer and schizophrenia
(18). In light of these previous
studies, the present study aimed to investigate whether
SCNN1B promoter DNA methylation was associated with the risk
of EH, and whether antihypertensive drug treatment would alter the
SCNN1B methylation status in patients with EH.
Materials and methods
Sample collection
A total of 282 individuals (94 incident cases, 94
prevalent cases and 94 normotensive controls) were recruited to the
present study from the Ningbo Seventh Hospital (Ningbo, China). All
individuals selected were Han Chinese that had resided in Ningbo
for ≥3 generations. Incident cases were hypertensive patients who
had previously never received any antihypertensive drug treatment.
Prevalent cases were hypertensive patients who had previously
received antihypertensive drug treatment. The diagnosis of
hypertension was made when the average of ≥2 diastolic blood
pressure (DBP) measurements on ≥2 subsequent visits was ≥90 mmHg,
or when the average of several systolic blood pressure (SBP)
readings on ≥2 subsequent visits was consistently ≥140 mmHg.
Isolated systolic hypertension is defined as SBP ≥140 mmHg and DBP
<90 mmHg. Individuals with SBP <120 mmHg and DBP <80 mmHg
were recruited as controls (19).
None of the control group had received antihypertensive therapy.
Furthermore, none of the recruited individuals suffered from other
diseases, including diabetes mellitus, secondary hypertension,
myocardial infarction, stroke, renal failure, and drug abuse. Blood
samples were collected in 3.2% citrate sodium-treated tubes and
were then stored at −80°C for DNA extraction. The protocol of the
present study was approved by the ethical committee of the Ningbo
Seventh Hospital. Written informed was obtained from all
subjects.
DNA isolation and bisulfite
treatment
Genomic DNA was extracted and underwent bisulfite
conversion for subsequent polymerase chain reaction (PCR)
amplification and pyrosequencing, as described previously (11,13–15,20).
Genomic DNA was extracted from peripheral blood samples using a
nucleic acid extraction analyzer (Lab-Aid 820; Xiamen Zeesan
Biotech Co., Ltd., Xiamen, China). The NanoDrop 1000
spectrophotometer (NanoDrop; Thermo Fisher Scientific, Inc.,
Wilmington, DE, USA) was used to measure the concentrations of
extracted DNA. Subsequently, ~500 ng genomic DNA isolated from
whole blood cells was bisulfite-treated using the EZ DNA
Methylation-Gold™ kit (Zymo Research Corp, Irvine, CA, USA),
according to the manufacturer's protocol. This treatment involves
converting unmethylated cytosines into uracil, whereas methylated
cytosines remain unchanged. Once converted, the methylation profile
of the DNA can be determined by PCR amplification followed by DNA
sequencing.
DNA methylation assay
Ten primer sets (including forward, reverse and
sequencing primers) were designed by PyroMark Assay Design software
v2.0.1.1 (Qiagen, Inc., Valencia, CA, USA) to amplify the CpG
island region in overlapping fragments and to sequence target DNA
fragments. Each primer set was given a score and these primer sets
were ranked from high to low. According to the rank, the top three
primer sets were synthesized and purified by high-performance
liquid chromatography by Sangon Biotech (Shanghai) Co., Ltd.
(Shanghai, China) for PCR. Subsequently, 2% agarose gel
electrophoresis was used to analyze the PCR products. The results
indicated that primer set one exhibited the best amplification
effects. Detailed information regarding this primer set is
presented in Table I. PCR was
conducted in a final volume of 20 µl containing 10 µl Zymo Taq™
PreMix, 1.5 µl forward primer (10 µM), 1.5 µl reverse primer (10
µM) and 40 ng DNA and water was used to raise volume to 20 µl. The
PCR process began with an initial denaturation step at 95°C for 10
min, followed by 40 cycles of denaturation at 95°C for 30 sec,
annealing at 58°C for 40 sec and extension at 72°C for 50 sec. The
final extension step was performed at 72°C for 7 min. Following
amplification, PCR products were maintained at 4°C for ≥4 min. The
target sequence was finally sequenced by synthesis assay (Pyromark
Gold Q24 Reagents; #970802; Qiagen, Inc.).
 | Table I.Oligonucleotides for bisulfite
sequencing. |
Table I.
Oligonucleotides for bisulfite
sequencing.
| Variable | Sequence | Nucleotides
(bp) |
|---|
| Forward primer |
5′-GGATGAGGGGTTTGTGGATA-3′ | −227 to −207 |
| Reverse primer |
5′-ACCTCCCTCCCCTCCCAATAAACT-3′ | −66 to −42 |
| Amplicon
sequence |
5′-GGATGAGGGGTTTGTGGATATATTCGTGGCGTATGTGGGTATCGTTGGTGTTTCGAGGTGGGGAGGGAGAATGCGGAGCGCGTGCGTGCGGGGGGCGTTTAGTGTTTTTGAATTTGGCGTGTGGGGGTTGGAGTTTATTGGGAGGGGAGGGAGGT-3′ | −227 to −42 |
| Sequencing
primer |
5′-GGTGGGGAGGGAGAA-3′ | −172 to −158 |
| Sequence to
analyze |
5′-TGCGGAGCGCGTGCGTGCGGGGGGCGTTTAGTGTTTTTGAATTTGGCG-3′ | −156 to −107 |
Statistical analysis
Statistical analyses were performed using PASW
Statistics 18.0 software (SPSS, Inc., Chicago, IL, USA). The mean
of continuous variables [including age, body mass index (BMI), DNA
methylation level, total cholesterol (TC), triglycerides (TG),
glucose and alanine aminotransferase (ALT)] between case and
control groups was analyzed using two-tailed unpaired t test or
one-way analysis of variance followed by least significant
difference multiple comparison tests. Either Pearson χ2
test or Fisher exact test were used to analyze the association
between EH and categorical variables including gender, and smoking
and drinking habits. Partial correlative test was used to analyze
the relationship between continuous variables. Logistic regression
was implemented for controlling the possible confounding factors to
analyze the correlation between independent variables and dependent
variables. Meanwhile, R software and GraphPad Prism (version 5.01;
GraphPad Software, Inc., La Jolla, CA, USA) were used for
statistical computing and graphical representation. P<0.05 was
considered to indicate a statistically significant difference.
Results
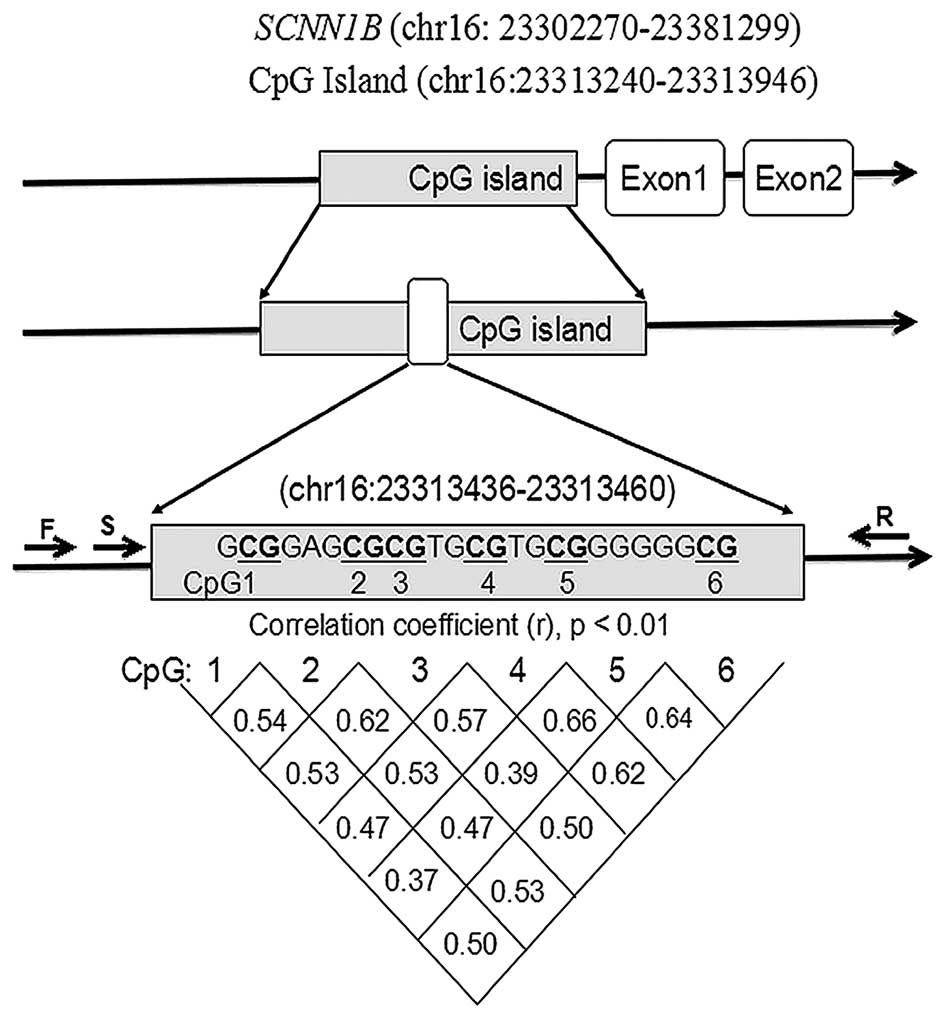
As presented in Fig.
1, PCR primers were designed for amplification of the CpG
island region of SCNN1B, whereas the sequencing primer was
set to sequence the fragment containing seven CpG dinucleotides
(Fig. 1 and Table I). Since the last CpG dinucleotide
(CpG7) was not well sequenced, the remaining six CpG sites (CpG1-6)
were analyzed in the present study (Fig. 1). Significant correlations were
found among CpG1-6 (Fig. 1;
r>0.30; P<0.01) in DNA methylation levels.
A series of EH risk factors (including BMI, TC, TG
and glucose) were compared among the three groups using the
variance analysis. As shown in Table
II, there were significant differences among the three groups
in the mean levels of BMI (F=12.478, P=6.45E-06), TG (F=4.631,
P=0.011) and glucose (F=3.982, P=0.02). In addition, alcohol and
smoking consumption were compared among the three groups, since
they are known to be associated with hypertension (21). The results demonstrated that the
ratio of alcohol drinking (χ2=9.189; P=0.011) was
inconsistent among the three groups using the χ2 test
(Table II).
 | Table II.Comparison of characteristics among
the three groups (n=282). |
Table II.
Comparison of characteristics among
the three groups (n=282).
|
| Non-EH | Incident cases | Prevalent
cases |
|
|
|---|
|
|
|
|
|
|
|
|---|
| Characteristic | (Mean ± SD) | (Mean ± SD) | (Mean ± SD) |
F/χ2 | P |
|---|
| Age (years) | 58.36±7.53 | 56.31±7.73 | 58.37±7.95 | 2.217 | 0.111 |
| Gender (M/F) | 29/65 | 34/60 | 33/61 | 0.537 | 0.642 |
| Smoking (Y/N) | 17/77 | 21/73 | 20/74 | 0.586 | 0.714 |
| Drinking (Y/N) | 7/87 | 21/73 | 20/74 | 0.007 | 0.011 |
| BMI
(kg/m2) | 22.752±3.721 | 24.331±2.543 | 24.962±3.021 | 12.478 |
6.45×10−6 |
| TC (mmol/l) | 5.231±1.002 | 5.381±1.043 | 5.491±1.032 | 1.456 | 0.235 |
| TG (mmol/l) | 1.521±0.892 | 1.893±1.071 | 1.591±0.702 | 4.631 | 0.011 |
| Glu (mmol/l) | 5.301±0.642 | 5.371±0.591 | 5.562±0.760 | 3.982 | 0.020 |
| ALT (IU/l) | 24.490±40.231 | 24.862±16.781 | 25.181±25.261 | 0.013 | 0.987 |
| CpG1 (%) | 16.211±4.082 | 17.371±4.090 | 16.072±2.628 | 3.555 | 0.030 |
| CpG2 (%) | 10.021±2.778 | 9.123±2.252 | 9.121±1.652 | 4.952 | 0.008 |
| CpG3 (%) | 6.391±1.904 | 6.550±2.040 | 6.361±1.490 | 0.298 | 0.743 |
| CpG4 (%) | 8.841±2.192 | 9.140±2.671 | 9.430±3.281 | 1.065 | 0.346 |
| CpG5 (%) | 11.104±4.250 | 10.961±2.870 | 11.060±4.041 | 0.035 | 0.966 |
| CpG6 (%) | 6.061±2.493 | 6.402±1.601 | 6.432±1.464 | 1.068 | 0.345 |
Among the six CpG sites, CpG1 (F=3.555, P=0.03) and
CpG2 (F=4.952, P=0.008) methylation levels exhibited a significant
difference among the three groups. Increased methylation levels of
CpG1 were detected in the incident cases compared with in the other
two groups. Conversely, lower methylation levels of CpG2 were
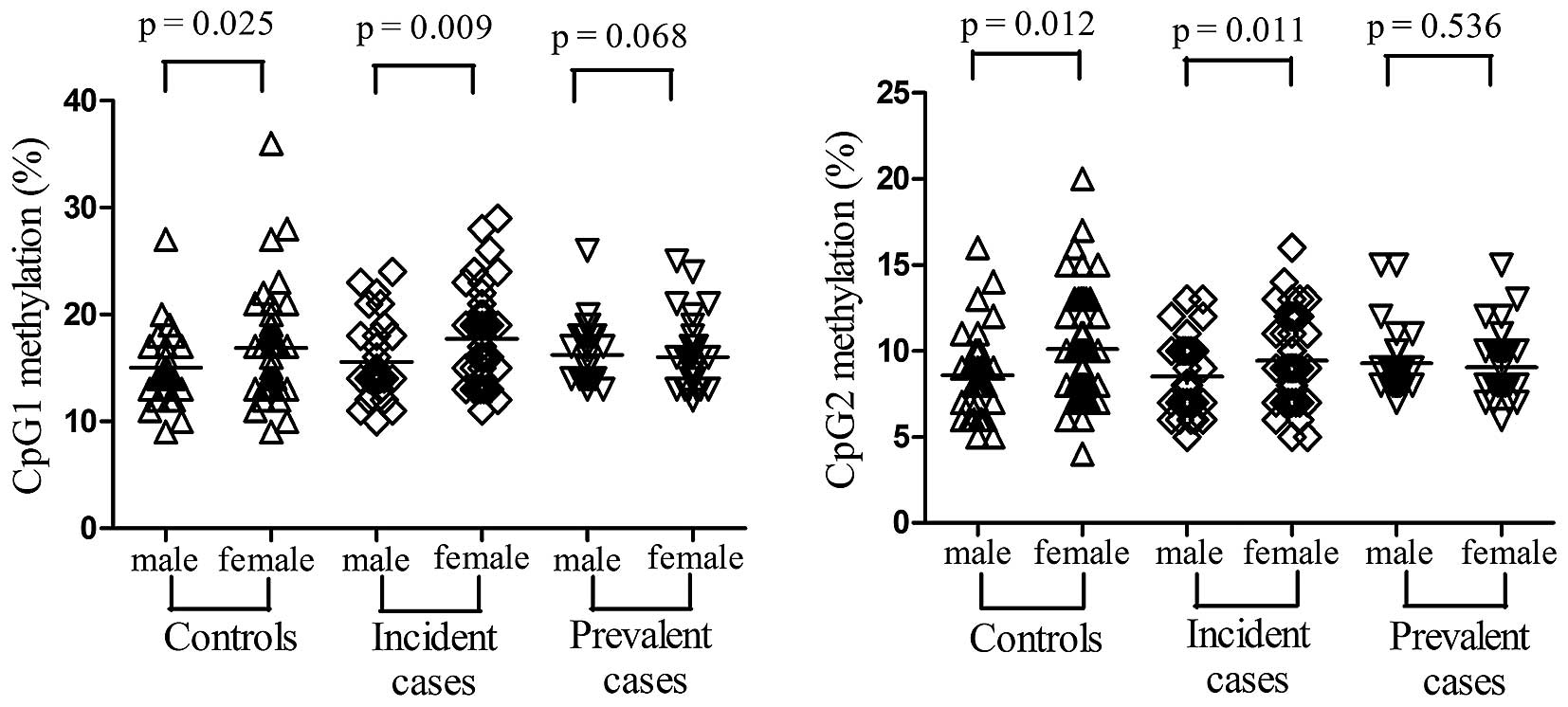
detected in the cases compared with in the controls (Table II). In addition, higher
methylation levels of the two CpG sites were detected in women
compared with in men for controls (CpG1: t=−2.283, P=0.025; CpG2:
t=−2.568, P=0.012, Fig. 2) and
incident cases (CpG1: t=−2.694, P=0.009; CpG2: t=−2.583, P=0.011,
Fig. 2). Notably, for the two CpG
sites no significant difference was observed between males and
females in the prevalent cases group (CpG1: t=0.409, P=0.068; CpG2:
t=0.621, P=0.536), thus suggesting a role for antihypertensive
therapy in the modification of DNA methylation.
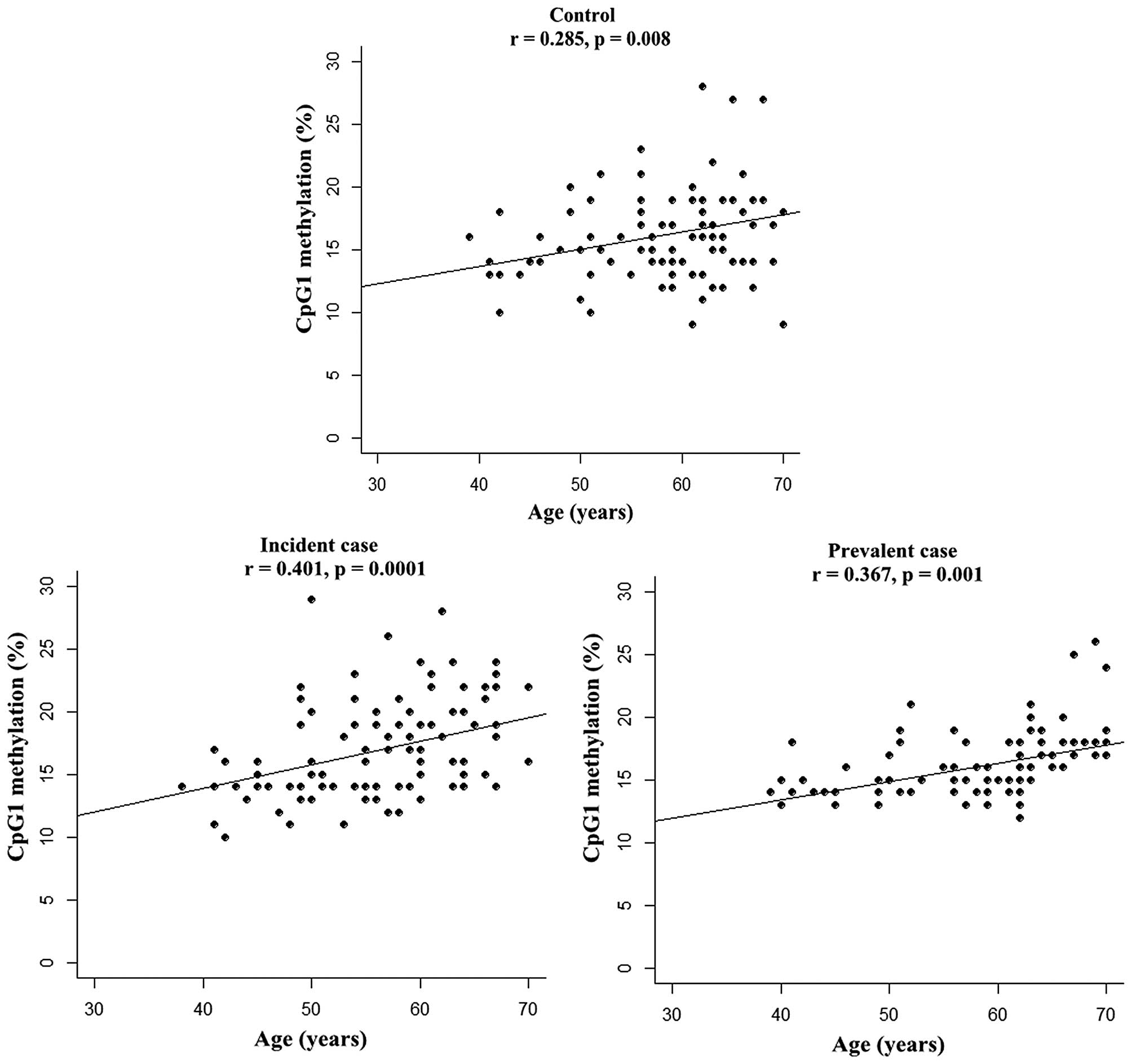
Since aging is able to alter levels of DNA
methylation (22), the present
study further explored the association of SCNN1B CpG
methylation with age using the partial correlative test.
Significant correlations were found between age and CpG1 (Controls:
r=0.285, P=0.008; Incident cases: r=0.401, P=0.0001; Prevalent
cases r=0.367, P=0.001, Fig. 3)
with an adjustment for other metabolic phenotypes (including BMI,
TC, TG, glucose, ALT, and smoking and alcohol drinking habits). In
addition, no correlations were detected between SCNN1B
methylation and these aforementioned metabolic phenotypes.
In the present study, EH cases were separated into
two groups: Incident cases without antihypertensive therapy and
prevalent cases with antihypertensive therapy, since drug treatment
may influence DNA methylation (23–26).
The current study compared the two case groups to explore whether
antihypertensive therapy may affect SCNN1B CpG methylation
status using a logistic regression test.
As shown in Table
III, the P-values and odds ratio (OR) values were all adjusted
by other parameters. When analyzing the association between a
specific independent variable and dependent variable, the other
independent variables are controlled in a logistic regression
model. When the P-value is <0.05, this suggests that the
variable has an effect on the dependent variable. When the OR value
in a logistic regression model is >1, this variable acts as a
risk factor for the dependent variable. Conversely, when the OR
value is <1, this variable acts as a protective factor. As
determined using this analytical model, methylation of CpG1
(β-standardized=0.17, OR=1.185, adjusted P=0.015) and CpG2
(β-standardized=−0.41, OR=0.663, adjusted P=0.001) were
significantly associated with EH, since both P-values were <0.05
in the logistic regression analysis of controls and incident cases.
In this model, the OR of CpG1 was 1.185, which suggests that
hypermethylation of CpG1 is associated with EH; however, the OR of
CpG2 was 0.663, which suggests that hypomethylation of CpG2 may
increase the probability of EH. In addition, according to this
statistical model, the methylation level of CpG1
(β-standardized=−0.252, OR=0.777, adjusted P=3.77E-04, Table III) was higher in the incident
cases compared with in the prevalent cases in the regression model
of the two cases. This result suggests that antihypertensive
therapy may lower the methylation level of CpG1. No significant
associations were detected in the remaining CpG sites.
 | Table III.Variables in the logistic regression
model. |
Table III.
Variables in the logistic regression
model.
| Variable | βa | ORa | Pa | βb | ORb | Pb | βc | ORc | Pc |
|---|
| Gender | 0.146 | 1.157 | 0.732 | 0.242 | 1.274 | 0.559 | 0.414 | 1.513 | 0.340 |
| Age | −0.066 | 0.936 | 0.019 | 0.062 | 1.064 | 0.014 | −0.044 | 0.957 | 0.136 |
| BMI | 0.136 | 1.146 | 0.028 | 0.062 | 1.064 | 0.321 | 0.246 | 1.279 |
2.89×10−4 |
| TC | 0.248 | 1.281 | 0.191 | 0.114 | 1.121 | 0.508 | 0.131 | 1.140 | 0.539 |
| TG | 0.500 | 1.649 | 0.012 | −0.558 | 0.573 | 0.008 | 0.029 | 1.030 | 0.905 |
| Glu | 0.338 | 1.403 | 0.275 | 0.479 | 1.615 | 0.103 | 1.320 | 3.743 | 0.001 |
| ALT | −0.004 | 0.996 | 0.554 | 0.002 | 1.002 | 0.845 | 0.004 | 1.004 | 0.442 |
| CpG1 | 0.170 | 1.185 | 0.015 | −0.252 | 0.777 |
3.77×10−4 | 0.005 | 1.005 | 0.934 |
| CpG2 | −0.410 | 0.663 | 0.001 | 0.183 | 1.201 | 0.176 | −0.530 | 0.589 |
1.66×10−4 |
| CpG3 | 0.084 | 1.088 | 0.523 | −0.095 | 0.909 | 0.527 | 0.052 | 1.054 | 0.738 |
| CpG4 | 0.075 | 1.078 | 0.526 | 0.137 | 1.147 | 0.191 | 0.397 | 1.487 | 0.080 |
| CpG5 | −0.054 | 0.948 | 0.473 | −0.073 | 0.930 | 0.349 | −0.207 | 0.813 | 0.057 |
| CpG6 | 0.186 | 1.204 | 0.145 | −0.004 | 0.996 | 0.981 | 0.346 | 1.413 | 0.078 |
| Smoking | −0.350 | 0.704 | 0.505 | 0.317 | 1.372 | 0.547 | −0.306 | 0.737 | 0.595 |
| Drinking | 1.345 | 3.838 | 0.034 | −0.064 | 0.938 | 0.897 | 1.291 | 3.638 | 0.048 |
| Constant | −4.423 | 0.012 | 0.067 | −5.152 | 0.006 | 0.032 | −11.002 | 0.001 |
5.84×10−5 |
Discussion
The present study observed opposite results in the
association of CpG1 and CpG2 methylation with EH. CpG2 methylation
was significantly lower in incident cases and prevalent cases
compared with in the controls. In addition, CpG2 methylation was
observed to be inversely correlated with blood pressure in controls
and incident cases. Notably, CpG2 methylation was not significantly
different between the incident cases and the prevalent cases, thus
suggesting that the antihypertensive therapy of EH did not affect
CpG2 methylation levels.
Previous studies have revealed the correlation
between DNA methylation and blood pressure (27–29).
The present study hypothesized that CpG1 hypermethylation and CpG2
hypomethylation may increase blood pressure by upregulating the
protein expression of SCNN1B, thus amplifying the function and
activity of the ENaC and leading to increased sodium reabsorption
and water retention. However, to what extent this change will alter
ENaC expression, and to what extent this change will affect sodium
reabsorption requires follow-up research. In our subsequent
studies, we aim to focus on functional research regarding the
effects of DNA methylation on ENaC expression.
Drug treatment may affect DNA methylation levels
(23,24,26).
Several CpG sites have been reported to be differentially
methylated between patients with Behçet's disease (BD) prior to and
following treatment (23),
providing strong evidence that DNA methylation was modified by BD
treatment. Furthermore, bisphosphonate treatment in 68 patients
with hypocalcaemia altered the DNA methylation levels of excision
repair cross-complementation group 8, prolyl 3-hydroxylase 2 and
syndecan 2 genes, thus affecting the cumulative bisphosphonate
exposure levels (24).
Glucocorticoid treatment has also been reported to induce acute and
long-term effects on DNA methylation states in the fetus and
offspring (26). Similarly, the
present study indicated that CpG1 methylation, rather than CpG2
methylation, was likely to be altered by antihypertensive drug
treatment. These results may provide novel information regarding
pharmaco-epigenomic research of EH.
Alterations in DNA methylation patterns are a
hallmark of aging (22). Previous
studies have demonstrated that DNA methylation levels are altered
alongside aging in several tissue types in mice and humans
(30–34). Aging is also known to be a risk
factor in the progression of hypertension (35,36).
The results of the present study revealed that a positive
correlation existed between aging and SCNN1B CpG methylation
levels. Furthermore, gender differences in DNA methylation levels
are frequently reported (37–44).
In the present study, DNA methylation levels of CpG1 and CpG2 in
SCNN1B were higher in women compared with in men. Epigenetic
changes associated with aging, and gender differences in DNA
methylation, may provide clues to elucidate the mechanisms
underlying hypertension.
In conclusion, the present study was the first, to
the best of our knowledge, to demonstrate a drug-, age-, and
gender-dependent association between SCNN1B promoter
methylation and EH.
Acknowledgements
The present study was supported by Zhejiang Province
Social Development Research Project (grant no. 2016C33178), K.C.
Wong Magna Fund in Ningbo University, Ningbo Social Development
Research Project (grant no. 2014C50051) and Ningbo Medical Science
and Technology Plan Project (grant no. 2013A39).
Glossary
Abbreviations
Abbreviations:
|
ALT
|
alanine aminotransferase
|
|
BD
|
Behçet's disease
|
|
BMI
|
body mass index
|
|
EH
|
essential hypertension
|
|
ENaC
|
epithelial sodium channel
|
|
SCNN1B
|
amiloride-sensitive sodium channel
beta subunit
|
|
TC
|
total cholesterol
|
|
TG
|
triglyceride
|
References
|
1
|
Pratt JH: Central role for ENaC in
development of hypertension. J Am Soc Nephrol. 16:3154–3159. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Garty H: Molecular properties of
epithelial, amiloride-blockable Na+ channels. FASEB J. 8:522–528.
1994.PubMed/NCBI
|
|
3
|
Loffing J and Schild L: Functional domains
of the epithelial sodium channel. J Am Soc Nephrol. 16:3175–3181.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang LP, Gao LG, Zhou XL, Wu HY, Zhang L,
Wen D, Li YH, Liu YX, Tian T, Fan XH, et al: Genetic diagnosis of
liddle's syndrome by mutation analysis of SCNN1B and SCNN1G in a
Chinese family. Chin Med J (Engl). 125:1401–1404. 2012.PubMed/NCBI
|
|
5
|
Sawathiparnich P, Sumboonnanonda A,
Weerakulwattana P and Limwongse C: A novel mutation in the
beta-subunit of the epithelial sodium channel gene (SCNN1B) in a
Thai family with liddle's syndrome. J Pediatr Endocrinol Metab.
22:85–89. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang W, Zhou W, Jiang L, Cui B, Ye L, Su
T, Wang J, Li X and Ning G: Mutation analysis of SCNN1B in a family
with liddle's syndrome. Endocrine. 29:385–390. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Noda Y: Liddle's syndrome. Nihon Jinzo
Gakkai Shi. 53:160–162. 2011.(In Japanese). PubMed/NCBI
|
|
8
|
Choy MK, Movassagh M, Goh HG, Bennett MR,
Down TA and Foo RS: Genome-wide conserved consensus transcription
factor binding motifs are hyper-methylated. BMC Genomics.
11:5192010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang LN, Liu PP, Wang L, Yuan F, Xu L,
Xin Y, Fei LJ, Zhong QL, Huang Y, Xu L, et al: Lower ADD1 gene
promoter DNA methylation increases the risk of essential
hypertension. PLoS One. 8:e634552013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang X, Falkner B, Zhu H, Shi H, Su S, Xu
X, Sharma AK, Dong Y, Treiber F, Gutin B, et al: A genome-wide
methylation study on essential hypertension in young african
american males. PLoS One. 8:e539382013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu L, Zheng D, Wang L, Jiang D, Liu H, Xu
L, Liao Q, Zhang L, Liu P, Shi X, et al: GCK gene-body
hypomethylation is associated with the risk of coronary heart
disease. Biomed Res Int. 2014:1517232014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jiang D, Zheng D, Wang L, Huang Y, Liu H,
Xu L, Liao Q, Liu P, Shi X, Wang Z, et al: Elevated PLA2G7 gene
promoter methylation as a gender-specific marker of aging increases
the risk of coronary heart disease in females. PLoS One.
8:e597522013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cheng J, Tang L, Hong Q, Ye H, Xu X, Xu L,
Bu S, Wang Q, Dai D, Jiang D and Duan S: Investigation into the
promoter DNA methylation of three genes (CAMK1D, CRY2 and CALM2) in
the peripheral blood of patients with type 2 diabetes. Exp Ther
Med. 8:579–584. 2014.PubMed/NCBI
|
|
14
|
Tang L, Wang L, Ye H, Xu X, Hong Q, Wang
H, Xu L, Bu S, Zhang L, Cheng J, et al: BCL11A gene DNA methylation
contributes to the risk of type 2 diabetes in males. Exp Ther Med.
8:459–463. 2014.PubMed/NCBI
|
|
15
|
Tang L, Ye H, Hong Q, Wang L, Wang Q, Wang
H, Xu L, Bu S, Zhang L, Cheng J, et al: Elevated CpG island
methylation of GCK gene predicts the risk of type 2 diabetes in
chinese males. Gene. 547:329–333. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tang LL, Liu Q, Bu SZ, Xu LT, Wang QW, Mai
YF and Duan SW: The effect of environmental factors and DNA
methylation on type 2 diabetes mellitus. Yi Chuan. 35:1143–1152.
2013.(In Chinese). View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Holliday R: The inheritance of epigenetic
defects. Science. 238:163–170. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cheng J, Wang Y, Zhou K, Wang L, Li J,
Zhuang Q, Xu X, Xu L, Zhang K and Dai D: Male-specific association
between dopamine receptor D4 gene methylation and schizophrenia.
PLoS One. 9:e891282014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
European Society of Hypertension-European
Society of Cardiology Guidelines Committee, . 2003 European society
of hypertension-european society of cardiology guidelines for the
management of arterial hypertension. J Hypertens. 21:1011–1053.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dai D, Cheng J, Zhou K, Lv Y, Zhuang Q,
Zheng R, Zhang K, Jiang D, Gao S and Duan S: Significant
association between DRD3 gene body methylation and schizophrenia.
Psychiatry Res. 220:772–777. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Altobelli E, Petrocelli R, Maccarone M,
Altomare G, Argenziano G, Giannetti A, Peserico A, Vena GA, Tiberti
S, Chimenti S and Peris K: Risk factors of hypertension, diabetes
and obesity in italian psoriasis patients: A survey on
socio-demographic characteristics, smoking habits and alcohol
consumption. Eur J Dermatol. 19:252–256. 2009.PubMed/NCBI
|
|
22
|
Mathers JC: Nutritional modulation of
ageing: Genomic and epigenetic approaches. Mech Ageing Dev.
127:584–589. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hughes T, Ture-Ozdemir F, Alibaz-Oner F,
Coit P, Direskeneli H and Sawalha AH: Epigenome-wide scan
identifies a treatment-responsive pattern of altered DNA
methylation among cytoskeletal remodeling genes in monocytes and
CD4+ T cells from patients with behcet's disease.
Arthritis Rheumatol. 66:1648–1658. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Polidoro S, Broccoletti R, Campanella G,
Di Gaetano C, Menegatti E, Scoletta M, Lerda E, Matullo G, Vineis
P, Berardi D, et al: Effects of bisphosphonate treatment on DNA
methylation in osteonecrosis of the jaw. Mutat Res. 757:104–113.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Laytragoon-Lewin N, Rutqvist LE and Lewin
F: DNA methylation in tumour and normal mucosal tissue of head and
neck squamous cell carcinoma (HNSCC) patients: New diagnostic
approaches and treatment. Med Oncol. 30:6542013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Crudo A, Petropoulos S, Moisiadis VG,
Iqbal M, Kostaki A, Machnes Z, Szyf M and Matthews SG: Prenatal
synthetic glucocorticoid treatment changes DNA methylation states
in male organ systems: Multigenerational effects. Endocrinology.
153:3269–3283. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guay SP, Brisson D, Lamarche B, Biron S,
Lescelleur O, Biertho L, Marceau S, Vohl MC, Gaudet D and Bouchard
L: ADRB3 gene promoter DNA methylation in blood and visceral
adipose tissue is associated with metabolic disturbances in men.
Epigenomics. 6:33–43. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bellavia A, Urch B, Speck M, Brook RD,
Scott JA, Albetti B, Behbod B, North M, Valeri L, Bertazzi PA, et
al: DNA hypomethylation, ambient particulate matter and increased
blood pressure: Findings from controlled human exposure
experiments. J Am Heart Assoc. 2:e0002122013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Alexeeff SE, Baccarelli AA, Halonen J,
Coull BA, Wright RO, Tarantini L, Bollati V, Sparrow D, Vokonas P
and Schwartz J: Association between blood pressure and DNA
methylation of retrotransposons and pro-inflammatory genes. Int J
Epidemiol. 42:270–280. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bell JT, Tsai PC, Yang TP, Pidsley R,
Nisbet J, Glass D, Mangino M, Zhai G, Zhang F, Valdes A, et al:
Epigenome-wide scans identify differentially methylated regions for
age and age-related phenotypes in a healthy ageing population. PLoS
Genet. 8:e10026292012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Teschendorff AE, Menon U, Gentry-Maharaj
A, Ramus SJ, Weisenberger DJ, Shen H, Campan M, Noushmehr H, Bell
CG, Maxwell AP, et al: Age-dependent DNA methylation of genes that
are suppressed in stem cells is a hallmark of cancer. Genome Res.
20:440–446. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Maegawa S, Hinkal G, Kim HS, Shen L, Zhang
L, Zhang J, Zhang N, Liang S, Donehower LA and Issa JP: Widespread
and tissue specific age-related DNA methylation changes in mice.
Genome Res. 20:332–340. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Issa JP, Ottaviano YL, Celano P, Hamilton
SR, Davidson NE and Baylin SB: Methylation of the oestrogen
receptor CpG island links ageing and neoplasia in human colon. Nat
Genet. 7:536–540. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wilson VL and Jones PA: DNA methylation
decreases in aging but not in immortal cells. Science. 220:1055–75.
1983. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Khattar RS, Swales JD, Dore C, Senior R
and Lahiri A: Effect of aging on the prognostic significance of
ambulatory systolic, diastolic and pulse pressure in essential
hypertension. Circulation. 104:783–789. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Grodzicki T, Michalewicz L and Messerli
FH: Aging and essential hypertension: Effect of left ventricular
hypertrophy on cardiac function. Am J Hypertens. 11:425–429. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kaz AM, Wong CJ, Dzieciatkowski S, Luo Y,
Schoen RE and Grady WM: Patterns of DNA methylation in the normal
colon vary by anatomical location, gender, and age. Epigenetics.
9:492–502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tapp HS, Commane DM, Bradburn DM,
Arasaradnam R, Mathers JC, Johnson IT and Belshaw NJ: Nutritional
factors and gender influence age-related DNA methylation in the
human rectal mucosa. Aging Cell. 12:148–155. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Burghardt KJ, Pilsner JR, Bly MJ and
Ellingrod VL: DNA methylation in schizophrenia subjects: Gender and
MTHFR 677C/T genotype differences. Epigenomics. 4:261–268. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang FF, Cardarelli R, Carroll J, Fulda
KG, Kaur M, Gonzalez K, Vishwanatha JK, Santella RM and Morabia A:
Significant differences in global genomic DNA methylation by gender
and race/ethnicity in peripheral blood. Epigenetics. 6:623–629.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hao Y, Huang W, Nielsen DA and Kosten TA:
Litter gender composition and sex affect maternal behavior and DNA
methylation levels of the oprm1 gene in rat offspring. Front
Psychiatry. 2:212011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Boks MP, Derks EM, Weisenberger DJ,
Strengman E, Janson E, Sommer IE, Kahn RS and Ophoff RA: The
relationship of DNA methylation with age, gender and genotype in
twins and healthy controls. PLoS One. 4:e67672009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Vaissiere T, Hung RJ, Zaridze D, Moukeria
A, Cuenin C, Fasolo V, Ferro G, Paliwal A, Hainaut P, Brennan P, et
al: Quantitative analysis of DNA methylation profiles in lung
cancer identifies aberrant DNA methylation of specific genes and
its association with gender and cancer risk factors. Cancer Res.
69:243–252. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
El-Maarri O, Becker T, Junen J, Manzoor
SS, Diaz-Lacava A, Schwaab R, Wienker T and Oldenburg J: Gender
specific differences in levels of DNA methylation at selected loci
from human total blood: A tendency toward higher methylation levels
in males. Hum Genet. 122:505–514. 2007. View Article : Google Scholar : PubMed/NCBI
|